Abstract
Identification of suitable candidate reference genes is an important prerequisite for validating the gene expression data obtained from downstream analysis of RNA sequencing using quantitative real time PCR (qRT-PCR). Though existence of a universal reference gene is myth, commonly used reference genes can be assessed for expression stability to confer their suitability to be used as candidate reference genes in gene expression studies. In this study, we evaluated the expression stability of ten most commonly used reference genes (GAPDH, ACTB, HSP90, HMBS, 18S rRNA, B2M, POLR2A, HPRT1, ACAC, YWHAZ) in fourteen different Peste des petits ruminants virus (PPRV) infected tissues of goats and sheep. RefFinder and RankAggreg software were used to deduce comprehensive ranking of reference genes. Our results suggested HMBS and B2M in goats and HMBS and HPRT1 in sheep can be used as suitable endogenous controls in gene expression studies of PPRV infection irrespective of tissues and condition as a whole, thus eliminating the use of tissue specific/ condition specific endogenous controls. We report for the first time suitable reference genes for gene expression studies in PPRV infected tissues. The reference genes determined here can be useful for future studies on gene expression in sheep and goat infected with PPRV, thus saving extra efforts and time of repeating the reference gene determination and validation.
Similar content being viewed by others
Introduction
In the era of high throughput sequencing, RNA–Sequencing (RNA-Seq) has been widely applied to evaluate global gene expression levels and composition1,2,3. RNA-Seq produces reproducible results with little technical variation due to its immense power4,5. It offers a clear-cut measure of gene expression over a wide dynamic range5,6. Validation is an important part in a RNA-Seq experiment7. The differentially expressed genes identified are often validated using quantitative RT-PCR (qRT-PCR)8,9.
qRT-PCR is the premier molecular biological technique to define accurate expression profiles of selected genes of interest10,11. It is highly sensitive, specific and reproducible12,13 and acts as a key factor in the systems biology based studies where both quality control and validation are essential criteria11. The major concern in qRT-PCR is a suitable endogenous control/reference gene to nullify variations that arise in the due course of experiment14. The variation can be introduced at any step starting from RNA extraction to quantification of qRT-PCR in terms of quality and quantity15,16. Endogenous control genes are assumed to be constitutively and uniformly expressed within the samples to be compared, irrespective of experimental conditions or treatments and tissue differentiation14,17. Housekeeping genes are the most commonly used endogenous control genes. These genes are used as reference control genes to normalize the variations in the qRT-PCR experiment18,19. However, varying expression of housekeeping genes under different experimental conditions has been reported in viral infections10,20,21,22,23,24,25,26,27, cancer research14,21 and heat stress response in sheep22. The use of an invalidated reference gene in normalization leads to unreliable conclusions especially when used with tissue samples15,18,23. This warrants for a need to identify suitable reference gene(s) for normalization for every gene expression experiment to do away with the hurdles in qRT-PCR24. Sometimes, validated endogenous controls for the desired experimental conditions can be derived from the literature describing the similar type of experiment.
Peste des petits ruminants (PPR) is one of the most economically important diseases of goats and sheep, characterized by acute febrile condition, erosive stomatitis, diarrhea and pneumonia25,26,27. Eradication of rinderpest (RP) has put PPR in spotlight to be the next eradicable disease due to similar nature of the causative agent28,29. PPR caused by Peste des petits ruminants virus (PPRV) belongs to genus Morbillivirus of family Paramyxoviridae. It is known that virus infection (e.g. SARS corona virus, yellow fever virus, human herpes virus, cytomegalovirus etc.) often results in modified or fluctuating gene transcription patterns of conventionally used housekeeping genes30,31. Recently, GAPDH was identified to be the most suitable reference gene for evaluating the gene expression in PPRV infected goats PBMCs in vitro9. No published data is currently available on the use of specific reference gene(s) in goats and sheep infected with PPRV in vivo. RNA-Seq experiments are being carried out in our laboratory to identify specific host gene expressions signatures in goats and sheep under PPRV infection. The indiscriminate use of any endogenous control to validate the RNA–Seq experiment may lead to erroneous conclusions. Therefore, in our study we used a panel of ten reference genes viz. GAPDH (Glyceraldehyde-3-phosphate dehydrogenase), 18S rRNA (18S ribosomal RNA), B2M (β 2 microglobulin), HSP 90 (heat shock protein 90), ACAC-alpha (Acetyl coenzyme carboxylase alpha), HMBS (Hydroxymethylbilane synthase), YWHAZ (Tyrosine 3-monooxygenase activation protein zeta polypeptide), POLR2A (Polymerase32 II (DNA directed) polypeptide A), ACTB (beta actin) and HPRT1 (Hypoxanthin Phosphoribosyl transferase 1) in fourteen different tissues obtained from healthy and PPRV infected goats and sheep to identify the best possible reference gene(s) for qRT-PCR normalization. We recommend different sets of reference genes based on the experimental condition.
Results
Performance of qRT-PCR primers
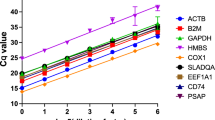
Gene specific amplification was confirmed by a single peak in the melting-curve analysis for all the genes (Supplementary Figs S1 and S2). The linear regression equation, correlation coefficient, PCR efficiency and standard curve for each gene are shown in Supplementary Figs S3 and S4. The efficiency of all the primers for the genes ranged from 93–107%.
Confirmation of viral infection in tissue samples
The tissue samples were found positive for PPRV by sandwich ELISA, N gene based RT-PCR and qRT-PCR (Supplementary Fig. S5) and histopathology and immunohistochemistry (data not shown). All tissue samples of both the species had OD value above the cut off indicating the presence of PPRV antigen (Supplementary Figs S5A and S5B). The standard curve generated for N gene had the efficiency of 100.29%, and R2 -0.968 with a slope of −3.315. Expression of PPRV - N gene was detected in all the infected tissues of goats and sheep by qRT-PCR (Supplementary Figs S5A and S5B). RT-PCR revealed positive N gene amplicon of 351 bp for all the tissues from infected animals (Supplementary Figs S5C and S5D).
Ct value of candidate reference genes
The mean Ct value of the genes in control, infected and combined groups is given in Supplementary Table S1. The mean Ct value of the reference genes ranged from 26.939 ± 0.153 (B2M) to 28.938 ± 0.191 (ACTB) in goats and 26.339 ± 0.19 (ACTB) to 28.332 ± 0.166 (POLR2A) in goats. Expression profile of all the 10 reference genes in both the species is represented by box whisker plots (Supplementary Fig. S6).
Stability of candidate reference genes under specific experimental conditions
The lower the M-value coefficient, higher is the stability ranking in geNorm and NormFinder. In control goats, infected goats and goats combined, B2M and HSP90; GAPDH and HMBS; and ACAC and HMBS, respectively, were the most stable candidate genes by geNorm analysis. Similarly, HMBS and HPRT1 were co-ranked as most stable genes in control sheep, infected sheep and sheep combined group by geNorm analysis (Table 1 and Supplementary Table S2). NormFinder and comparative delta Ct method analysis ranked HMBS as the stable gene for control goats, infected goats, goats combined, control sheep and sheep combined groups, and HSP90 for infected sheep group (Table 1 and Supplementary Tables S3 and S4). The stability of a gene is inversely proportional to the standard deviation value in the BestKeeper algorithm. HMBS was found to be most stable reference gene in all groups of goats as well as for infected sheep and sheep combined groups and B2M for control sheep group (Table 1 and Supplementary Table S5).
Comprehensive ranking of reference genes
RefFinder is a comprehensive program that integrates all four above mentioned software tools to rank the candidate reference genes based on their stability. The overall ranking suggested HMBS (Fig. 1A–F and Table 1 and Supplementary Table S6) to be the most stable among all groups of goats, control sheep and sheep combined groups while HSP 90 (Fig. 1E) was found to be the most stable reference genes in infected sheep group. Tissue specific studies among various goat tissues revealed HMBS as the most stable gene in spleen (Fig. 2A), caecum (Fig. 2B), small intestine (Fig. 2C), lower lip (Fig. 2D), large intestine (Fig. 2E) and trachea (Fig. 2F); GAPDH in rectum (Fig. 2G), prescapular lymph node (Fig. 2H), mesenteric lymph node (Fig. 2I) and abomasum (Fig. 2J); POLR2A in lung (Fig. 2K) and liver (Fig. 2L); B2M in upper lip (Fig. 2M) and ACAC in tongue (Fig. 2N). Comprehensive ranking among tissue specific studies in sheep suggested HMBS as the most stable gene in caecum (Fig. 3A), lower lip (Fig. 3B) and trachea (Fig. 3C); B2M, in lung (Fig. 3D) and rectum (Fig. 3E); ACTB in spleen (Fig. 3F) and liver (Fig. 3G); HPRT1 in mesenteric lymph node (Fig. 3H) and abomasum (Fig. 3I); GAPDH in small intestine (Fig. 3J); YWHAZ in tongue (Fig. 3K); ACAC in prescapular lymph node (Fig. 3L); HSP90 in upper lip (Fig. 3M) and POLR2A in large intestine (Fig. 3N).
Comprehensive ranking pattern of ten candidate reference genes by RefFinder in fourteen different goat tissues. Most stable reference genes for each tissue are as follows- Spleen, Caecum, Small intestine, Lower lip, Large intestine, Trachea- HMBS (A–F); Rectum, Prescapular lymph node, Mesenteric lymph node, Abomasum- GAPDH (G–J); Lung and Liver- POLR2A (K,L); Upper lip- B2M (M); Tongue- ACAC (N).
Comprehensive ranking pattern of ten candidate reference genes by RefFinder in fourteen different sheep tissues. Most stable reference genes for each tissue are as follows: Caecum, Lower lip, Trachea- HMBS (A–C); Lung, Rectum- B2M (D,E); Spleen, Liver- ACTB (F,G); Mesenteric lymph node, Abomasum- HPRT1 (H,I); Small intestine- GAPDH (J); Tongue- YWHAZ (K); Prescapular lymph node- ACAC (L); Upper lip- HSP90 (M); Large intestine- POLR2A (N).
RankAggreg provides the consensus ranking by BruteAggreg function of the package. RankAggreg suggested HMBS to be the most stable endogenous control gene in all groups of goats and sheep (Table 1 and Supplementary Table S6). Similarly, for tissues specific studies, RankAggreg suggested HMBS for caecum, large intestine, lower lip, small intestine, spleen and trachea; GAPDH for abomasum, mesenteric lymph node, prescapular lymph node and rectum; POLR2A for liver and lung; ACAC for tongue and B2M for upper lip as the most stable candidate reference genes in goats (Supplementary Table S7). In sheep, RankAggreg suggested HMBS for caecum, lower lip and trachea; HPRT1 for abomasum and mesenteric lymph node; GAPDH for large intestine and small intestine; ACTB for liver and spleen; B2M for lung and rectum; ACAC for prescapular lymph node; YWHAZ for tongue and HSP90 for upper lip, as the most stable genes (Supplementary Table S8). Final consensus ranking was obtained for each condition and tissue from RankAggreg results. The consensus ranking was obtained for each condition and tissue by considering the results of both RefFinder and RankAggreg. A detailed list of recommended endogenous control genes for individual tissues is given in Table 2.
Validation of suitable endogenous control genes
The amplification efficiency was 100.04% with R2 of 0.998 and slope of −3.3209 for ISG15, and amplification efficiency of 104.8%, R2 of 0.999 and slope of −3.2121 for IRF7 (Supplementary Fig. S4). Significant (p ≤ 0.05) difference between the expression values (delta Ct values) of ISG15 and IRF7 in lung and spleen tissues of control and infected groups of both species was observed when the two best stable candidate reference genes were used as calibrator (Figs 4 and 5A,C,E,G), and no significant difference was obtained when two least stable endogenous controls were used as calibrator (Figs 4 and 5B,D,F,H).
Expression of ISG15 in lung and spleen tissues of both goats and sheep with two most stable reference genes (HMBS and B2M in goats; HMBS and HPRT1 in sheep) and two least stable reference genes (ACTB and YWHAZ in goats; ACTB and POLR2A in sheep). ISG15 expression in control and infected lung tissues of goats with the two most stable reference genes (A) and two least stable reference genes (B). ISG15 expression in control and infected lung tissues of sheep with the two most stable reference genes (C) and two least stable reference genes (D). ISG15 expression in control and infected spleen tissues of goats with two most stable reference gene (E) and two least stable reference genes (F). ISG15 expression in control and infected spleen tissues of sheep with two most stable reference genes (G) and two least stable reference genes (H). The expression was calculated as delta Ct value (Ct(ISG15) − Ct(geometric mean of Ct of the best endogenous control genes) or Ct(geometric mean of the least stable endogenous control genes)). Significance (p < 0.05) of difference in expression between the control and infected groups was tested using t-test. Levels not connected by the same superscript are significantly (p < 0.05) different.
Expression of IRF7 in lung and spleen tissues of both goats and sheep with two most stable reference genes (HMBS and B2M in goats; HMBS and HPRT1 in sheep) and two least stable reference genes (ACTB and YWHAZ in goats; ACTB and POLR2A in sheep). IRF7 expression in control and infected lung tissues of goats with the two most stable reference genes (A) and two least stable reference genes (B). IRF7 expression in control and infected lung tissues of sheep with the two most stable reference genes (C) and two least stable reference genes (D). IRF7 expression in control and infected spleen tissues of goats with two most stable reference gene (E) and two least stable reference genes (F). IRF7 expression in control and infected spleen tissues of sheep with two most stable reference genes (G) and two least stable reference genes (H). The expression was calculated as delta Ct value (Ct(IRF7) − Ct(geometric mean of Ct of the best endogenous control genes) or Ct(geometric mean of the least stable endogenous control genes)). Significance (p < 0.05) of difference in expression between the control and infected groups was tested using t-test. Levels not connected by the same superscript are significantly (p < 0.05) different.
Discussion
Due to its high specificity and sensitivity, qRT-PCR dominated the world of gene expression studies among all other contemporary techniques. It is extremely useful in gene expression studies to document host cell responses to virus infection9,20,31,33,34,35,36,37,38,39,40,41. Elucidation of molecular pathogenesis from global gene expression profile by high-throughput omics study ultimately ends up in a number of candidate genes. qRT-PCR provides the simplest platform for its validation. In spite of these facts, qRT-PCR requires a robust normalization of the data to overcome the variability introduced at any of the steps in an experiment42,43. An ideal reference gene should be stably expressed in tissues under varied experimental conditions. However, this constant expression of any reference gene only refers to a specific condition under certain environment and the expression level in different cell types and tissues significantly varies under different experimental systems41,44,45,46. Thus identification and validation of reference genes for expression studies in an experiment is widely supported and practiced23,47.
The data in the study was analyzed using geNorm, NormFinder, BestKeeper and Comparative ΔCt method. geNorm calculates the standard deviation of the expression ratio of two candidate reference genes, which are not co-regulated as a pairwise variation23. The stability value (M) is calculated as the average pairwise variation of a specific gene compared with all other reference genes. Genes with the highest M values have the least stable expression. geNorm also identifies the number of reference genes required for the normalization of a particular experiment48. The NormFinder allows comparison of intra- and inter-group variation and calculates expression stability value (M)49,50. BestKeeper uses repeated pairwise correlation analysis to determine the optimal reference genes51. The comparative delta Ct method52 evaluates the average of standard deviation values derived from comparison of relative expression between a reference gene with other reference genes. The difference in ranking results obtained from different software programs in our study may be attributed to the use of different algorithms by different softwares to determine gene expression stability41,43,53. Most of the reports recommend consensus comprehensive ranking for use as best endogenous control41,43,53. Therefore, we recommend the candidate reference genes obtained through comprehensive ranking method in all the three different experimental conditions i.e. control, infected and combined. In control goats and control sheep, HMBS and HSP90, and HMBS and B2M are recommended as the most stable endogenous controls. These genes can be used as suitable reference genes in studies where basal expression of target genes across healthy tissues is compared in goats and sheep. In PPRV infected tissue studies for comparing across tissues, we recommend the use of HMBS and ACAC in goats and HMBS and HSP90 in sheep. In comparative studies of PPRV infected with uninfected tissues as a whole, we recommend the use of HMBS and B2M in goats and HMBS and HPRT1 in sheep, thus eliminating the use of multiple tissue specific endogenous controls. The purpose of the combined analysis was to demonstrate the stability of reference genes with respect to different conditions and tissues. We recommend to use that reference gene which shows highest stability in the combined groups for studies under PPRV infection.
ISG15 plays a key role in the innate immune response to viral infection either via its conjugation to a target protein (ISGylation) or via its action as a free or unconjugated protein. ISGylation involves a cascade of enzymatic reactions to alter host immune system. It exhibits antiviral activity towards both DNA and RNA viruses, including influenza A, HIV-1 and Ebola virus27,54,55,56. IRF7, a key innate immune modulator controlling the induction of type I interferons during viral infections57,58. Upon activation, phosphorylated IRF7 induce expression of genes responsible for type I interferon production inside the nucleus in virus infection58. ISG15 and IRF7 were chosen as the target gene of interest as these genes have been identified and predicted as important antiviral molecules by RNA–sequencing data analysis of PPRV infection studies in our lab (data not shown). The significant difference in expression of ISG15 and IRF7 on use of the most stable reference genes in goats and sheep corroborated with the findings of the RNA-Seq experiment conducted in the laboratory. The reference genes determined herein can be used by other researchers of the same field, thereby, saving the cost, effort and time of repeating the endogenous control determination and validation.
In conclusion, this study is the first attempt to establish suitable reference genes for gene expression studies in PPRV infection in fourteen different tissue samples of goats and sheep. Our results provide the starting point to validate candidate gene(s) expression obtained from transcriptome studies in PPRV infection.
Materials and Methods
Ethics Statement
The vaccine potency testing experiment was carried out at ICAR - Indian Veterinary Research Institute Mukteshwar Campus as per the guidelines of Indian Pharmacopia-2014. The study was done after obtaining permission from Indian Veterinary Research Institute Animal Ethics Committee (IVRI - IAEC) under the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), India. The protocols were approved vide letter no 387/CPCSEA. Apparently healthy, non-descriptive hill goats (local Rohilkhand breed) and sheep (Muzaffarnagri breed) between 6 months to 1 year of age were used in the present study. Virulent PPRV (Izatnagar/94- lineage IV, accession number KR140086.1)59 isolate was used as a challenge virus for infection. The tissue samples - upper lip, lower lip, tongue, trachea, lung, pre-scapular lymph node, mesenteric lymph node, spleen, liver, small intestine, large intestine, abomasum, caecum and rectum were collected from PPRV infected sheep and goats (n = 6 for each of the species). The counterpart healthy tissues were collected from apparently healthy animals (negative for PPRV antibody by competitive ELISA and serum neutralization test) housed separately. The apparently healthy animals are referred as control. These animals were handled in a humane manner and euthanized as per the CPCSEA guidelines. The graphical abstract of this study is represented in Supplementary Fig. S7.
Selection of Candidate Reference genes
A total of ten candidate reference genes were selected based on, their use as reference genes in diverse studies on gene expression in goats and sheep, availability of their sequences in databases and their function in the cell (Table 3). The ten reference genes used were GAPDH, ACTB, B2M, HSP 90, ACAC-α, HMBS, YWHAZ, POLR2A, HPRT1 and 18S rRNA9,31,35,38,39,40,41,43,53,60. Primers for YWHAZ, POLR2A, ACTB and HPRT1 were designed based on the sequence obtained from NCBI with the help of software Primer3Plus61. The quality parameters for these primers were checked in Oligo Analyzer and NCBI Primer BLAST62. The primers for rest of the genes were obtained from already published literature9,41,53.
RNA extraction and cDNA synthesis
About 30–50 mg tissues were cut into slices and dipped in TRIzol at 4 °C. The tissues were homogenized with tissue ruptor (QIAgen, Cat No: 9001271) and total RNA was isolated from the homogenized tissue using Ribozol™ RNA extraction reagent (Amresco LLC, Solon, USA) as per manufacturer’s protocol. Total RNA was dissolved in 30 μl ddH2O and quantified using NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA). One microgram of total RNA from each sample was treated with DNAse I, RNAse free enzyme (Thermo Fisher Scientific Inc., Cat No. EN0521) in the presence of 40U of RNAse inhibitor (RNAseOUT, Invitrogen), following the instructions of the manufacturer. One microgram of treated RNA was reverse transcribed using a RevertAid™ First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc., Wilmington, DE, USA, Cat No. K1622) according to the manufacturer’s protocol. Briefly, 12 μl of a mixture 1 was prepared taking 1000 ng of RNA along with 1 μl of random primers and volume adjusted by NFW (Nuclease free water, Himedia, Cat No. TCL018). Mixture 1 was incubated at 65 °C for 5 min and then snap chilled at 25 °C for 5 min. To this 12 μl of mixture 1, 8 μl of mixture 2 (4 μl of 5X RT buffer, 2 μl of 10 mM dNTPs, 1 μl each of Ribolock inhibitor and Reverse Transcriptase enzyme) was added and the final mixture of 20 μl was incubated initially at 25 °C for 10 min, then at 42 °C for 1 hour followed by 72 °C for 10 min and finally at 40 °C for 10 min. The cDNA thus obtained was stored at −20 °C till further use.
Confirmation of PPRV Infection in Tissue Samples
Sandwich ELISA (sELISA) of the Tissue Samples
PPR sandwich-ELISA kit for PPRV antigen detection was obtained from National Morbilivirus referral laboratory, Division of Virology, IVRI, Mukteshwar63. sELISA for the tissue samples was carried out as per the instructions provided with the kit.
RT-PCR of N gene in Tissue samples
Diagnostic PCR for N gene was carried out with the help of published primers PPRV-NP3-FP 5′-TCTCGGAAATCGCCTCACAGACTG-3′ and PPRV NP4-RP 5′-CCTCCTCCTGGTCCTCCAGAATCT-3′64. 25 μl of reaction mixture was prepared with 2.5 μl of 10X DreamTaq buffer, 1 μl of 10 mM dNTPs (Thermo Fisher Scientific Inc., Wilmington, DE, USA, Cat No. R0181), 0.5 μl of 10 pM each of forward and reverse primers, 0.25 μl of 1.25 U DreamTaq DNA polymerase (Thermo Fisher Scientific Inc., Wilmington, DE, USA, Cat. No. EP0702), 2 μl of 100 ng template cDNA and 18.25 μl of NFW. PCR was carried out with an initial denaturation at 95 °C for 5 min followed by 35 cycles of denaturation at 95 °C for 30 sec, annealing at 60 °C for 30 sec, renaturation at 72 °C for 30 sec with a final extension step at 72 °C for 5 min. PCR product was visualized on 1.5% agarose gel.
Absolute quantification of N gene in all tissues
Absolute quantification of N gene for viral load by qRT-PCR for infected tissues was performed using primers specific to PPRV N gene. Primers – PPRV N-FP: ATCTGCAGGAAAGGTCAGCT-3′ and PPRV N-RP: TCCCTCTCCTGTTTTGTGCT-3′ were designed using Primer3plus. The standard curve was generated using a series of 10-fold dilutions of gel purified PCR product of N gene. The amplification efficiency was calculated from the slope of the standard curve using the formula E = 10(−1/slope). Copy number was calculated from the standard curve (Supplementary Fig. S5). Ct values greater than 35 were considered negative.
Reverse transcriptase-quantitative polymerase chain reaction (qRT-PCR) of reference genes
Gene specific primers (Table 3) were used in PCR reactions of 10 µl containing 5 µl of 2X Maxima SYBR Green qPCR MasterMix (Thermo Fisher Scientific Inc., Wilmington, DE, USA, Cat. No. K0251), 0.1 µl of 10 pm/µl of forward and reverse primers, 0.1 µl of ROX dye and 2.7 µl of Nuclease free water with 10 ng (2 µl) of template cDNA. The reactions were set up in MicroAmp Fast 96 well reaction plate (0.1 ml) (Applied Biosystems) in triplicates. Reactions were performed in a 7500 Fast Real Time PCR detection system (Applied Biosystem, USA) for all the tissues of control (uninfected) and PPRV infected animals. The efficiency of Real-Time PCR and slope values were determined for each primer. The standard curve was generated using a series of 10 fold dilutions. The amplification efficiency was calculated from the slope of the standard curve using the formula E = 10(−1/slope). A melting curve analysis confirmed the presence of a single gene specific peak and the absence of primer dimers. Melting curve analysis consisted of 95 °C for 15 sec, 60 °C for 1 min, followed by 95 °C for 30 sec and a final step of 60 °C for 15 sec.
Data analysis
The Ct values for the control (uninfected), and PPRV infected samples were initially analyzed for each of the species to determine the best possible endogenous control(s) for healthy and PPRV infected conditions separately. Then, the data (Ct values) from the control and infected were combined for each of the species to identify the best endogenous control for the case where PPRV infected samples were compared with control healthy samples. The data was analyzed for six groups: Goats – control goats, infected goats (PPRV infected) and goats combined (combining both the control and infected Ct values); Sheep – control sheep, infected sheep and sheep combined. To determine tissue specific endogenous controls, both infected and control tissue Ct values were taken into consideration for each species. Stability of the 10 candidate reference genes were evaluated by algorithms geNorm23, NormFinder49, BestKeeper51, and the comparative Δ delta Ct method52 in RefFinder (http://leonxie.esy.es/RefFinder/). A comprehensive overall ranking of the stability by integrating all four algorithms was provided in the RefFinder.
The final consensus ranking was obtained with RankAggreg package65 by considering the results obtained from all the above analysis. The RankAggreg package of R software combines the stability measurements obtained from softwares (geNorm, NormFinder, BestKeeper, comparative delta Ct method and RefFinder) and establishes a consensus rank of reference genes65. A weighted rank aggregation was applied by using BruteAggreg function of the package. This function performs rank aggregation using the brute force approach. The aim of rank aggregation is to find an aggregated ranking that minimizes the distance to each of the ranked lists in the input set.
Validation of reference genes identified
The stability of the identified best reference genes was validated by evaluating the expression of ISG15 and IRF7 as target genes in the PPRV infected lung and spleen tissues with respect to the control tissues in both the species. ISG15 and IRF7 were chosen as the target genes of interest as these genes have been identified and predicted as important antiviral molecules by RNA–sequencing data analysis of PPRV infection studies in our lab. Forward primer 5′-CAGTTCATCGCCCAGAAGAT-3′ and reverse primer 5′-GTCGTTCCTCACCAGG ATGT-3′ were used for ISG15. Similarly, for IRF7 5′-GACACGCCCATCTTTGACTT-3′ and 5′- ACTGTCCAGGGAGGACACAC-3′ were used as primers. The amplification efficiency was calculated from the standard curve generated by 7 point, 10 fold serial dilutions. The Ct values for control and infected tissue samples with two most stable endogenous control genes (HMBS and B2M in goats, and HMBS and HPRT1 in sheep) and two least stable endogenous control gene (ACTB and YWHAZ for goats and ACTB and POLR2A for sheep) were estimated. Expression for control and infected groups was represented by delta Ct value (Ct(target genes) − Ct(geometric mean of Ct of the two best endogenous control genes) or Ct(geometric mean of Ct of two least stable endogenous control genes)). t- test in GraphPad Prism 7 was used to compare the expression of ISG15 and IRF7 in infected relative to control.
MIQE guidelines
This enables the researcher to evaluate the technical quality of the qRT-PCR experiments66,67. All the experiments were carried out as per the MIQE guidelines. A summary sheet of MIQE guideline of this experiment is provided in Supplementary Table S9.
References
Fang, Z. & Cui, X. Design and validation issues in RNA-seq experiments. Briefings in bioinformatics 12, 280–287, https://doi.org/10.1093/bib/bbr004 (2011).
Marioni, J. C., Mason, C. E., Mane, S. M., Stephens, M. & Gilad, Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome research 18, 1509–1517, https://doi.org/10.1101/gr.079558.108 (2008).
Pickrell, J. K. et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464, 768–772, https://doi.org/10.1038/nature08872 (2010).
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods 5, 621–628, https://doi.org/10.1038/nmeth.1226 (2008).
Wang, Z., Gerstein, M. & Snyder, M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews. Genetics 10, 57–63, https://doi.org/10.1038/nrg2484 (2009).
Rajkumar, A. P. et al. Experimental validation of methods for differential gene expression analysis and sample pooling in RNA-seq. BMC genomics 16, 548, https://doi.org/10.1186/s12864-015-1767-y (2015).
Hughes, T. R. ‘Validation’ in genome-scale research. Journal of biology 8, 3, https://doi.org/10.1186/jbiol104 (2009).
Nanda, S. K. et al. Infection of bovine dendritic cells by rinderpest or measles viruses induces different changes in host transcription. Virology 395, 223–231, https://doi.org/10.1016/j.virol.2009.09.031 (2009).
Siddappa Manjunath et al. Identification of suitable reference gene in goat peripheral blood mononuclear cells (PBMCs) infected with peste des petits ruminants virus (PPRV). Livestock Science 181, 150–155, https://doi.org/10.1016/j.livsci.2015.09.010 (2015).
Bustin, S. A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. Journal of molecular endocrinology 25, 169–193 (2000).
Bustin, S. A. et al. The need for transparency and good practices in the qPCR literature. Nature methods 10, 1063–1067, https://doi.org/10.1038/nmeth.2697 (2013).
Nolan, T., Hands, R. E. & Bustin, S. A. Quantification of mRNA using real-time RT-PCR. Nature protocols 1, 1559–1582, https://doi.org/10.1038/nprot.2006.236 (2006).
Liu, D. et al. Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PloS one 7, e46451, https://doi.org/10.1371/journal.pone.0046451 (2012).
McNeill, R. E., Miller, N. & Kerin, M. J. Evaluation and validation of candidate endogenous control genes for real-time quantitative PCR studies of breast cancer. BMC molecular biology 8, 107, https://doi.org/10.1186/1471-2199-8-107 (2007).
Huggett, J., Dheda, K., Bustin, S. & Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes and immunity 6, 279–284, https://doi.org/10.1038/sj.gene.6364190 (2005).
Han, X. et al. Selection of reliable reference genes for gene expression studies using real-time PCR in tung tree during seed development. PloS one 7, e43084, https://doi.org/10.1371/journal.pone.0043084 (2012).
Huth, A., Vennemann, B., Fracasso, T., Lutz-Bonengel, S. & Vennemann, M. Apparent versus true gene expression changes of three hypoxia-related genes in autopsy derived tissue and the importance of normalisation. International journal of legal medicine 127, 335–344, https://doi.org/10.1007/s00414-012-0787-2 (2013).
Guenin, S. et al. Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. Journal of experimental botany 60, 487–493, https://doi.org/10.1093/jxb/ern305 (2009).
Cinar, M. U. et al. Evaluation of suitable reference genes for gene expression studies in porcine PBMCs in response to LPS and LTA. BMC research notes 6, 56, https://doi.org/10.1186/1756-0500-6-56 (2013).
Kuchipudi, S. V. et al. 18S rRNA is a reliable normalisation gene for real time PCR based on influenza virus infected cells. Virology journal 9, 230, https://doi.org/10.1186/1743-422X-9-230 (2012).
Jacob, F. et al. Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PloS one 8, e59180, https://doi.org/10.1371/journal.pone.0059180 (2013).
Serrano, M. et al. Use of Maximum Likelihood-Mixed Models to select stable reference genes: a case of heat stress response in sheep. BMC molecular biology 12, 36, https://doi.org/10.1186/1471-2199-12-36 (2011).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology 3, RESEARCH0034 (2002).
Modesto, P. et al. Evaluation of internal reference genes for quantitative expression analysis by real-time reverse transcription-PCR in somatic cells from goat milk. Journal of dairy science 96, 7932–7944, https://doi.org/10.3168/jds.2012-6383 (2013).
Banyard, A. C. et al. Global distribution of peste des petits ruminants virus and prospects for improved diagnosis and control. The Journal of general virology 91, 2885–2897, https://doi.org/10.1099/vir.0.025841-0 (2010).
Parida, S. et al. Peste des petits ruminants. Veterinary microbiology 181, 90–106, https://doi.org/10.1016/j.vetmic.2015.08.009 (2015).
Wani, S. A. et al. Expression kinetics of ISG15, IRF3, IFNgamma, IL10, IL2 and IL4 genes vis-a-vis virus shedding, tissue tropism and antibody dynamics in PPRV vaccinated, challenged, infected sheep and goats. Microb Pathog 117, 206–218, https://doi.org/10.1016/j.micpath.2018.02.027 (2018).
de Haan, N. C., Kimani, T., Rushton, J. & Lubroth, J. In Peste des Petits Ruminants Virus (ed. Munir, M.) Ch. 12, 195–226 (Springer, 2015).
FAO. Supporting Livelihoods and Supporting Livelihoods and Peste Des Petits Ruminants (PPR) and Small Ruminant Diseases Control (2013).
Radonic, A. et al. Reference gene selection for quantitative real-time PCR analysis in virus infected cells: SARS corona virus, Yellow fever virus, Human Herpesvirus-6, Camelpox virus and Cytomegalovirus infections. Virology journal 2, 7, https://doi.org/10.1186/1743-422X-2-7 (2005).
Xue, J. L. & Cheng, X. W. Using host 28S ribosomal RNA as a housekeeping gene for quantitative real-time reverse transcription-PCR (qRT-PCR) in virus-infected animal cells. Current protocols in microbiology Chapter 1, Unit1D 2, https://doi.org/10.1002/9780471729259.mc01d02s19 (2010).
Bonnet, M., Bernard, L., Bes, S. & Leroux, C. Selection of reference genes for quantitative real-time PCR normalisation in adipose tissue, muscle, liver and mammary gland from ruminants. Animal: an international journal of animal bioscience 7, 1344–1353, https://doi.org/10.1017/S1751731113000475 (2013).
Baron, J. et al. Early changes in cytokine expression in peste des petits ruminants disease. Veterinary research 45, 22, https://doi.org/10.1186/1297-9716-45-22 (2014).
Dhanasekaran, S. et al. Toll-like receptor responses to Peste des petits ruminants virus in goats and water buffalo. PloS one 9, e111609, https://doi.org/10.1371/journal.pone.0111609 (2014).
Jarczak, J., Kaba, J. & Bagnicka, E. The validation of housekeeping genes as a reference in quantitative Real Time PCR analysis: application in the milk somatic cells and frozen whole blood of goats infected with caprine arthritis encephalitis virus. Gene 549, 280–285, https://doi.org/10.1016/j.gene.2014.07.063 (2014).
Mackay, I. M., Arden, K. E. & Nitsche, A. Real-time PCR in virology. Nucleic acids research 30, 1292–1305 (2002).
Patel, A. et al. Cytokines expression profile and kinetics of Peste des petits ruminants virus antigen and antibody in infected and vaccinated goats. Virologica Sinica 27, 265–271, https://doi.org/10.1007/s12250-012-3240-2 (2012).
Yang, F., Lei, X., Rodriguez-Palacios, A., Tang, C. & Yue, H. Selection of reference genes for quantitative real-time PCR analysis in chicken embryo fibroblasts infected with avian leukosis virus subgroup J. BMC research notes 6, 402, https://doi.org/10.1186/1756-0500-6-402 (2013).
Yin, R. et al. Systematic selection of housekeeping genes for gene expression normalization in chicken embryo fibroblasts infected with Newcastle disease virus. Biochemical and biophysical research communications 413, 537–540, https://doi.org/10.1016/j.bbrc.2011.08.131 (2011).
Yue, H., Lei, X. W., Yang, F. L., Li, M. Y. & Tang, C. Reference gene selection for normalization of PCR analysis in chicken embryo fibroblast infected with H5N1 AIV. Virologica Sinica 25, 425–431, https://doi.org/10.1007/s12250-010-3114-4 (2010).
Zhang, Y. et al. Reference gene screening for analyzing gene expression across goat tissue. Asian-Australasian journal of animal sciences 26, 1665–1671, https://doi.org/10.5713/ajas.2013.13199 (2013).
Hruz, T. et al. RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC genomics 12, 156, https://doi.org/10.1186/1471-2164-12-156 (2011).
Zhu, W., Lin, Y., Liao, H. & Wang, Y. Selection of reference genes for gene expression studies related to intramuscular fat deposition in Capra hircus skeletal muscle. PloS one 10, e0121280, https://doi.org/10.1371/journal.pone.0121280 (2015).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical chemistry 55, 611–622, https://doi.org/10.1373/clinchem.2008.112797 (2009).
Lanoix, D. et al. Quantitative PCR pitfalls: the case of the human placenta. Molecular biotechnology 52, 234–243, https://doi.org/10.1007/s12033-012-9539-2 (2012).
Radonic, A. et al. Guideline to reference gene selection for quantitative real-time PCR. Biochemical and biophysical research communications 313, 856–862 (2004).
De Spiegelaere, W. et al. Reference gene validation for RT-qPCR, a note on different available software packages. PloS one 10, e0122515, https://doi.org/10.1371/journal.pone.0122515 (2015).
Hellemans, J. & Vandesompele, J. Selection of reliable reference genes for RT-qPCR analysis. Methods in molecular biology 1160, 19–26, https://doi.org/10.1007/978-1-4939-0733-5_3 (2014).
Andersen, C. L., Jensen, J. L. & Orntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer research 64, 5245–5250, https://doi.org/10.1158/0008-5472.CAN-04-0496 (2004).
Wang, Y. et al. Validation of Suitable Reference Genes for Assessing Gene Expression of MicroRNAs in Lonicera japonica. Frontiers in plant science 7, 1101, https://doi.org/10.3389/fpls.2016.01101 (2016).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnology letters 26, 509–515 (2004).
Silver, N., Best, S., Jiang, J. & Thein, S. L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC molecular biology 7, 33, https://doi.org/10.1186/1471-2199-7-33 (2006).
Najafpanah, M. J., Sadeghi, M. & Bakhtiarizadeh, M. R. Reference genes selection for quantitative real-time PCR using RankAggreg method in different tissues of Capra hircus. PloS one 8, e83041, https://doi.org/10.1371/journal.pone.0083041 (2013).
Harty, R. N., Pitha, P. M. & Okumura, A. Antiviral activity of innate immune protein ISG15. Journal of innate immunity 1, 397–404, https://doi.org/10.1159/000226245 (2009).
Morales, D. J. & Lenschow, D. J. The antiviral activities of ISG15. Journal of molecular biology 425, 4995–5008, https://doi.org/10.1016/j.jmb.2013.09.041 (2013).
Skaug, B. & Chen, Z. J. Emerging role of ISG15 in antiviral immunity. Cell 143, 187–190, https://doi.org/10.1016/j.cell.2010.09.033 (2010).
Honda, K. et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777, https://doi.org/10.1038/nature03464 (2005).
Zhou, S., Cerny, A. M., Fitzgerald, K. A., Kurt-Jones, E. A. & Finberg, R. W. Role of interferon regulatory factor 7 in T cell responses during acute lymphocytic choriomeningitis virus infection. J Virol 86, 11254–11265, https://doi.org/10.1128/JVI.00576-12 (2012).
Sahu, A. R. et al. Genome sequencing of an Indian peste des petits ruminants virus isolate, Izatnagar/94, and its implications for virus diversity, divergence and phylogeography. Archives of virology 162, 1677–1693, https://doi.org/10.1007/s00705-017-3288-2 (2017).
Puech, C., Dedieu, L., Chantal, I. & Rodrigues, V. Design and evaluation of a unique SYBR Green real-time RT-PCR assay for quantification of five major cytokines in cattle, sheep and goats. BMC veterinary research 11, 65, https://doi.org/10.1186/s12917-015-0382-0 (2015).
Untergasser, A. et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic acids research 35, W71–74, https://doi.org/10.1093/nar/gkm306 (2007).
Ye, J. et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC bioinformatics 13, 134, https://doi.org/10.1186/1471-2105-13-134 (2012).
Singh, R. P., Sreenivasa, B. P., Dhar, P. & Bandyopadhyay, S. K. A sandwich-ELISA for the diagnosis of Peste des petits ruminants (PPR) infection in small ruminants using anti-nucleocapsid protein monoclonal antibody. Archives of virology 149, 2155–2170, https://doi.org/10.1007/s00705-004-0366-z (2004).
Couacy-Hymann, E. et al. Rapid and sensitive detection of peste des petits ruminants virus by a polymerase chain reaction assay. Journal of virological methods 100, 17–25 (2002).
Pihur, V., Datta, S. & Datta, S. RankAggreg, an R package for weighted rank aggregation. BMC bioinformatics 10, 62, https://doi.org/10.1186/1471-2105-10-62 (2009).
Bustin, S. A. Why the need for qPCR publication guidelines?–The case for MIQE. Methods 50, 217–226, https://doi.org/10.1016/j.ymeth.2009.12.006 (2010).
Taylor, S., Wakem, M., Dijkman, G., Alsarraj, M. & Nguyen, M. A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 50, S1–5, https://doi.org/10.1016/j.ymeth.2010.01.005 (2010).
Acknowledgements
This study was supported in part by Department of Biotechnology (BT/PR7729/AAQ/1/542/2013), Government of India and by Centre for Agricultural Bioinformatics (ICAR-IASRI). We also thank Department of Biotechnology for providing fellowship and contingency for students – Amit Ranjan Sahu (DBT Fellow No. DBT/2014/IVRI/170) and Sajad Ahmad Wani (DBT Fellow No. DBT/2014/IVRI/171).
Author information
Authors and Affiliations
Contributions
R.K.S., B.P.M., A.K.T. and R.K.G. conceived and designed the research. A.R.S., S.A.W., S.S., D.C. and A.K. conducted the wet lab work. A.R.S., A.P. and R.K.G. analyzed the data. A.R.S. and R.K.G. wrote the manuscript. A.R.S., P.M., W.A.M., R.I.N.K., R.K.G., A.P.S., K.K.R. and D.M. helped in manuscript drafting and editing. R.K.S., B.P.M., A.K.T. and R.K.G. proof read the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sahu, A.R., Wani, S.A., Saxena, S. et al. Selection and validation of suitable reference genes for qPCR gene expression analysis in goats and sheep under Peste des petits ruminants virus (PPRV), lineage IV infection. Sci Rep 8, 15969 (2018). https://doi.org/10.1038/s41598-018-34236-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-34236-7
Keywords
This article is cited by
-
Comprehensive evaluation and validation of optimal reference genes for normalization of qPCR data in different caprine tissues
Molecular Biology Reports (2024)
-
QuilA® adjuvanted Coxevac® sustains Th1-CD8+-type immunity and increases protection in Coxiella burnetii-challenged goats
npj Vaccines (2023)
-
Selection of species specific panel of reference genes in peripheral blood mononuclear cells of native livestock species adapted to trans-Himalayan region of Leh-Ladakh
Scientific Reports (2022)
-
Modulation of mouse laryngeal inflammatory and immune cell responses by low and high doses of mainstream cigarette smoke
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.