Abstract
Rhizobacteria is an important ingredient for growth and health of medicinal herbs, and synthesis of pharmacological effective substances from it. In this study, we investigated the community structure and composition of rhizobacteria in Baphicacanthus cusia (NeeS) Bremek via 16S rRNA amplicon sequencing. We obtained an average of 3,371 and 3,730 OTUs for bulk soil and rhizosphere soil samples respectively. Beta diversity analysis suggested that the bacterial community in the rhizosphere was distinctive from that in the bulk soil, which indicates that B.cusia can specifically recruit microbes from bulk soil and host in the rhizosphere. Burkholderia was significantly enriched in the rhizosphere. Burkholderia is a potentially beneficial bacteria that has been reported to play a major role in the synthesis of indigo, which was a major effective substances in B. cusia. In addition, we found that Bacilli were depleted in the rhizosphere, which are useful for biocontrol of soil-borne diseases, and this may explain the continuous cropping obstacles in B. cusia. Our results revealed the structure and composition of bacterial diversity in B. cusia rhizosphere, and provided clues for improving the medicinal value of B. cusia in the future.
Similar content being viewed by others
Introduction
Plant roots grow into the soil and are continuously in contact with the microbes living in the soil. Rhizosphere is a narrow interface between plant roots and soils for energy and material exchange. Rhizosphere microbes are affected by root exudation. This area contains up to 1011 microbial cells per gram root1. The microbes living in this narrow play a vital role in plant growth and health. Microbes help to increase the bioavailability of important mineral nutrients such as N, P and K2. Another beneficial function of rhizobacteria is to suppress soil-borne diseases3,4. In terms of medicinal herb research, the rhizosphere bacteria also influence the synthesis of effective substances5. The plants in turn feed the microbes in the rhizosphere with carbohydrates derived from photosynthesis in the form of rhizodeposition6. It has been reported that about 17% of photoassimilates are released into the rhizosphere in the form of rhizodeposition7, which results in the recruitment and enrichment of beneficial or detrimental soil bacteria from bulk soil8. Hence, the microbes living in the rhizosphere of the plant can be divided into beneficial microbes, neutral microbes and detrimental microbes. The neutral microbes are harmless to plants. Beneficial microbes can dissolve some insoluble minerals, and promote plant growth or provide phytohormones such as IAA, while the detrimental microbes can cause plant diseases by producing toxic substances. The rhizosphere bacteria are dominated by bacteria, fungi, actinomycetes, algae, protozoa, etc9. Bacteria are the most abundant microorganisms in the soil10. The analysis of abundance of microbes in the rhizosphere of Paris polyphylla var. yunnanensis showed the following relationship11: bacteria > actinomycetes > fungi. The classification and identification of bacteria are developed from phenotypic characteristics identification to genetic characteristics classification. In addition, the composition of rhizosphere community is determined by the soil type and plant genotype12. Hence, understanding the composition of bacterial community in the nature is important for the utilization of beneficial bacteria to improve the production and quality of medicinal herbs.
The rhizosphere is one of the most complex ecological niches in the nature13, which makes it difficult to investigate the composition and function of the bacteria in it. High-throughput sequencing has facilitated major advances in the understanding of microbial ecology. The 16S rRNA gene of bacteria and archaea are frequently used to characterize the taxonomic composition, phylogenetic diversity and microbial community composition14. 16S rRNA is located in prokaryotic small subunit ribosome, and includes ten conserved regions and nine hypervariable regions15. This technology has been used to investigated the microbial composition in different plants, such as Arabidopsis accessions16,17,18, maize19, Populus deltoids20,21 and rice22. However, there are very few reports on the bacterial community in medicinal herb roots or rhizosphere.
Baphicacanthus cusia (Nees) Bremek (Figure S1) is a common medicinal herb in China, which is usually used in Traditional Chinese Medicine (TCM). Its underground roots are often used as raw materials to produce radix isatidis that has been listed in the Chinese Pharmacopoeia23. As an important medicinal herb, it is widely cultivated in Southern and Eastern China24,25. B. cusia, with its antibacterial and antiviral properties26, is often used to treat colds, fever, meningitis, and other symptoms27. Its leaves and stems are important source of Qing Dai, which is useful to treat diseases such as ulcerative colitis28, leukemia29, and psoriasis30. Indigo, indirubin and tryptanthrin are reported to be the major effective substances of B. cusia responsible for its anti-inflammatory and anti-tumor effects31,32,33,34,35,36. Tryptanthrin can inhibit multi-drug resistance gene expression, and exhibits anti-inflammatory effect by inhibiting nitric oxide (NO) synthesis37. However, due to continuous cropping obstacle, B. cusia has to be transplanted after every three years or else has the risk of poor growth38,39,40. Many plants have various degrees of continuous cropping obstacles. Related researches have focused on the cause of continuous cropping obstacle in the deterioration of physicochemical properties, microbial community structure and diversity imbalances and the changes in enzyme activity of continuous cropping soil41. The problem of continuous cropping obstacle is very common in medicinal plants, especially in rhizomatous medicinal plants, such as Pseudostellaria heterophylla(Miq)Pax., Angelica sinensis (Oliv.) Diels., Ligulariaduciformis (C.Winkl.) Hand.-Mazz., Coptischinensis Franch. and Panaxginseng C. A. Mey.42. B. cusia, it is less likely to be attacked by pathogens and pests, but is susceptible to root rot43. Root rot is closely related to rhizomatic pathogenic bacteria44, which is usually caused by breaking the homeostasis of rhizobacterial community. Therefore, revealing the bacterial community composition of B. cusia is essential for understanding the underlying mechanism. In this study, we investigated the structure and composition of B. cusia rhizosphere soil and bulk soil by employing an Illumina-based sequencing approach targeting the V4 hypervariable regions of the 16S rRNA gene. This research provided the theoretical basis for exploring the relationship between B. cusia and rhizobacteria, which can uncover the relationship between continuous cropping obstacle and rhizobacteria in B. cusia. All these can lead to finding a new way to improve the yield and quality of B. cusia.
Results
Overall analysis of bacterial community in bulk soil and B. cusia rhizosphere
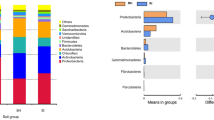
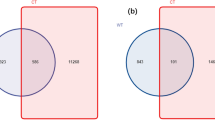
Through 16S rRNA sequencing of all bulk soil and rhizosphere soil samples, we obtained a total of 420,599 total tags. After quality control, a total of 397,699 taxon tags were obtained. We picked the operational taxonomic units (OTUs) to create an OTUs table. Sequences with 97% similarity were assigned to the same OTU. We obtained an average of 3,371 OTUs for bulk soil and 3,730 OTUs for rhizosphere soil samples (Fig. 1). The rarefaction curve of observed species showed that the sequencing depth was sufficient to cover detectable species in both bulk soil and rhizosphere soil samples, since the curve had almost plateaued (Fig. 2a). In addition, the rarefaction curve of Shannon index was consistent with the observed species (Fig. 2b). We also analyzed the bacterial composition at the phylum taxonomic level, which showed that B. cusia rhizosphere and bulk soils were dominated by Proteobacteria, Acidobacteria, Chloroflexi, Actinobacteria, Firmicutes, Planctomycetes, Verrucomicrobia, Gemmatimonadetes, Bacteroidetes and Cyanobacteria (Fig. 2c). The Venn map showed that 3463 OTUs existed in bulk soil and B. cusia rhizosphere, while 642 OTUs were only enriched in bulk soil and 976 OTUs in rhizosphere (Fig. 2d). As shown in Table 1, these indices showed that the diversities of bacterial communities in rhizosphere soil were higher than in bulk soil. Among the Observed species, ACE, Chao1 index, Shannon index and Simpson’s index, only ACE showed significant difference between the rhizosphere soil and bulk soil, but no significant difference were found in other indices.
Overall analysis of bacterial communities in B. cusia rhizosphere soil and bulk soil. (a) Rarefaction curve of observed species between B. cusia rhizosphere soil and bulk soil. (b) Rarefaction curve of Shannon index between B.cusia rhizosphere soil and bulk soil. (c) The bacterial composition of B. cusia rhizosphere soil and bulk soil at the phylum taxonomic level. (d) The Venn map of bacterial communities in B. cusia rhizosphere soil and bulk soil. There were 3463 OTUs both shown in bulk soil and B.cusia rhizosphere, and 642 OTUs were only in bulk soil and 976 OTUs were only shown in the rhizosphere samples. R: rhizosphere soil, CK: bulk soil.
Bacterial diversity in B. cusia rhizosphere
To analyze the bacterial structure and composition, we examined the bacterial relative abundance in rhizosphere soil at different taxonomic levels. We mainly presented the top 30 relative abundance of bacteria. At the class taxonomic level, the top five bacteria with relative high abundance were: Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, Ktedonobacteria and Gammaproteobacteria. Anerolineae, Chloroplast, Sphingobacteriia and Gammaproteobacteria were significantly enriched in rhizosphere soil. In contrast, the TK10, Deltaproteobacteria, Nitrospira and Clostridia were significantly depleted in the rhizosphere as compared to the bulk soil. Bacilli were also depleted in the rhizosphere as compared to the bulk soil, but was not significant (Fig. 3a). At the genus taxonomic level, the top five bacteria with relative high abundance were Acidothermus, Acidibacter, Bacillus, Bradyrhizobium and Bryobacter. In addition, Kitasatospora, Anaeromyxobacter, Gemmatimonas, Acidibacter, Burkholderia, Variibacter, Variovorax, Gemmata and Telmatobacter were significantly enriched in the rhizosphere. Bacilus was also depleted in the rhizosphere as compared to the bulk soil, but the difference was not significant (Fig. 3b).
The top 30 relative abundance of bacteria in B. cusia rhizosphere soil and bulk soil at different taxonomic level. (a) The top 30 relative abundance of bacteria at the class taxonomic level. (b) The top 30 relative abundance of bacteria at the genus taxonomic level. R: rhizosphere soil, CK: bulk soil (**p < 0.01; *p < 0.05).
B.cusia recruits special microbes from bulk soil and hosts a distinctive bacterial community in the rhizosphere
In order to analyze the differences in bacterial communities between rhizosphere soil and bulk soil, we performed Principal Coordinates Analysis (PCoA) and Nonmetric Multidimensional Scaling (NMDS). The results of the unweighed and weighed PCoA showed that bulk soil samples were clearly separated from rhizosphere soil samples by PC1 (unweighed PC1 = 41.04%, weighed PC1 = 50.48%). The NMDS analysis showed similar result as PCoA, which indicated that the bacterial communities in the rhizosphere were significantly different from that of bulk soil (Fig. 4). In addition, both bray_Curtis distance matrix and UPGMA clustering analysis based on weighted and unweighted unifrac distance showed that the bacterial communities in the rhizosphere were different from that of bulk soil based on their cluster pattern (Fig. 5). To further identify the microbes that were significantly enriched or depleted in rhizosphere, we analyzed the significant microbes between rhizosphere soil and bulk soil at both the family and genus levels. At the family taxonomic level, Burkholderiaceae, Comamonadaceae, Xanthomonadaceae, Anaerolineaceae, Chitinophagaceae, Cytophagaceae, Intrasporangiaceae, Pseudonocardiaceae, Chthoniobacteraceae, Micrococcaceae, Methylobacteriaceae, Alcaligenaceae, Catenulisporaceae, Gaiellaceae, Actinospicaceae, SubsectionIIIf and Lactobacillaceae were significantly enriched in the rhizosphere (Fig. 6a). At the genus taxonomic level, Acidibacter, Burkholderia, Gemmatimonas, Variovorax, Telmatobacter, Variibacter, Phenylobacterium, Kitasatospora, Gemmata, Chthoniobacter, Aquincola, Labrys, Intrasporangium, Catenulispora, Rhizobium, Gaiella, Pseudonocardia, Granulicella, Actinospica, Ralstonia, Ktedonobacter and Lactobacillus were significantly enriched in the rhizosphere (Fig. 6b). The LEfSe analysis showed that the biomarkers of bulk soil were Firmicutes, Deltaproteobacteria, Bacillales, Bacilli, DA111, and the biomarkers for B. cusia rhizosphere were Betaproteobacteria, Burkholderiales, Xanthomonadales and Gammaproteobacteria. The result of LEfSe analysis was consistent with the previous results indicating that Burkholderia was significantly enriched in the rhizosphere of B. cusia, while Bacilli exhibited low abundance in the rhizosphere as compared to the bulk soil, although not significant (Fig. 7). Taken together, these results suggested that the bacterial community in the rhizosphere of B. cusia was distinctive from the bulk soil and the Burkholderia was significantly enriched in the rhizosphere indicating that it is an important biomarker for the rhizosphere.
Principal Coordinates Analysis (PCoA) and Nonmetric Multidimensional Scaling (NMDS) of bacterial communities in B. cusia rhizosphere soil and bulk soil. (a) Unweighted unifrac PCoA of bacterial communities in B.cusia rhizosphere soil and bulk soil. PC1 explained 41.04% of the variation while PC2 explained 23.94%. (b) Weighted unifrac PCoA of bacterial communities in B. cusia rhizosphere soil and bulk soil. PC1 explained 50.48% of the variation while PC2 explained 37.25%. (c) NMDS of bacterial communities in B. cusia rhizosphere soil and bulk soil. R: rhizosphere soil, CK: bulk soil.
Correlation of bacterial communities between B. cusia rhizosphere soil and bulk soil. (a) Bray_Curtis distance matrix of bacterial communities in B. cusia rhizosphere soil and bulk soil. (b) UPGMA clustering analysis using unweighted unifrac distances of bacterial communities between B. cusia rhizosphere soil and bulk soil at the phylum taxonomic level. (c) UPGMA clustering analysis using weighted unifrac distances of bacterial communities between B. cusia rhizosphere soil and bulk soil at the phylum taxonomic level. R: rhizosphere soil, CK: bulk soil.
Comparison of the significant microbes between rhizosphere soil and bulk soil at different taxonomic levels. (a) The histogram of significant microbes between rhizosphere soil and bulk soil at the family taxonomic level. (b) The histogram of significant microbes between rhizosphere soil and bulk soil at the genus taxonomic level. R: rhizosphere soil, CK: bulk soil (***p < 0.001; **p < 0.01; *p < 0.05).
The LEfSe analysis of bacterial communities in B. cusia rhizosphere soil and bulk soil. (a) Cladogram of bacterial communities in B. cusia rhizosphere soil and bulk soil. (b) LDA scores of biomarker bacteria. LDA scores are shown as horizontal bars for the biomarker bacteria with an LDA score >4. R: rhizosphere soil, CK: bulk soil.
Discussion
Soil has known to be one of the environments with the most diverse microbes45. Soil biodiversity is a key determinant of the ecological and evolutionary responses of terrestrial ecosystems to current and future environmental changes46. The function of soil is mainly dependent on the diversity of microbes living in it. These microbes are essential for plant nutrition and health. Rhizosphere is an important exchange interface of material and energy between plants and microbes. The rhizosphere of medicinal herbs is similar to other crops, since it is a nutrient-rich zone, where soil bacteria compete for the limited nutrients derived from plants. Furthermore, the plant-associated microbial community, which is also referred to as the second genome of the plant is crucial for plant health and growth47. Previous studies have shown about 75 × 105 CFU of cultivable microbes per gram rhizosphere soil in burdock48. Zhang et al.49 also found 1549 OTUs in rhizosphere soil of Cypripedium macranthum. In this study, we obtained an average of 3,730 OTUs for B. cusia rhizosphere using 16S rRNA sequencing. This suggested that there were numerous bacteria in the rhizosphere of B. cusia, and investigation of their function requires extensive work in the future.
Taxonomic identification based molecular methods, which are independent of microbial cultivation, are widely used to investigate the composition of soil bacterial community. The basic structure of bacteria includes a cell wall that can maintain inherent shape a cell membrane that underlines the cell wall cytoplasm that plays a major role in determining its size and structural integrity50 and karyoplasm that controls the various bacterial genetic traits. There are three ribosomal RNAs in the bacterial cytoplasm: 16S rRNA, 23S rRNA, and 5S rRNA that they participate in bacterial protein synthesis. 16S rRNA sequence is used for microbial taxonomic identification. Recently, molecular identification methods combined with high-throughput sequencing have been widely applied in the study of bacterial community composition51. The application of high-throughput sequencing in the study of microbial community is based on culture independent method. For 16S rRNA amplicon sequencing, total microbial DNA was extracted to study the low abundance of microbes. To a certain extent, the relative abundance and diversity of sequence reflect relative microbial abundance and diversity in the sample52. Several plant microbes have been studied, including those in rice53,54, soybean55, cotton56 and many other plants. There are very few reports on medicinal herb microbes using high-throughput sequencing. However, there are some studies on the rhizosphere microbes of medicinal herbs. In this study, we examined a common and important medicinal herb B. cusia by the 16S rRNA amplicon sequencing method, and found that the diversity of bacterial community in the rhizosphere is higher than in bulk soil. However, this was only shown to be significant in the ACE index (p = 0.0325), but not in Observed species, Chao1 index, Shannon index and Simpson’s index. This result was similar to a previous study57 on poplar plantation which found that the bacterial community diversity of rhizosphere soil was higher than that of bulk soil, but the difference was not significant.
Plants can determine the composition of the roots, and rhizosphere microbial community secretion of root exudates can specifically stimulate or repress the microbes58. This phenomenon is known as the rhizosphere effect. The microbes can release soil enzymes59, degrade pollutants and catalyze oxidation reduction. Therefore, microbes are beneficial for nutrient cycling in soil60,61. Soil microbial structural stability and functional diversity played an important role in maintaining soil system health62,63. Rhizosphere microbes in turn exert strong effect on plant growth and development by nitrogen fixation64,65, phosphate solubilization66,67, hormone production68,69, and forming a plant-rhizosphere microbe interaction environment. Studies showed that roots with selectivity for rhizosphere microbes57,70 can attract both beneficial and detrimental microbes8. In this study, at the class taxonomic level, Bacilli were depleted in the rhizosphere as compared to the bulk soil. At the genus taxonomic level, Burkholderia was significantly enriched in the rhizosphere. Bacillus was also depleted in the rhizosphere as compared to the bulk soil. The results were similar to the LEfSe analysis. This suggests that B. cusia recruits special microbes from bulk soil and hosts a distinctive bacterial community in the rhizosphere. A previous study demonstrated that Burkholderia can present resistances to multiple heavy metals and antibiotics. It can also produce indole-3-acetic acid, 1-aminocyclopropane-1-carboxylic acid deaminase and siderophores. Inoculation with Burkholderia improved germination of seeds of the investigated vegetable plants in the presence of Cu, promoted elongation of roots and hypocotyledonary axes, enhanced the dry weights of the plants grown in the soils polluted with Cu and/or Pb, and increased activity of the soil urease and the rhizobacteria diversity71. Indigo-producing gene from Burkholderia sp. was cloned72. Indigo is the primary effective substances in B. cusia. Multiple Bacillus species are known to promote plant growth, in addition to the beneficial N2− fixing activity, which can promote drought resistance in various plant models, including Arabi-dopsis73, Brachypodium74, pepper75 and rice76. Burkholderia may be related to effective substances in B. cusia, and the decrease of Bacillus may be related to the continuous cropping obstacle of B. cusia. Further studies are needed to confirm this hypothesis. We can potentially design a management approach to control the presence of bacterial species in the soil and improve production and quality of B. cusia based on detrimental or beneficial species.
Materials and Methods
Sampling and Material Processing
Rhizosphere soil samples of B. cusia were collected from Shufeng domination Farm in Fujian, China (25°25′N 118°39′E). Sampling site: B. cusia field management measures were consistent. Bulk soil samples (CK) were collected from fifteen different sites away from B. cusia cultivation in the same field, and five sites were combined to form one biological replicate77. At each sampling site, soil samples were collected from five points within the 0–30 cm topsoil layer after the litter layer was removed. For rhizosphere sampling (R), B. cusia was dug out, and the roots with attached soils were gently shaken to remove loose soil until only firmly attached soil remained. This attached soil was collected as the rhizosphere soil using sterilized brushes. Rhizosphere soils from five strains of B. cusia were mixed to form one biological replicate77. In total, three biological bulk soil samples and three biological rhizosphere soil samples were analyzed. In addition, the rhizosphere soil samples were subjected to a more precise method for collecting rhizosphere soils through centrifugation of root washings according to Bulgarelli et al.16,78.
DNA Extraction and PCR Amplification
Total genomic DNA from samples was extracted using CTAB method79,80, with minor modification. DNA concentration and purity were monitored on 1% agarose gels. V4 region of the 16S rRNA gene was amplified using 515-F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806-R (5′-GGACTACHVGGGTW TCTAAT-3′) primers81. PCR reactions (30 μL) included 15 μL of Phusion Master Mix (2x), 3 μL of primer (2 μM), 10 μL of gDNA (1 ng/μL), 2 μL of H2O. The PCR cycling program consisted of an initial denaturation step at 98 °C for 1 min, followed by 30 cycles of 98 °C for 10 s, 50 °C for 30 s, and 72 °C for 30 s, and a final 5 min extension at 72 °C.
PCR Products Mixing and Purification
The PCR products were detected with 2% agarose gels electrophoresis82. PCR products with bright band between 300 and 400 bp were mixed in equal density ratios. Then, the mixture of PCR products was purified with gel extraction kit (Qiagen, Germany).
Library Preparation and Sequencing
Sequencing libraries were generated using TruSeq® DNA PCR-free sample preparation kit (Illumina, USA) as per manufacturer’s recommendations and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on an Illumina HiSeq. 2500 platform and 250 bp paired-end reads were generated (completed by Beijing Novogene Science and Technology Co., Ltd)
Data Analysis
Paired-end reads were merged using FLASH83. Quality filtering on the raw tags were performed under specific filtering conditions to obtain the high-quality clean tags84 according to the QIIME85 (http://qiime.org/index.html) quality controlled process. The tags were compared with the reference database using UCHIME algorithm86 (http://www.drive5.com/usearch/manual/uchime_algo.html) to detect chimera sequences, and then the chimera sequences were removed87. Then, we used pick_de_novo_otus.py to pick OTUs by creating an OTU table. Sequences with ≥97% similarity were assigned to the same OTUs. Representative sequence for each OTU was screened for further annotation. For each representative sequence, the Green Gene Database88 (http://greengenes.lbl.gov/cgi-bin/nph-index.cgi) was used based on RDP classifier89 algorithm to annotate taxonomic information. Observed-species, ACE, Chao1, Shannon, Simpson were calculated with QIIME. ACE and Chao1 were selected to identify community richness. Shannon and Simpson were used to identify community diversity. Principal coordinates analysis (PCoA), Nonmetric Multidimensional Scaling (NMDS) and Unweighted Pair Group Method with Arithmetic mean (UPGMA) clustering were conducted by QIIME software. Linear discriminant analysis effect size (LEfSe) was performed using the online LEfSe program (http://huttenhower. sph.harvard. edu/galaxy/root/index)90. The significant difference was calculated using t-test.
References
Egamberdieva, D. et al. High incidence of plant growth stimulating bacteria associated with the rhizosphere of wheat grown on salinated soil in Uzbekistan. Environmenta Microbiology 10, 1–9 (2008).
Atkinson, D. & Watson, C. A. The beneficial rhizosphere: a dynamic entity. Applied Soil Ecology 15, 99–104 (2000).
Nihorimbere, V. et al. Impact of rhizosphere factors on cyclic lipopeptide signature from the plant beneficial strain Bacillus amyloliquefaciens S499. Fems Microbiology Ecology 79, 176–191 (2012).
Haney, C. H., Samuel, B. S., Bush, J. & Ausubel, F. M. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nature Plants 1, 1–9 (2015).
Guo, F. X., Liu, Y., Tang, L., Chen, C. C. & Pei, D. N. Research status and prospect on rhizosphere microbiome of medicinal plants. Journal of Agricultural Science and Technology 19, 12–21 (2017).
Hirsch, A. M. et al. Complete genome sequence of micromonospora strain L5, a potential plant-growth-regulating actinomycete, originally isolated from casuarina equisetifolia root nodules. Genome Announcements 5, e00759 (2013).
Jones, D. L., Nguyen, C. & Finlay, D. Carbon flow in the rhizosphere: carbon trading at the soil-root interface. Plant and Soil 321, 5–33 (2009).
Dohrmann, A. B. et al. Importance of rare taxa for bacterial diversity in the rhizosphere of Bt- and conventional maize varieties. The ISME Journal 7, 37–49 (2013).
Hao, D. C., Chen, S. L. & Xiao, P. G. Study of rhizosphere microbe based on molecular biology and genomics. Microbiology 36, 892–899 (2009).
Han, W. Y., Wang, W. M., Guo, Y., Yang, M. Z. & Jia, Z. J. Bacterial abundance of tea garden soils and its influencing factors. Journal of Tea Science 33, 147–154 (2013).
Zhou, N. et al. Correlation between distribution of rhizospheric microorganisms and contents of steroidal saponins of Paris polyphylla var. yunnanensis.China. Journal of Chinese Materia Medica 40, 1055–1060 (2015).
Garbeva, P., Elsas, J. D. V. & Veen, J. A. V. Rhizosphere microbial community and its response to plant species and soil history. Plant Soil 302, 19–32 (2008).
Zeng, M. J., Zhong, Y. J. & Diao, Y. Promoting mechanism of plant growth-promoting rhizobacteria in medicinal plants. Biotechnology Bulletin 33, 13–18 (2017).
Langille, M. G. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology 31, 814–821 (2013).
Yang, X., Chen, L. & Wang, C. Q. Advance in application of 16S rRNA gene in bacteriology. Journal of Northwest A & F University (Natural Science Edition) 36, 55–60 (2008).
Bulgarelli, D. et al. Revealing structure and assembly cues for Arabidopsis root inhabiting bacterial microbiota. Nature 488, 91–95 (2012).
Lundberg, D. S. et al. Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–9 (2012).
Bai, Y. et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369 (2015).
Peiffer, J. A. et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proceedings of the National Academy of Sciences USA 110, 6548–6553 (2013).
Gottel, N. R. et al. Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Applied and Environmental Microbiology 77, 5934–5944 (2011).
Shakya, M. et al. A multifactor analysis of fungal and bacterial community structure in the root microbiome of mature Populus deltoides trees. PLoS ONE 8, e76382 (2013).
Edwards, J. et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proceedings of the National Academy of Sciences USA 112, 911–920 (2015).
National Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China Vol 1. National Pharmacopoeia Committe, Beijing, 2015.
Hu S. L. Original color figures of Chinese typical medicine materials. 86, 498, 16 (Shandong Scientific Technology Press, Jinan, 1998).
Flora of Fujian Editors. Flora of Fujian (Volume 5). 132–133 (Fuzhou Scientific Technology Press, Fuzhou, 1993).
Sun, X. B., Sheng, J. R. & Wang, D. P. Research progress of chemical constituents and pharmacological activities for Baphicacanthus cusia (Nees)bremek. Journal of Guangxi Teachers Education University 25, 66–69 (2008).
Zhen, Z. H., Dong, Z. H. & Yu, J. Morden research and application of Chinese tradititional patent medicine. 3166 (The Academy Press, Beijing, 1993).
Fan, H. et al. Intervention effects of QRZSLXF, a Chinese medicinal herb recipe, on the DOR-β-arrestin1-Bcl2 signal transduction pathway in a rat model of ulcerative colitis. Journal of ethnopharmacology 154, 88–97 (2014).
Hu, X. M. et al. Arsenic disulfide induced apoptosis and concurrently promoted erythroid differentiation in cytokine-dependent myelodysplastic syndrome-progressed leukemia cell line F-36p with complex karyotype including monosomy 7. Chinese Journal of Integrative Medicine 20, 387–393 (2014).
Lin, Y. K. et al. Comparison of refined and crude indigo naturalis ointment in treating psoriasis: randomized, observer-blind, controlled, intrapatient trial. Archives of Dermatology 148, 397–400 (2012).
Zeng, M. J. & Diao, Y. Secondary metabolites of Baphicacanthus cusia (NeeS) Bremek. Chinses Agricultural Science Bulletin 32, 30–34 (2016).
Ma, Q., Qu, Y. Y., Zhang, X. W., Xu, B. W. & Zhou, J. T. Recent advances in microbial synthesis of indigo. Chinese Journal of Applied & Environmental Biology 18, 344–350 (2012).
Yang, M., Liu, Z. Y., Su, T. T. & Zhou, W. Q. Study on mechanism of precursors transforming into indigo and indirubin in blue-genera plants. China Journal of Chinese Materia Medica 35, 928–931 (2010).
Spink, B. C., Hussain, M. M., Katz, B. H., Eisele, L. & Spink, D. C. Transinet induction of cytohromes P450 1A1 and 1B1 in MCF-7 human breast cancer cells by indirubin. Biochemical Pharmacology 66, 2313–2321 (2003).
Mak, N. K. et al. Inhibtion of RANTES expression by in influenza virus-infected humanbronchial epithelial cell. Biochemical Pharmacology 67, 167–174 (2004).
Wu, X. X. et al. Characterization of anti-leukemia components from indigo naturalis using comprehensive two-dimensional K562/cell membrane chromatography and in silico target identification. Scientific Reports 6, 30103 (2016).
Miao, S., Sun, J. Y., Xie, Y. H., Wang, J. B. & Wang, S. W. Research progress of tryptanthrin. Chinese Pharmacological Bulletin 24, 152–155 (2008).
Wang, S. M., Pan, D. R. & Zhu, Q. Q. Contents of medicinal ingredients of Baphicacanthus cusia (NeeS) Bremek in different planting-age. Guizhou Agricultural Sciences 42, 184–186 (2014).
Zhang, Y. Q. & Gao, J. M. The cultivation technique of Strobilanthes cusia planted in paddy fields and its processing technology. Guizhou Agricultural Sciences 37, 66–67 (2009).
Li, Q. J. The planting technology of Baphicacanthus cusia (NeeS) Bremek. China Agricultural information 9, 28 (2007).
Liu, L. et al. Relationship between soil microbial quantity, enzyme activity and soil fertility in hot pepper greenhouse soils of different continuous cropping years. Soil and Fertilizer Sciences in China 2, 5–10 (2013).
Chen, H., Yang, Z. L., Yuan, Z. L., Yuan, X. & Liu, X. F. Changes of physicochemical property and microflora in rhizosphere soil of continuous cropping of Atractylodes macrocephala. Journal of Plant Resources and Environment 23, 24–29 (2014).
Li, Q. J. Cultivation techniques of Baphicacanthus cusia(NeeS) Bremek. China Agricultural Information 9, 28–29 (2007).
Gao, F., Ren, X. X., Wang, M. L. & Qin, X. M. Research progress in root rot diseases of Chinese herbal medicine and control strategy by antagonistic microorganisms. China Journal of Chinese Materia Medica 40, 4122–4126 (2015).
Fierer, N. & Lennon, J. T. The generation and maintenance of diversity in microbial communities. American Journal of Botany 98, 439–448 (2011).
Bardgett, R. D. & Putten, W. H. V. D. Belowground biodiversity and ecosystem functioning. Nature 515, 505–511 (2014).
Berendsen, R. L., Pieterse, C. M. & Bakker, P. A. The rhizosphere microbiome and plant health. Trends in Plant Science 17, 478–486 (2012).
Hou, J. H., Fan, J. Q., Wang, F. W., Cai, K. & Kong, W. G. Priliminary analysis of cultivable bacteria diversity and Cd2+ resistance in burdock rhizosphere. Biotechnology Bulletin 28, 158–162 (2012).
Zhang, J., Hou, X. Q. & Fu, Y. J. Rhizospheric Bacteria Diversity of Cypripedium macranthum Estimated via High Throughput Sequencing. Southwest China Journal of Agricultural Sciences 30, 811–816 (2017).
Wong, K. K. L. Three-dimensional discrete element method for the prediction of protoplasmic seepage through membrane in a biological cell. Journal of Biomechanics 65, 115–124 (2017).
Akinsanya, M. A., Goh, J. K., Lim, S. P. & Ting, A. S. Metagenomics study of endophytic bacteria in Aloe vera using next-generation technology. Genomics Data 6, 159–163 (2015).
Huang, J. Y. & Zhou, W. Ecological effect on soil microbes on the basis of 16S rRNA/DNA analyses. Chinese Agricultural Science Bulletin 22, 291–294 (2006).
Liu, B., Hu, G. P., Zheng, X. F., Zhang, J. F. & Xie, H. A. Analysis on microbial diversity in the rhizosphere of rice by phospholipid fatty acids biomarkers. Chinese Journal of Rice Science 24, 278–288 (2010).
Björn, B., Judith, P. & Dumont, M. G. Microbial community structure in the rhizosphere of rice plants. Frontiers in Microbiology 6, 1537 (2015).
Zhang, J. F. et al. Effects of saline alkali stress on diversity of bacterial community in rhizosphere soil of soybean. Journal of Jilin Agricultural University 39, 262–269 (2017).
Gu, M. Y. et al. Microbial community diversity of rhizosphere soil in continuous cotton cropping system in Xinjiang. Acta Ecologica Sinica 32, 3031–3040 (2012).
Wang, Q. T. et al. Comparison on bacterial community of rhizosphere and bulk soil of poplar plantation based on pyrosequencing. Chinese Journal of Applied & Environmental Biology 21, 967–973 (2015).
Doornbos, R. F., Loon, L. C. V. & Bakker, P. A. H. M. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agronomy for Sustainable Development 32, 227–243 (2012).
Liu, E. K. et al. Biological properties and enzymatic activity of arable soils affected by long-term different fertilization systems. Journal of Plant Ecology 32, 176–182 (2008).
Jiao, Y. J. et al. Effects of continuous tobacco on soil microbial diversity and enzyme activities. Soil and Crop 3, 56–62 (2014).
Dai, Y. T. et al. Soil bacteria diversity in rhizosphere under two types of vegetation restoration based on high throughput sequencing. Acta Pedologica Sinica 54, 735–748 (2017).
Jacobsen, C. S. & Hjelms, M. H. Agricultural soils, pesticides and microbial diversity. Current Opinion in Biotechnology 27, 15–20 (2014).
Rutigliano, F. A. et al. Soil activities related to nitrogen cycle under three plant cover types in Mediterranean environment. Applied Soil Ecology 43, 40–46 (2009).
Zhang, Q. L., Lin, M. & Ping, S. Z. Biological nitrogen fixation and its application in sustainable agriculture. Biotechnology Bulletin 18, 1–4 (2008).
Hayat, R., Ali, S., Amara, U., Khalid, R. & Ahmed, I. Soil beneficial bacteria and their role in plant growth promotion: a review. Annal of Microbiology 60, 579–598 (2010).
Bhattacharyya, P. N. & Jha, D. K. Plant growth promoting rhizobacteria (PGPR): emergence in agriculture. World journal of microbiology & biotechnology 28, 1327–1350 (2012).
Ma, C. Y., Zhang, Y., Ma, W. B., Li, J. H. & Yao, T. Identification of plant growth promoting rhizobacteria Astragalus membranaceus and their effectives. Acta Prataculturae Sinica 26, 149–159 (2017).
Li, X. Q. Studies and prospection plant growth-promoting rhizobacteria of tea rhizosphere (camellia sinensis). Journal of Shandong Agricultural University 40, 301–303 (2009).
Rodriguez, M. T. F., Valverde, N. B., Lagurara, P., Revale, S. & Vilaro, M. D. R. Soil and rhizosphere bacterial diversity in maize agro-ecosystem. Sustainable Agriculture Research 6, 35–51 (2017).
Hartmann, A., Schmid, M., Tuinen, D. V. & Berg, G. Plant-driven selection of microbes. Plant Soil 321(1–2), 235–257 (2009).
Huang, G. H. et al. Characterization of plant-growth-promoting effects and concurrent promotion of heavy metal accumulation in the tissues of the plants grown in the polluted soil by Burkholderia strain LD-11. International Journal of Phytoremediation 15, 991–1009 (2013).
Liu, Z. Y. et al. Cloning, expression and application of an indigo-producing gene from Burkholderia sp. IDO3. Microbiology China 44, 2634–2643 (2017).
Zhou, C. et al. Rhizobacterial strain Bacillus megaterium BOFC15 induces cellular polyamine changes that improve plant growth and drought resistance. International Journal of Molecular Sciences 17, 976 (2016).
Gagné-Bourque, F. et al. Accelerated growth rate and increased drought stress resilience of the model grass Brachypodium distachyon colonized by Bacillus subtilis B26. PLoS One 10, e0130456 (2015).
Marasco, R. et al. A drought resistance-promoting microbiome is selected by root system under desert farming. PLo S One 7, e48479 (2012).
Kakar, K. U. et al. A consortium of rhizobacterial strains and biochemical growth elicitors improve cold and drought stress tolerance in rice (Oryza sativa L.). Plant Biology 18, 471–483 (2016).
Inceoglu, O., Salles, J. F., van Overbeek, L. & Elsas, J. D. Effects of plant genotype and growth stage on the betaproteobacterial communities associated with different potato cultivars in two fields. Applied and Environmental Microbiology 76, 3675–3684 (2010).
Bulgarelli, D. et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host & Microbe 17, 392–403 (2015).
Xu, P., Li, W. J., Xu, L. H. & Cheng, L. A microwave-based method for genomic DNA extraction from actionmycetes. Microbiology 30, 82–84 (2003).
Wang, A. F. & Hong, K. CTAB method for genomic DNA extraction from Nonomuraea. Microbiology 37, 1211–1215 (2010).
Qin, S. J. et al. Analysis of the bacterial community structures diversity in rhizosphere of Cerasus sachalinensis Kom. Journal of Jilin Agricultural University. 33, 643–648 (2011).
Yang, J. H. & Kong, W. Q. Rhizosphere bacteria diversity of Morus mongolica revealed based on 16S rRNA sequencing. Genomics and Applied Biology 34, 2161–2168 (2015).
Magoč, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Nicholas, A. B. et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature methods 10, 57–59 (2013).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods 7, 335–336 (2010).
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011).
Haas, B. J. et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome research 21, 494–504 (2011).
DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology 72, 5069–5072 (2006).
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and environmental microbiology 73, 5261–5267 (2007).
Zettler, E. R., Mincer, T. J. & Amaral-Zettler, L. A. Life in the plastisphere: microbial communities on plastic marine debris. Environmental Science & Technology 47, 7137–7146 (2013).
Acknowledgements
This work was supported by the Natural Science Foundation of China (Association of Science and technology cooperation across the Taiwan straits, Project No. U1405215).
Author information
Authors and Affiliations
Contributions
M.J.Z., Y.J.Z. and Y.D. conceived and designed the study. M.J.Z. and Y.J.Z. performed the experiments. M.J.Z. and Y.J.Z. performed data analysis. M.J.Z. wrote the manuscript. M.J.Z., Y.J.Z., S.J.C. and Y.D. revised the paper. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zeng, M., Zhong, Y., Cai, S. et al. Deciphering the bacterial composition in the rhizosphere of Baphicacanthus cusia (NeeS) Bremek. Sci Rep 8, 15831 (2018). https://doi.org/10.1038/s41598-018-34177-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-34177-1
Keywords
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.