Abstract
Epigenetic regulators have been shown to influence breast cancer progression. However, the detailed mechanism by which TET2 plays the suppressive role in tumorigenesis remains not completely understood. We employed RT-qPCR and westernblot to examine genes expression. Next, the bisulphite sequencing PCR was used to determine the methylation level at CASP4 promoter in the cells. Phenotypically, we utilized growth curve analysis, colony formation in soft agar and xenograft tumor assay to assess tumorigenesis of MCF-7 cell. We found that TET2 knockout enhanced colony formation ability and in vivo tumor formation ability of MCF-7 cell, whereas TET2 depletion not affected the growth rate of MCF-7 cell in the culture. Mechanistically, TET2 loss led to a significant decrease in caspase-4 expression possibly via increasing DNA methylation of CASP4 promoter in MCF-7 cell. To validate, TET2 overexpression led to higher level of caspase-4 in MDA-MB-231 and 293T cells, which was dependent on TET2 enzymatic activity. Finally, we observed that caspase-4 could revert, at least partially, TET2 deletion-induced tumorigenesis of MCF-7. In summary, we reveal a novel mechanism that TET2 suppresses tumorigenesis of breast cancer cells through caspase-4. Our findings will facilitate development of new diagnostic markers or therapeutical therapies for breast cancer.
Similar content being viewed by others
Introduction
Breast cancer is one of the most malignant and highly risky diseases in women. Similar to other types of cancer, breast cancer is also caused by a number of genetic and epigenetic factors. Among which, DNA methylation is reported to be one of the primary factors involved in breast cancer progression. However, to our knowledge, the detailed mechanism of how DNA methylation regulates breast cancer tumorigenesis remains not fully understood.
Previous studies have been shown that ten eleven translocation (TET) proteins, a well studied DNA methylation dioxygenase, are closely associated with the malignancy of tumors1,2. Indeed, the expression levels of TETs in tumors are greatly lower than that in normal tissues3,4. In addition, a variety of loss-of-function mutations of TET2 has been found in myelodysplastic syndromes (MDS) and acute myeloid leukaemias (AML), as well as low frequency of mutations in solid tumors, including breast tumor5. More importantly, TET2 was significantly downregulated in various types of cancers6,7,8. Although TET2 have recently been demonstrated to inhibit invasiveness and metastasis of breast cancer9, the molecular mechanism of TET2 regulating tumorigenesis of breast cancer are still required to be further investigated.
Caspase-4 has been shown to be implicated in inflammation, immunity and cell death (i.e., Pyroptosis)10,11,12. Interestingly, loss-of-function mutations of CASP4 were observed in colorectal cancer13. Furthermore, pro-apoptotic caspases are downregulated in certain cancers. For example, CASP4 expression is suppressed and associated with poor prognosis in esophageal squamous cell carcinoma and head and neck squamous cell carcinoma14. However, it remains unknown whether caspase-4 is involved in breast cancer progression.
Here, we report that caspase-4 acts as a primary downstream target of TET2 to exert the suppressive role in the tumorigenesis of breast cancer cells. TET2 loss results in decrease in caspase-4 expression and regulates DNA methylation level at CASP4 promoter. For the first time, We utilize colony formation assay and xenograft tumor experiment to prove that caspase-4 acts as a brake for breast cancer. Furthermore, caspase-4 overexpression largely reverts TET2 null-enhanced tumor phenotypes of MCF-7, suggesting that caspase-4 is essential for tumor suppressive role of TET2 in breast cancer cells. Collectively, our findings provide deeper understandings of breast cancer progression and help develop novel diagnostic markers and therapeutical strategies for breast cancer.
Results
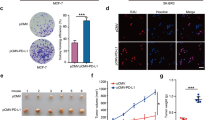
TET2 loss enhances tumorigenesis of MCF-7 cell
In order to investigate the role of TET2 in breast cancer tumorigenesis, we generated TET2 knockout MCF-7 cells by CRISPR approach (Fig. 1a). First, we examined cell proliferation of wildtype and TET2 KO MCF-7 in culture. The growth curve analysis showed that TET2-depleted MCF-7 cells (TET2 KO1, TET2 KO2) exhibited comparable growth rate to the wildtype cells over the period of 10 days, which suggested that TET2 had no evident effect on MCF-7 cell growth (Fig. 1b).
TET2 loss enhances tumorigenesis of MCF-7 cell. (a) Westernblot analysis of TET2 level in MCF-7 (WT, TET2 KO1, TET2 KO2) cultured in normal media, laminB1 as loading control. WT denotes wildtype. (b) Growth curve analysis of MCF-7 (WT, TET2 KO1, TET2 KO2) treated with EtOH or 1 nM E2 over a period of 10 days. WT denotes wildtype. (c) Colony formation assay of MCF-7 (WT, TET2 KO1, TET2 KO2) treated with EtOH or 1 nM E2. This assay was performed in 6-well plate, after 2 weeks, the cell colonies were harvested and stained. Then, the colony number was counted. WT denotes wildtype. (d) Statistical analysis of colony number shown in Fig. 1c. (e) Xenograft tumor assay of MCF-7 cells (WT, TET2 KO1, TET2 KO2) in NOD-SCID female mice, tumors were excised at day 30 after initial injection, n = 4 for each group. WT denotes wildtype. (f) Weight measurement of tumors shown in Fig. 1e. All data are presented as mean ± SD from three biological replicates. **p < 0.01; ***p < 0.001.
Next, we attempted to explore whether TET2 knockout influenced anchorage-independent growth of MCF-7 cells. We performed colony formation assay of wildtype and TET2-null MCF-7 cells in soft agar, and found that, expectedly, E2 could greatly stimulate anchorage-independent growth rate of MCF-7 cells compared to cells treated with EtOH. More interestingly, the TET2 null MCF-7 cells formed significantly more colonies than wildtype cells treated with both EtOH and E2, indicating a primary role of TET2 in anchorage-independent growth of MCF-7 cells (Fig. 1c,d).
To confirm that TET2 also exerted the tumor suppressive role in other breast cancer cell lines, we conducted colony formation assay of MDA-MB-231 stably expressing mock or TET2 in soft agar (Supplementary Fig. 1a). The results showed that the colony number of TET2-overexpressed MDA-MB-231 cells was less than half of mock cells (Supplementary Fig. 1b and c), suggesting the role of TET2 in repressing anchorage-independent growth of various breast cancer cells.
To determine whether the critical role of TET2 in detached MCF-7 cells could be observed in vivo, we carried out the xenograft tumor experiment of wildtype and TET2 KO MCF-7 cells in immunodeficient NOD-SCID mice (n = 4 per group). We observed TET2 knockout of MCF-7 cells led to remarkably larger tumor size compared to wildtype MCF-7 cells, the statistical analysis of tumor weight confirmed the observation (Fig. 1e,f). Taken together, TET2 had the ability to suppress tumorigenesis of MCF-7 cell under suspended condition.
Caspase-4 is specifically regulated by TET2
As TET2 knockout only resulted in tumorigenesis of MCF-7 under suspended condition, we hypothesized that TET2 might exert its pro-apoptotic effect via regulating apoptosis-related genes expression. To search for the downstream target responsible for TET2 knockout-induced phenotypes of MCF-7 cell, we focused on the genes (Bcl-2 and Caspase family) directly involved in classical apoptosis pathway (besides CASP3), because MCF-7 do not express caspase-3. The RT-qPCR result demonstrated that only CASP4 mRNA level was downregulated (decreased by ~60–70%) in TET2-deficient MCF-7 cells relative to wildtype cells (Fig. 2a). As a validation, westernblot result showed markedly decreased caspase-4 protein level in TET2-null MCF-7 (Fig. 2b). To investigate whether CASP3 was the downstream target of TET2, we determined its expression level in stable MDA-MB-231 cell lines (mock, flag-TET2). No evident change in CASP3 expression was observed in MDA-MB-231 cells overexpressing TET2 compared with mock cells (Supplementary Fig. 2).
Caspase-4 is specifically regulated by TET2. (a) RT-qPCR analysis shows mRNA levels of Bcl-2 and caspase family genes in MCF-7 (WT, TET2 KO1, TET2 KO2). WT denotes wildtype. (b) Westernblot analysis shows protein level of endogeneous caspase-4 in MCF-7 (WT, TET2 KO1, TET2 KO2), laminB1 as loading control. WT denotes wildtype. (c) Westernblot analysis shows protein level of exogeneously overexpressed Flag-TET2 and endogeneous caspase-4 in stable MDA-MB-231 (mock, Flag-TET2) cell lines, laminB1 as loading control. (d) Westernblot analysis shows protein level of exogeneously overexpressed Flag-TET2 and Flag-TET2 mutant, as well as endogeneous caspase-4 in stable 293T (mock, Flag-TET2, Flag-TET2-mutant) cell lines, laminB1 as loading control.
To test whether the regulation of caspase-4 expression by TET2 is specifically occurred in MCF-7 cells, we generated stably TET2 overexpressed, triple-negative human breast cancer cell MDA-MB-231, and found that caspase-4 level was greatly upregulated (Fig. 2c). Similarly, we also observed this regulation existed in 293T cell (Fig. 2d). More interestingly, TET2 mutant (without enzymatic activity) has no ability to upregulate caspase-4 expression. To conclude, caspase-4 can be significantly regulated by TET2 in various types of cells.
TET2 regulates DNA methylation of CASP4 promoter
As caspase-4 expression was modulated by TET2 in an enzymatic activity-dependent manner, which prompted us to hypothesize that TET2 might affect the methylation status of regulatory regions in the vicinity of CASP4 gene. Therefore, we used bisulphite PCR approach to evaluate the DNA methylation level at CASP4 promoter containing several CpG sites, locating at ~500 bp upstream of transcription start site (TSS) (Fig. 3a). The results revealed that the methylation level of CASP4 promoter in wildtype MCF-7 cells (65%) was obviously lower than that in TET2 KO MCF-7 cells (90% and 91.7%) (Fig. 3b).
TET2 regulates DNA methylation of CASP4 promoter. (a) Schematic diagram indicates DNA methylation of CASP4 promoter influenced by TET2. TSS indicates transcription start site, the detected methylation region was marked in blue, the red filled circles indicate CpGs whose methylation levels were examined by Bisulphite sequencing PCR. (b) Bisulphite sequencing PCR analysis shows methylation level at promoter region (TSS upstream of ∼500 bp) in MCF-7 (WT, TET2 KO1, TET2 KO2). Six circles in row indicate six CpGs at the analysed region, ten circles in column indicate ten clones picked to sequence. The filled circle denotes methylated CpG and the blank circle denotes unmethylated CpG. WT denotes wildtype, TSS denotes transcription start site. (c) Bisulphite sequencing PCR analysis shows methylation level at promoter region (TSS upstream of ∼500 bp) in stable MDA-MB-231 (mock, Flag-TET2) cell lines. Six circles in row indicate six CpGs at the analysed region, ten circles in column indicate ten clones picked to sequence. The filled circle denotes methylated CpG and the blank circle denotes unmethylated CpG. TSS denotes transcription start site. (d) Bisulphite sequencing PCR analysis shows methylation level at promoter region (TSS upstream of ∼500 bp) in stable 293T (mock, Flag-TET2, Flag-TET2-mutant) cell lines. Six circles in row indicate six CpGs at the analysed region, ten circles in column indicate ten clones picked to sequence. The filled circle denotes methylated CpG and the blank circle denotes unmethylated CpG. TSS denotes transcription start site.
In addition, TET2 overexpression led to a remarkable decrease in methylation level of CASP4 promoter in stable MDA-MB-231 cells (3.3%) compared to mock cells (98.3%), which was consistent with the result observed in 293T cells (Fig. 3c,d). Expectedly, the 293T cell stably expressing mutant TET2 exhibited methylation level (58.3%) at CASP4 promoter comparable to that in mock 293T cells (61.7%), suggesting that the methylation status at CASP4 promoter was modulated by TET2 in an enzymatic activity-dependent fashion (Fig. 3d). Based on the results described above, we propose a possible mechanism that TET2 regulates caspase-4 expression via impacting DNA methylation of CASP4 promoter.
Caspase-4 reverts TET2 loss-induced tumorigenesis of MCF-7 cell
To test whether caspase-4 mediated TET2-induced tumor phenotype of MCF-7 cell, we established caspase-4 knockdown MCF-7 (shCASP4-2, shCASP4-5) by short hairpin RNAs (Fig. 4a), and then performed colony formation in soft agar to determine whether caspase-4 had a role in anchorage-independent growth of MCF-7 cells. The result demonstrated that caspase-4 knockdown resulted in remarkable increase in colony number of MCF-7 cells compared to scramble cells in the presence of E2, indicating caspase-4 played a suppressive role in tumorigenesis of MCF-7 cell (Fig. 4b,c).
Caspase-4 reverts TET2 loss-induced tumorigenesis of MCF-7 cell. (a) Westernblot analysis shows protein level of endogeneous caspase-4 in MCF-7 cells (SCR, shCASP4-2 and shCASP4-5), shCASP4-2 and shCASP4-5 were two shRNAs against CASP-4, SCR was scramble shRNA. LaminB1 as loading control. (b) Colony formation assay of MCF-7 (SCR, shCASP4-2 and shCASP4-5) treated with EtOH or 1 nM E2, shCASP4-2 and shCASP4-5 were two shRNAs against CASP4, SCR was scramble shRNA. This assay was performed in 12-well plate, after 10 days, the cell colonies were harvested and stained. Then, the colony number was counted. (c) Statistical analysis of colony number shown in Fig. 4b. (d) RT-qPCR analysis shows inducible expression level of caspase-4 in stable MCF-7 cells (WT-mock, TET2 KO1-mock, TET2 KO1-CASP4) were treated with vehicle or 2 μg/mL Dox for 1 day. WT-mock denotes wildtype MCF-7 transduced with empty vector, KO1-mock denotes TET2 KO1 MCF-7 transduced with empty vector, KO1-CASP4 denotes TET2 KO1 MCF-7 transduced with caspase-4-inducible vector. (e) Colony formation assay of stable MCF-7 cells (WT-mock, TET2 KO1-mock, TET2 KO1-CASP4) treated with 1 nM E2 plus vehicle or 2 μg/mL Dox for ∼2 weeks. This assay was performed in 12-well plate, after 2 weeks, the cell colonies were harvested and stained. Then, the colony number was counted. WT-mock denotes wildtype MCF-7 transduced with empty vector, KO1-mock denotes TET2 KO1 MCF-7 transduced with empty vector, KO1-CASP4 denotes TET2 KO1 MCF-7 transduced with caspase-4-inducible vector. (f) Statistical analysis of colony number shown in Fig. 4e. (g) Xenograft tumor assay of stable MCF-7 cells (WT-mock, TET2 KO1-mock, TET2 KO1-CASP4) in NOD-SCID female mice administered with vehicle or 2 mg/mL Dox. Tumors were excised at day 30 after initial injection. n = 6 for each group. WT-mock denotes wildtype MCF-7 transduced with empty vector, KO1-mock denotes TET2 KO1 MCF-7 transduced with empty vector, KO1-CASP4 denotes TET2 KO1 MCF-7 transduced with caspase-4-inducible vector. (h) Weight measurement of tumors shown in Fig. 4g. All data are presented as mean ± SD from three biological replicates. *p < 0.05; **p < 0.01; ***p < 0.001.
As a validation, we inducibly expressed caspase-4 in TET2 KO1 MCF-7 cells (KO1-CASP4) and carried out colony formation assay of the cell in soft agar with the treatment of E2 (Fig. 4d). We found that TET2 KO1-CASP4 MCF-7 cell displayed significantly reduced colony number relative to TET2 KO1-mock cell without dox treatment, which might be due to leaky expression of caspase-4. When induced by dox, TET2 KO1-CASP4 cells formed much lower number of colonies than TET2 KO1-mock and WT-mock MCF-7 cells (Fig. 4e,f).
More importantly, xenograft tumor experiment showed that induced expression of caspase-4 greatly attenuated tumor formation ability of TET2 KO1 MCF-7 cell, compared to TET2 KO1-mock MCF-7 cell (Fig. 4g,h). Taken together, these data suggested that caspase-4 could reverse, at least partially, TET2 knockout-enhanced tumorigenic phenotype of breast cancer cell MCF-7.
Discussion
This study identifies a novel TET2/caspase-4 pathway antagonistic to breast cancer tumorigenesis (Fig. 5). We used the most common breast cancer cell lines MCF-7 and MDA-MB-231 as research models, which demonstrated the non-cell type-specific role of TET2 in human breast cancer. Mechanistic and biological studies help us reach several conclusions: first of all, TET2 loss enhanced anchorage-independent growth and xenograft tumor growth ability of MCF-7. Secondly, we identify a key pro-apoptotic gene CASP4 as downstream target of TET2, and this regulation is dependent on enzymatic activity of TET2. Thirdly, TET2 regulates caspase-4 expession probably through alteration of DNA methylation at CASP4 promoter region. Finally, TET2 regulates tumorigenesis of MCF-7 cells partially, if not fully, via caspase-4. These conclusions provide a solid foundation for explaining the suppressive role of TET2 in breast cancer.
TET2, an oxidative enzyme during active erasure of 5mC, has been shown to be affected via abolishing its enzymatic activity with loss-of-function mutations in MDS and AML15. Whether and how TET2 exerts its effects on initiation and progression of human solid tumors are still required to be further investigated, in spite of advances in understanding of mechanism underlying regulation of tumorigenesis.
In addition to hematological malignancies, recently, increasing evidences demonstrate that TET2 is implicated in progression of solid tumors, including prostate cancer, gastric cancer, epithelial ovarian cancer, melanoma and breast cancer7,8,16,17,18. Consistent with our study, TET2 is also identified as a brake for breast cancer in some researches17,19. To date, there are two, even more, well-established molecular mechanisms underlying TET2-mediated phenotype of breast cancer. Besides miR-200, J-Y Chen et al. revealed TET2 had the ability to silence tumor suppressor genes by modulating DNA methylation, thereby inhibiting migration and invasion of breast cancer9,17. Beyond these findings, we placed caspase-4 in the downstream of TET2 to mediate the suppressive role in breast cancer cells.
In spite of suppressive role of TET2 in a variety of cancers, some studies reveal that TET2 exerts the tumor-promoting effect. For instance, deletion of TET2 in tumor-infiltrating myeloid cells reduces melanoma progression via IL-1R-MyD88 axis20. Apart from this, another fact is that TET2 knockdown in osteosarcoma cells (OS) downregulates IL-6, then modulating MEK/ERK/HIF-1α pathway and finally decreasing lung metastasis of OS cells21. Based on these conclusions, we speculate that TET2 displays tumor-contributory role, mostly via affecting tumor microenviroment. To conclude, double-edged sword roles of TET2 in cancer initiation and progression necessitate more investigations to identify the respective mechanisms.
Although caspase-4 has been largely shown to mediate pyroptotic cell death in response to gram-negative bacterial infection and cytoplasmic lipopolysaccharide (LPS), the association of caspase-4 with cancer remains scarce22,23. Young Hwa Soung et al. reported low frequency (0.6%) of CASP4 mutation in 343 tumor samples, indicating that somatic mutations of CASP4 genes were rare in common solid tumors13. In consideration of this observation, it was very likely that dysregulated expression of caspase-4, not mutation, occurred in cancers. To confirm, we found that caspase-4 was strictly modulated by TET2 knockout in MCF-7 cells, and served as a key brake for tumorigenesis and progression of breast cancer. Presumably, caspase-4 may also play an important role in other types of tumors, which is required to be further investigated.
In summary, we propose a novel mechanism by which TET2 regulates tumorigenesis of breast cancer cell through caspase-4. This finding provides a rationale for diagnosis and prognosis of breast cancer, even other types of cancers, according to TET2 and caspase-4 expression level. However, some questions are required to be addressed: (1) whether TET2 regulates tumorigenesis of breast cancer cells through other pathways or downstream targets? (2) If exist, what are the pathways or targets? (3) What is the detailed mechanism by which caspase-4 inhibits tumorigenesis of the breast cancer cells? (4) Are there other factors or partner proteins involved in caspase-4-mediated phenotypes?
Materials and Methods
Cell culture
MDA-MB-231 and 293T were purchased from ATCC and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Hyclone) supplemented with 10% fetal bovine serum (FBS, Gibco) and 100 U/mL penicillin/streptomycin (Invitrogen). Before inducing estrogen signaling, MCF-7 cells were hormone-stripped for 5 days by culturing in phenol red-free RPMI 1640 medium (HyClone) plus 10% charcoal-depleted FBS (Biological Industries). To make complete 2 × phenol red-free DMEM media, powdered DMEM (Hyclone) was dissolved in ddH2O supplemented with charcoal-depleted FBS, Sodium Pyruvate (Gibco) and sodium bicarbonate, then sterilize through 0.22 μm filter (Millipore).
Construction and stable knockdown cells generation
CASP4 was amplified from human cDNA and then subcloned into pcw57 inducible plasmid. For knockdown, shRNA against CASP4 were ligated into pLKO.1 vector, and then co-transfected with lentiviral packaging plasmids (pPAX2 and pVSVG) into 293T cells. Forty-eight hours later, Lentiviruses were harvested, and used for cell infection. Stable cell lines were selected with 2 mg/mL puromycin within 7 days. Two shRNAs for CASP4 (shCASP4-2, GCAACGTATGGCAGGACAAAT; shCASP4-5, CAAGAGAAGCAACGTATGGCA) were used in our study. Hairpin sequence of scramble shRNA is CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG.
RT-qPCR
All these experiments were carried out strictly as previously described24. The real-time quantitative PCR was performed using ABI 7500 real-time machine. The relative amounts of the mRNA were calculated by 2ΔΔCt method.
The primers used for detecting mRNAs level are listed as follows: CASP1, AGCACAAGACC-TCTGACAGC/TAAACCACACCACACCAGGG; CASP2, GGTCCACCTTCCAGCACAAG/GCT-CCCTCATTTCCAAGGTGA; CASP3, AACCCGGGTAAGAATGTGCA/AAATACCAGTGGAGG-CCGAC; CASP4, AGTTTGACCATCTGCCTCCG/CACAGTTCCGCAGATT-CCCT; CASP5, CAT-GGGGAACTCTGGGTCAG/TGAAGATGGAGCCCCTTGTG; CASP6, GTGTGTGTCTTCCTGA-GCCA/CCCCGACATGCCTGAATGAT; CASP7, TGCGATCCATCAAGACCACC/TCACGTCAA-AACCCAGGCTT; CASP8, CCTCAAGTTCCTGAGCCTGG/TGCCTGGTGTCTGAAGTTCC; CASP9, TGAACTTCTGCCGTGAGTCC/AGAGAATGACCACCACGCAG; CASP10, TCTTGGA-AGCCTTACCGCAG/ACAGAACACGAAGCAGTCCC; BAD, AGAGTTTGAGCCGAGTGAGC/CATCCCTTCGTCGTCCTCC; BAK, GACGACATCAACCGACGCTA/GTAGACGTGTAGGGCC-AGAC; BAX, AAGGTGCCGGAACTGATCAG/AAAGTAGGAGAGGAGGCCGT; BID, ACTGGT-GTTTGGCTTCCTCC/ATGCTACGGTCCATGCTGTC; PUMA, GACCTCAACGCACAGTACGA/ATGGTGCAGAGAAAGTCCCC; BIK, CTTTGGAATGCATGGAGGGC/TGATGTCCTCAGTCTG-GTCG; BCL2L11, CTGAAGGCAATCACGGAGGT/CACTGGAGGATCGAGACAGC; BMF, GG-AACCCCAGCGACTCTTTT/ATCTGCCACCACACACGATT; BOK, CCACATCTTCTCTGCAGG-CA/CCAGGTTGCCAGGGTCTTG; GAPDH, GAGTCAACGGATTTGGTCGT/TTGATTTTGGAG-GGATCTCG.
Westernblot
This experiment was carried out as previously described25. The following antibodies were used: anti-TET2 (Diagenode, C15200179), anti-caspase-4 (MBL, M029-3), anti-GAPDH (Abmart, M20005M), anti-LaminB1 (Proteintech, 66095-1-Ig), anti-Flag (Abmart, M20008M). anti-mouse secondary IgG antibody (SAB, 3032), anti-rabbit secondary IgG antibody (SAB, 3012). All pictures were taken by digital Bio-Rad machine (ChemiDocTM) and processed by its built-in software (image lab).
Bisulphite sequencing PCR (BSP)
Genomic DNA were extracted from cells using standard protocol and then subjected gDNA to bisulphite treatment using EZ DNA Methylation-GoldTM Kit (ZYMO RESEARCH, D5005) according to manufacturer instruction. All primers used in this experiment were designed by Methyl Primer Express v1.0 software. We used two pairs of primers to perform nest-PCR, the products were ligated into pMD19-T vector, and then sequence. Two pairs of primers are used for nest-PCR: first round PCR (CASP4-m1-F: GTTGGATTAGAATTTTTATTAG; CASP4-m1-R: TAATACTCTTCAA-AACCAAC) and second round PCR (CASP4-m2-F: TTAGTT-AGTATAGTAGTTTGGAG; CASP4-m2-R: TA-CAAACATTTCTTACCGAA).
Growth curve measurement
The growth curve was measured by counting cells using Countess automated cell counter (Life Technologies) as previously described26.
Soft agar assay
This experiment was perfomed as previously published26 with minor modification, we seeded 1000 cells per well of 12-well plate and 3000 cells per well of 6-well plate. All these experiments were carried out with 2 × complete phenol red-free DMEM media. Before treated with EtOH or 1 nM E2 (Sigma), the MCF-7 cells were subject to hormone-deprivation for 5 days. All pictures were acquired through the scan machine.
Xenograft tumor experiment
We employed NOD-SCID female mice at the age of ~6 week-old in this experiment. Prior to cell injection, every mouse was planted with estrogen pill subcutaneously. After 2 days, 10 million cells were injected subcutaneously per mouse. All mice were euthanized 4 weeks after subcutaneous injection. Tumors were then excised and photographed by digital camera. Tumor weights were measured and subject to statistical analyses. The animal protocols were approved by the Animal Welfare Committee of Shanghai Medical College, Fudan University and all the methods were performed in accordance with the relevant guidelines and regulations.
Statistical analysis
All experiments were performed for three biological replicates, two comparisons were performed with graphpad prism 6 software using the two-tailed unpaired student’s t-test. Multiple comparisons were performed by one-way analysis of variance (ANOVA) with repeated measures, followed by a post hoc Fisher’s least signifcant diferences test. All values were presented as mean ± SD. *P < 0.05 was considered statistically significant.
Data Availability
No datasets were generated or analysed during the current study.
References
Ko, M. et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468, 839–843 (2010).
Wu, H. & Zhang, Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68 (2014).
Cheng, J. et al. An extensive network of TET2-targeting MicroRNAs regulates malignant hematopoiesis. Cell reports 5, 471–481 (2013).
Yang, H. et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 32, 663–669 (2013).
Huang, Y. & Rao, A. Connections between TET proteins and aberrant DNA modification in cancer. Trends in genetics: TIG 30, 464–474 (2014).
Yang, L., Yu, S. J., Hong, Q., Yang, Y. & Shao, Z. M. Reduced Expression of TET1, TET2, TET3 and TDG mRNAs Are Associated with Poor Prognosis of Patients with Early Breast Cancer. PloS one 10, e0133896 (2015).
Zhang, L. Y., Li, P. L., Wang, T. Z. & Zhang, X. C. Prognostic values of 5-hmC, 5-mC and TET2 in epithelial ovarian cancer. Archives of gynecology and obstetrics 292, 891–897 (2015).
Deng, W. et al. TET2 regulates LncRNA-ANRIL expression and inhibits the growth of human gastric cancer cells. IUBMB life 68, 355–364 (2016).
Song, S. J. et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 154, 311–324 (2013).
Jorgensen, I. & Miao, E. A. Pyroptotic cell death defends against intracellular pathogens. Immunological reviews 265, 130–142 (2015).
McIlwain, D. R., Berger, T. & Mak, T. W. Caspase functions in cell death and disease. Cold Spring Harbor perspectives in biology 5, a008656 (2013).
Hitomi, J. et al. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. The Journal of cell biology 165, 347–356 (2004).
Soung, Y. H. et al. Mutational analysis of caspase 1, 4, and 5 genes in common human cancers. Human pathology 39, 895–900 (2008).
Shibamoto, M. et al. The loss of CASP4 expression is associated with poor prognosis in esophageal squamous cell carcinoma. Oncology letters 13, 1761–1766 (2017).
Rasmussen, K. D. et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes & development 29, 910–922 (2015).
Nickerson, M. L. et al. TET2 binds the androgen receptor and loss is associated with prostate cancer. Oncogene 36, 2172–2183 (2017).
Chen, J. Y., Luo, C. W., Lai, Y. S., Wu, C. C. & Hung, W. C. Lysine demethylase KDM2A inhibits TET2 to promote DNA methylation and silencing of tumor suppressor genes in breast cancer. Oncogenesis 6, e369 (2017).
Deng, M. et al. TET-Mediated Sequestration of miR-26 Drives EZH2 Expression and Gastric Carcinogenesis. Cancer research 77, 6069–6082 (2017).
Wu, M. Z. et al. Hypoxia Drives Breast Tumor Malignancy through a TET-TNFalpha-p38-MAPK Signaling Axis. Cancer research 75, 3912–3924 (2015).
Pan, W. et al. The DNA Methylcytosine Dioxygenase Tet2 Sustains Immunosuppressive Function of Tumor-Infiltrating Myeloid Cells to Promote Melanoma Progression. Immunity 47, 284–297 e285 (2017).
Itoh, H. et al. TET2-dependent IL-6 induction mediated by the tumor microenvironment promotes tumor metastasis in osteosarcoma. Oncogene (2018).
Casson, C. N. et al. Human caspase-4 mediates noncanonical inflammasome activation against gram-negative bacterial pathogens. Proceedings of the National Academy of Sciences of the United States of America 112, 6688–6693 (2015).
Shi, J. et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 (2014).
Shen, H. et al. Suppression of Enhancer Overactivation by a RACK7-Histone Demethylase Complex. Cell 165, 331–342 (2016).
Kong, L. et al. A primary role of TET proteins in establishment and maintenance of De Novo bivalency at CpG islands. Nucleic acids research 44, 8682–8692 (2016).
Zhu, J. et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 525, 206–211 (2015).
Acknowledgements
We thank Li Tan and Yujiang Geno Shi for helpful suggestions and discussions. In addition, we also thank, Zhennan Shi for providing wildtype and TET2 knockout MCF-7 cells, Lingchun Kong for providing mock, TET2 WT or TET2 mutant-overexpressed stable MDA-MB-231 and stable 293T cells. This work was supported by the grants from Fudan University (IDH1340012).
Author information
Authors and Affiliations
Contributions
X.Z. designed, performed and analysed the experiments. S.L. and X.Z. conducted animal experiments. X.Z. wrote the manuscript and prepared figures. All authors read and approved this manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhu, X., Li, S. RETRACTED ARTICLE: TET2 inhibits tumorigenesis of breast cancer cells by regulating caspase-4. Sci Rep 8, 16167 (2018). https://doi.org/10.1038/s41598-018-34462-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-34462-z
Keywords
This article is cited by
-
Association of tet methylcytosine dioxygenase 2 and 5-hydroxymethylcytosine in endometrioid adenocarcinoma and its clinical significance
BMC Women's Health (2024)
-
The role of TET2 in solid tumors and its therapeutic potential: a comprehensive review
Clinical and Translational Oncology (2024)
-
MiR-196a promotes the proliferation and migration of esophageal cancer via the UHRF2/TET2 axis
Molecular and Cellular Biochemistry (2022)
-
Epigenetic induction of tumor stemness via the lipopolysaccharide-TET3-HOXB2 signaling axis in esophageal squamous cell carcinoma
Cell Communication and Signaling (2020)
-
BTG1 inhibits malignancy as a novel prognosis signature in endometrial carcinoma
Cancer Cell International (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.