Abstract
Extracellular vesicles (EVs) are reported to be involved in stem cell maintenance, self-renewal, and differentiation. Due to their bioactive cargoes influencing cell fate and function, interest in EVs in regenerative medicine has rapidly increased. EV-derived small non-coding RNA mimic the functions of the parent stem cells, regulating the maintenance and differentiation of stem cells, controlling the intercellular regulation of gene expression, and eventually affecting the cell fate. In this study, we used RNA sequencing to provide a comprehensive overview of the expression profiles of small non-coding transcripts carried by the EVs derived from human adipose tissue stromal/stem cells (AT-MSCs) and human pluripotent stem cells (hPSCs), both human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSC). Both hPSCs and AT-MSCs were characterized and their EVs were extracted using standard protocols. Small non-coding RNA sequencing from EVs showed that hPSCs and AT-MSCs showed distinct profiles, unique for each stem cell source. Interestingly, in hPSCs, most abundant miRNAs were from specific miRNA families regulating pluripotency, reprogramming and differentiation (miR-17-92, mir-200, miR-302/367, miR-371/373, CM19 microRNA cluster). For the AT-MSCs, the highly expressed miRNAs were found to be regulating osteogenesis (let-7/98, miR-10/100, miR-125, miR-196, miR-199, miR-615-3p, mir-22-3p, mir-24-3p, mir-27a-3p, mir-193b-5p, mir-195-3p). Additionally, abundant small nuclear and nucleolar RNA were detected in hPSCs, whereas Y- and tRNA were found in AT-MSCs. Identification of EV-miRNA and non-coding RNA signatures released by these stem cells will provide clues towards understanding their role in intracellular communication, and well as their roles in maintaining the stem cell niche.
Similar content being viewed by others
Introduction
Stem cells are responsible for the development and regeneration of tissues and maintaining steady-state of organ homeostasis. Stem cells of various types exist; pluripotent stem cells (PSCs), such as embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) have the potential to differentiate into all types of adult human tissues, while stem cells residing in the adult individual, such as mesenchymal stem/stromal cell (MSCs) have a more limited differentiation capacity1. Tissue development and regeneration involves cell activities such as recruitment, proliferation and differentiation, which are mediated by autocrine or paracrine effectors2. Therapeutic activities mediated by paracrine signalling in stem cells have been well documented.
The paracrine effectors of stem cells, such as extracellular vesicles (EVs), which mimic stem cell properties, could represent a relevant therapeutic option in regenerative medicine. EVs are important mediators of intercellular communication and regulate bidirectional transfer of proteins, lipids and nucleic acids between cells via specific receptor-mediated interactions3. The contribution of stem cell-derived EVs in lineage commitments, maintenance of self-renewal, differentiation, maturation, efficiency of cellular reprogramming and cell fate determination are largely regulated by non-coding RNA (ncRNA)4. Small ncRNA (<200 nucleotides) includes microRNA (miRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), piwi-interacting RNA (piRNA), transfer RNA (tRNA), small ribosomal RNA (rRNA), and small cytoplasmic RNA (Y RNA). These are involved in various biological processes and maintain the equilibrium between pluripotency and differentiation in stem cells, thereby aiding in governing stem cell potency and lineage-specific fate decisions5,6. Furthermore, the ncRNAs are known to be sorted into EVs thus modulating cellular processes7,8. Therefore, EV-derived ncRNAs are potential mediators of paracrine effects of stem cells.
Small ncRNAs, particularly microRNAs (miRNAs) which are central to gene regulation and cellular fate determination, can also mediate their regulatory effects via EVs9. miRNAs are small endogenous non-coding RNAs that function as posttranscriptional regulators of gene expression through translational inhibition or by promoting the degradation of mRNA. They are important regulators of reprogramming processes, maintenance of pluripotency and differentiation of stem cells10. EV-derived miRNAs thereby are mediators of the extended paracrine effects of stem cells11,12,13. Thus, it could be concluded that intercellular communication mediated by transfer of EV-derived miRNAs coordinate the intercellular regulation of gene expression, which eventually affects the fate of the stem cells and their surrounding niches.
The primary goal of this study was to characterize the EV-derived miRNAs and other small ncRNAs of AT-MSCs and hPSCs cultured in vitro, and to explore their biological relevance. Identification of EV-miRNA and other small ncRNA signatures released by these stem cells will provide clues towards understanding the role of these extracellular RNAs in intercellular communication and their role in regulation of stemness in vitro.
Taken together, our data indicates that the miRNAs previously reported to be the important regulators of stem cells at cellular level are also present in their EVs, indicating a regulatory role that can be mediated via EVs. Therefore, transfer of these miRNAs and small ncRNA to other cells may promote lineage commitment in stem cells, and subsequently enhance differentiation in a cell therapy setting.
Results
hPSCs and AT-MSCs showed characteristics typical of respective stem cell type
Both stem cell types (MSC and PSC) were characterized with standard methods for the respective cell type in conjunction with media collections for EV extraction. The cells showed characteristic morphology, marker expression, and differentiation capacity.
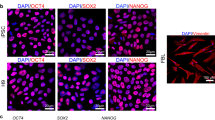
Human PSCs grew as well-defined colonies (Fig. 1A) and further as smooth monolayers reaching confluence, typical to undifferentiated hPSCs on laminin-521. The cells showed uniform expression of core pluripotency transcription factors Nanog, and octamer-binding transcription factor-3/4 (OCT-3/4), as well as typical hPSC surface marker expression of stage-specific embryonic antigens (SSEA) -3 and -4, tumor rejection antigens (TRA)-1-60 and -1-81, as well as lack of expression of early differentiation marker SSEA-1 (Fig. 1B). Further, the cells showed normal diploid karyotypes (Fig. 1C). Pluripotency was confirmed in vitro as differentiation capacity to derivative cells of all three embryonic germ layers (Fig. 1D). Characterisation of the hPSC-1 line is shown in Fig. 1 and hPSC2 line in Supplemental 1.
PSC characterisation. (A) Human PSC-1 characterized for A) typical undifferentiated colony morphology in phase contrast image and (B) expression of pluripotency markers Nanog, OCT-3/4, SSEA-3, SSEA-4, TRA-1-60, and TRA-1-81, and lack of expression of early differentiation marker SSEA-1 after immunofluorescence staining. Corresponding nuclei stains with DAPI shown. (C) Cells showed normal female (46, XX) karyotype after 28 passages in total (9 passages in feeder-free culture). The results of the KaryoLite BoBs assay are shown as signal relative to karyotypically normal female (/F, red) and male (/M, blue) genomic DNA used as a reference (equal to 1) for each of the probes covering both p and q arms of all chromosomes. Software threshold for changes shown as a green lines and deviations in red. (D) Pluripotency shown after spontaneous differentiation in vitro as expression of markers for mesoderm, endoderm, and ectoderm. All scale bars 200 µm.
The characterization of AT-MSCs conformed with previous results14,15 and the standards defined by the International Society for Cellular Therapy (ISCT)16, by cells being successfully isolated and expanded as adherent fibroblast-like cells. Further, the AT-MSCs cultured in FBS-supplemented medium expressed surface markers CD54, CD73, CD90 and CD105, while lacking the expression of hematopoietic markers CD14, CD19, CD45 and HLA-DR (Fig. 2A). In addition, AT-MSCs showed moderate expression of CD34, as previously reported for AT-MSCs14,15,17. Furthermore, CD34 and CD54 showed donor variability18.
AT-MSC characterisation. Surface marker expression (%) of undifferentiated AT-MSCs (n = 3) analyzed by flow cytometric analysis (A). Quantitative data of early and later phase response of osteogenesis by (B) alkaline phosphatase activity (early response) and (C) Alizarin red staining (late response). (D) Alizarin red staining for assessment of mineralized matrix formation. Results show no detection of mineralized matrix in control condition (MM), while osteogenically induced cultures (OM) show enhanced mineralized matrix formation after 21 days. Dots represent biological and technical replicates, and bars represent means. MM; maintenance medium, OM; osteogenic medium, ARS; alizarin red staining, ALP; alkaline phosphatase. Scale bar 100 μm.
The osteogenic differentiation capacity was analyzed for the AT-MSC donor cell lines, assessing both early differentiation capacity by alkaline phosphatase activity and later phase by alizarin red staining for mineralized matrix formation. The early response showed that the osteogenic differentiation capacity gradually increased up to 21 days in osteogenic induction cultures (OM), while control conditions (MM) showed low or no response (Fig. 2B). The alkaline phosphatase activity was normalized to the total DNA. Similarly, the later phase response in terms of mineralized matrix formation, showed gradual increase of matrix accumulation up to 21 days in OM, with no matrix formation seen in control conditions (Fig. 2C,D).
hPSCs and AT-MSCs showed robust secretion of EVs with a cell type specific size distribution
EVs were extracted from the culture media of AT-MSCs and hPSCs using ultracentrifugation15. Both cell types secreted abundant amount of EVs and their presence was confirmed by NTA, EM and WB. Characterization of EVs by NTA (Fig. 3A,B) supported the WB analysis with EV markers such as TSG101, Hsp70, CD63, CD90 (Fig. 3C). EM indicated the presence of heterogeneous population of EVs in both cell types (Fig. 3D,E). WB results indicated that AT-MSC EVs showed stronger signals of CD63 and CD90 than hPSC EVs. TSG101 expression was seen in both cell lines EVs. hPSC EVs showed a clear band of Hsp70, whereas due to technical reasons Hsp70 signal was not detectable (N.D) in AT-MSC EVs. Differences in the expression level of EV markers between cell types in our study could be explained by fact that the EV biogenesis pathways could be different among these cell lines. The purity of EVs was confirmed with the absence of the endoplasmic reticulum marker calnexin in all our EV samples. NTA results showed that AT-MSCs contained 2–4 × 104 of the 100–200 nm sized particles as the major group with a distribution of other sizes in smaller quantities, while hPSCs secreted 1.8–2.3 × 105 of 100–200 nm sized particles (101–200 nm) and very small quantities of larger particles. The media used for EV collections were analyzed parallel to samples. EV-depleted FBS used in AT-MSC culture media was analyzed by NTA displayed large number of particles (101–200 nm), hPSC medium on the other hand as expected contained insignificant amounts of EV sized particles. Particle size determination by NTA is not entirely accurate as it cannot differentiate EVs from other particles such as protein aggregates and lipoproteins19. This results in false positives and impairs the reliability of the analysis. Therefore a combination of TEM, which was also used in this study is needed to visualize and confirm the presence of vesicles.
EV characterisation. (A,B) EV concentration and size distribution measured by nanoparticle tracking analysis (NTA). On y-axis concentrations (particles 106/ml) are shown and on x-axis EV samples of hPSC, AT-MSC and their respective medias; hPSC media, and dAT-MSC media (EV-depleted AT-MSC media). In general. hPSCs contained more particles than AT-MSCs. (C) Western blotting showed presence of stronger signals of EV markers CD63 and CD90 in AT-MSC EVs as compared to hPSC EVs. TSG101 expression was seen in both hPSC and AT-MSC EVs. hPSC EVs showed clear band of hps70, whereas Hsp70 signal was not detectable (N.D) in AT-MSC EVs due to technical reasons. Absence of Calnexin protein in all the EV samples indicated purity of the samples. (D,E) TEM images of hPSC and AT-MSCs. Scale bar 500 nm.
hPSCs and AT-MSCs EVs have unique miRNA signature
Next generation sequencing was used to determine the small ncRNA expression profile of five EV samples (3 × AT-MSC and 2 × hPSC) as well as the corresponding media. Mapping of the reads from the sequencing is shown in Fig. 4, 71% and 43% of the reads of hPSC and AT-MSC samples, respectively, failed to align to the reference genome or were discarded, since they were outmapped, ie. mapped to abundant sequences such as polyA and polyC homopolymers, ribosomal RNA, the mitochondrial chromosome or ΦX174-genome. Most outmapped reads (>80% in all samples) were mapped to 28 S, 18 S, or 5.8 S ribosomal RNA. Out of the remaining reads, approximately half were mapped to small RNAs. On average, 13% of small RNA reads in hPSC samples were mapped to miRNA, while tRNA fragments (69%), Y RNA (7%), snRNA (4%), snoRNA (7%) and piRNA (<1%) made up the rest of reads. In AT-MSC samples, only miRNA (44%), tRNA (47%) and Y RNA (8%) were detected, while other small RNA types comprised <1% or total reads.
Length distribution of reads is shown in Fig. 5. An average of 21.5 million and 8.5 million raw reads were obtained from hPSC and AT-MSC samples, respectively, whereas 7.2 million (hPSC) and 9.3 million (AT-MSC) reads were obtained from the corresponding media. AT-MSC medium showed similar levels of mapped reads in the miRNA (20–25 nucleotides) and other small RNA (30–35 nucleotides) ranges as AT-MSC samples, except for sample AT-MSC-1, which had very high levels of miRNA reads. In contrast, hPSC medium had very low number of reads in the miRNA and smRNA range, resulting in a lower background signal in the hPSC samples compared to AT-MSC samples. The higher read count of hPSC samples compared to AT-MSC samples is explained by a large peak of 35–40 nucleotide reads, which were mostly mapped to ribosomal RNA.
hPSCs and AT-MSCs have distinct miRNA and small RNA profiles
Unsupervised clustering was performed on EV-miRNAs in order to assess the variability among the samples (Fig. 6A). hPSC and AT-MSC samples showed clearly different EV-miRNA expression patterns, but strong similarity within sample groups was observed. Expression patterns of AT-MSC-1 was distinctively different from the other two replicates. This could be due to the medical history of this donor (gastric bypass, antidepressant medication). hPSCs showed a wider range of expressed EV-miRNAs compared to AT-MSCs, with 159 out of 463 miRNAs only present in hPSC EVs. Unsupervised clustering analysis on EV-derived small ncRNA also separated AT-MSCs from hPSCs (Fig. 6B). SNORD family was exclusively present in hPSC-EVs, whereas Y and tRNA fragments were highly expressed in AT-MSCs EVs (Supplemental 2).
EV-miRNAs were divided into groups based on significant (p < 0.01) differential expression or expression in both sample types (|FC| < 2) and a cutoff of 32 mean counts per million (CPM) (Tables 1 and 2). AT-MSCs and hPSCs had unique miRNA profiles at their EV level. Finally, 32 EV-miRNA, majority of which are positive and negative regulators of osteogenic differentiation was exclusively present in MSC, whereas 77 miRNA involved in maintaining pluripotency (including mir families: mir-371/372, mir-302/367, 200, 17/92 and C19MC) were highly represented in hPSC.
Comparison of miRNA and other small RNAs detected in hPSC and AT-MSC medias
Due to the relatively high number of reads obtained from the AT-MSC medium, we compared each type of samples to their respective media. Various normalization strategies are used for EV-RNA data, such as library size, geometric mean, endogenous references, or external spike-ins20. Since practically no similarity in RNA content can be assumed to exist between the unconditioned media and cell-derived EV samples, we normalized the reads to external spike-ins, as has been done previously to compare RNAs between different types of samples8. As can be seen in Fig. 7, considerable amounts of both miRNAs and other small RNAs were identified in the media (for full list, see Supplemental 3). Especially in AT-MSC medium, levels of several RNAs were similar to AT-MSC samples. To take this into account, RNAs were required to have at least 2 times higher normalized read count compared to media in all samples to be included in unsupervised clustering and isomiR analysis (see Materials and Methods for details).
Comparison of sample miRNA and small RNA to the respective media. Read counts normalized to external spike-ins for 38 miRNAs and other small RNAs with highest counts in each medium (blue) and mean counts in corresponding samples (red). Error bars depict the range of reads (min-max). miRNA and small ncRNA identified in hPSC media (A,B) and AT-MSC media (C,D).
IsomiR identification
It is known that majority of miRNA genes encode isomiRs with 3′ or 5′ modifications. We restricted the isomiR identification to the major miRNA families in both stem cell types most abundant miRNAs present in both stem cell types (Tables 1 and 2). Our analysis showed that both hPSC and AT-MSC expressed isomiRs as shown in Fig. 8, where the frequency of different types of modifications in the identified miRNA families are highlighted. Some type of variation was present in a high proportion (72%) of the miRNA reads. In hPSC samples, majority (66%) of miRNAs were mostly comprised of the canonical isomiR and variation in the 3′ end was the most common isomiR type. In contrast, majority (53%) of AT-MSC miRNAs were mostly present as isomiRs with 3′ variation, and only 34% of miRNA were mostly present as the canonical isomiR. However, these common modifications are not as likely to have functional importance as the rarer variations in the 5′ end or in the miRNA seed region. Interestingly, some miRNAs in hPSC EVs (miR-302a-5p, miR-520f-3p, miR-523–3p) were predominantly present as isomiRs with 5′-end modifications.
Discussion
Understanding the exact composition of EV-derived small ncRNA of defined stem cell type will increase our knowledge about their role in intercellular communication as well as their potential as diagnostic biomarkers. Studies with in-depth characterization of EV-derived ncRNA from clinical grade hPSCs are lacking, and only few studies have explored EV-derived ncRNA of AT-MSCs. The present study utilized high-throughput RNA sequencing to provide an in-depth characterization of small ncRNAs in EVs derived from human AT-MSC and hPSC. This approach identified stem cell specific differences in ncRNA including: miRNA, miRNA isoforms, tRNA, yRNA, snoRNA, and snRNA. Limitation of the study is the low number of samples per group (n = 2–3), as is the case for most sequencing studies on EVs until now. To overcome this, we have reported the miRNAs with the highest expression levels and included seq data from fresh media as controls, to account for the background signal from the media. Some of the miRNA and other small ncRNA identified in our AT-MSC data are in the line with previously published reports, which not only confirm our findings but also indicates that signature EV RNAs are not affected by the donor variations. Our data validated the previously identified cellular genes at EV level, thus highlighting the regulatory potential of EV-ncRNAs and the potential stem cell specific EV biomarkers for future studies.
With all EV isolation methods, a mixed population of vesicles with varying yield is generally obtained. There is no consensus regarding the isolation methods as of date. Therefore, method of choice depends on the downstream applications. Initially, we selected miRCURY EV isolation kit for both EV characterization and EV isolation for RNA sequencing. Despite the dispute about residual cellular RNA in several EV precipitation methods21, miRCURY precipitation kit has been reported having better performance. The isolated fraction exhibited specfic surface markers, size distribution of the particles and microRNAs characteristic for EVs22. Due to high background with miRCURY precipitation kit, technical problems were encountered at all EV characterization steps. Therefore, we performed EV characterization using ultracentrifugation (UC). In our experience, for EV characterization, isolation of EVs using UC technically stands out as a better option than commercially available precipitation kits15,23. Our approach of using two different EV isolation methods may have yielded different and overlapping EV populations. This observation is supported by a recent study where five different EV isolation methods (including UC and precipitation based methods) were compared for profiling miRNAs by sequencing. They concluded that each method captured distinctive but partially overlapping EV populations, along with varying degrees of non-EV contamination24.
EV isolation and RNA sequencing was done according to the Exiqon’s workflow. miRCURY precipitation kit was used to purify EVs. Before precipitation, two centrifugation steps were done to remove cellular components and two supernatant removal steps to get as much of the un-precipitated material away before the next step of EVs lysis followed by RNA extraction. Despite these critical attempts to remove non-EV contaminants, we still cannot rule out the possibility that our data is not only based on EV associated RNA, but also RNA co-isolated from the non-vesicular compartment could be also involved.
Small ncRNA profiles of AT-MSCs and hPSCs were unique and highly distinct from each other, which was consistent with previous publications. At transcriptomic and proteomic level, embryonic stem cell and bone marrow stem cells derived EVs displayed different EV-RNA and proteomic profiles25. Stem cells and stem cell derived EVs have been reported to exhibit several similar features which affects the stem cell fates, for instance similar miRNA expression profiles between MSCs and MSC-EVs has been documented26. In our dataset, 1.4–1.5% of total reads mapped to miRNA in hPSCs and 2.5–4% of reads mapped to miRNA in AT-MSC-2 and AT-MSC-3. However, AT-MSC-1 displayed a different distribution of reads, with markedly greater proportion of miRNA reads (37%). A possible explanation for the discrepancy between AT-MSC-1 and other samples is the gastric bypass surgery and the anti-depression medication used by the donor of AT-MSC-1 sample.
A large fraction of identified EV-miRNAs in our data were also previously reported to be expressed at cellular level7,26. miRNAs shuttled by MSC-derived EVs were shown to be involved in the control of transcription, cell survival, multi-organ development, differentiation and immune system regulation27.
A low percentage of miRNA reads is typical to extracellular vesicle samples8,28,29,30,31,32,33. Although the number of miRNA reads per sample was relatively low, miRNA reads were of high quality and mostly close to full length (Supplemental 5), as well as showed robust expression in both sample types (Fig. 6).
Several studies have shown that miRNAs are involved in regulating the unique ESC cell cycle, and also the balance between pluripotency and differentiation of hESCs34,35,36. At cellular level, there are generally subtle differences in the miRNA profiles between the two pluripotent cell types (hESCs and hiPSCs)37. Our data showed very similar EV-miRNA patterns for the two pluripotent cell types indicating that irrespective of the derivation source, hPSCs release their characteristic miRNA content. In previous studies, human PSCs have shown characteristic miRNA profiles undetectable in adult organs such as miR-302, miR-372, mir-17-92, mir-200, and C19MC families38. Our EV-miRNA data showed that hPSC also secreted these aforementioned hPSC specific miRNA clusters (Table 1).
The mir-200 and mir-302 are two large miRNA families involved in maintaining pluripotency by regulating the core regulatory circuitry of pluripotency genes (Nanog, OCT4, SOX2, KLF4). miR-302-driven cellular reprogramming coordinates stem cell division by regulating targets in the cell cycle, particularly at the G1/S restriction point35,38,39,40. Moreover miR-302 and miR-372 cluster miRNAs promote human somatic cell reprogramming to iPSCs41,42. Furthermore, these miRNAs repress multiple target genes regulating various other cellular processes, including epithelial-mesenchymal transition (EMT), epigenetic regulation and vesicular transport.
The chromosome 19 miRNA cluster (C19MC) is one of the largest miRNA gene clusters in the human genome. Expression of mir-516a, b-5p, 517a, b-3p, 518b, 520c, f, g-3p, 522-3p, 523-3p, 526b-5p, 1323 from C19MC was detected in our analysis. This primate-specific miRNA cluster is restrictively expressed in pluripotent ESCs40,43,44 and, later during embryonic and fetal development, only in the placenta but not in other adult organs and tissues45. C19MC miRNAs represent the majority of miRNAs not only in the trophoblast but also in EVs derived from it. The target genes are strongly associated with developmental processes and various cancers. It is suggested that these miRNAs may play critical roles in regulating differentiation and growth during the early development and in maintaining the pluripotency of hESCs46.
ESC-EVs have shown to promote the survival and improve the expansion of hematopoietic progenitor cells47 and to induce de-differentiation and alterations in gene expression on cultured retinal progenitor Müller cells48. EV-treated Müller cells showed up-regulation of genes and miRNAs associated with cellular proliferation and induction of pluripotency and down-regulation of genes important to differentiation and cell cycle arrest. These findings suggest that by transferring stem cell-specific molecules, EVs can induce the activation of endogenous, adult, quiescent progenitor cells, increasing their pluripotency and possibly their ability to repair damaged tissues. Our data confirmed that PSC-specific miRNA previously identified on cellular level are also found in EVs. Furthermore, additional EV-specific miRNAs were also identified, but deciphering their role need further studies.
In order to establish a global miRNA expression profile of MSCs, the consensus miRNAs in MSCs detected by different methods and from various sources have been previously reported49. Among which mir-199a, 152, 125a, b, 143,100 and let-7b, c, e, f, were also identified in our AT-MSCs derived EVs (Table 2). Altogether our results strengthen the hypothesis that signature MSC miRNA are also expressed in their EV cargoes. Nine out of the highly expressed EV-miRNAs in our AT-MSC samples (let-7a-5p, let-7b-5p, let-7i-5p, miR-125-5p, miR-199a-3p, miR-199b-3p, miR-100-5p, miR-144-3p and miR-22-3p) have previously been also found to be present in bone-marrow-derived MSC EVs50. This clearly highlights that independent of cell of origin, MSCs share certain characteristic miRNA-EVs which are involved in regulating differentiation. mir-10a-5p, mir-10b-5p, mir-22-3p, mir-143-3p, mir-100-5p, let-7a, f-5p identified in our data have also previously been listed as most abundant miRNA in AT-MSCs EVs12. These results indicate that expression of these miRNA is unaffected by donor variability.
There are substantial numbers of experimental data confirming that miRNAs have multiple roles in regulating bone remodeling51 as well as osteogenesis. miRNAs are known to regulate osteoblast differentiation positively by targeting negative regulators of osteogenesis or negatively by targeting osteogenic factors52. Recently it has been reported that MSCs secrete miRNA and other small RNAs via EVs12. Our data revealed that in AT-MSCs, EV-miRNAs that modulate osteogenic commitments of AT-MSC, for example; let-7a and c, mir-22, 199a, 196a, 199b and negative regulators of osteogenic differentiation such as mir-27, 98, 100, 615, 125b and 195 were enriched exclusively in AT-MSCs as compared to hPSCs (Table 2). This indicates that these EV-miRNAs have the potential to modulate bone differentiation pathways of the target cell.
Specifically, let-7 family, which is classically involved in stem cell differentiation, is considered as a global regulator of differentiation53. let-7c is known to enhance osteogenesis and thus bone formation while repressing adipogenesis of human MSCs by targeting HMGA254. let-7a and mir-199b were detected in the EVs during osteogenic differentiation of BM-MSCs55. miR-199b-5p displayed positive role in osteoblast differentiation as it was significantly up-regulated during the osteogenesis in human BM-MSCs. Its functions as a positive regulator of osteogenesis is likely due to its involvement in GSK-3β/β-catenin signaling pathway56. miR-199a has been shown to increase osteogenic differentiation in vitro and to enhance ectopic bone formation in vivo. Expression of this miRNA is controlled by a HIF1α and Twist1 cyclic pathway57. mir-22 has been reported to enhance osteogenic differentiation of AT-MSCs by repressing its target HDAC6 and also by acting as a crucial regulator of switch between adipogenic and osteogenic differentiation58. EV delivery significantly enhances bone formation in vivo and bone regeneration has been speculated to be regulated by EV-derived mir-196a59. This miRNA has been shown to inhibit proliferation and enhance the differentiation process by regulating HOXC860.
Negative regulators of osteogenic differentiation (miR-100-5p, miR-125b-1-3p, miR-98-5p, miR-195-3p, miR-24-3p, miR-27a-3p, and miR-615-3p) were exclusively expressed in our AT-MSC samples as opposed to hPSC samples. Many of these miRNAs affect well-known BMP induced osteogenic differentiation61. In AT-MSCs overexpression of miR-100 inhibited osteogenic differentiation of stem cells by targeting BMPR262. mir125b directly targeting BMPR1b was identified as negative regulator of osteogenic differentiation of human BM-MSC63. mir-98 in BM-MSCs regulated osteogenic differentiation by targeting BMP264. mir-195 not only acts as a negative regulator of osteogenesis in human BM-MSCs, but also as an inhibitor of stem cell proliferative capacity and as an anti-angiogenic by targeting VEGF65. mir-27, whose promoter is negatively regulated by RUNX2, functionally inhibits osteogenesis by downregulating SATB2, which is an important regulator of osteogenic differentiation66. mir-615 negatively regulates the osteogenic differentiation by suppressing osteogenic regulators GDF5 and FOXO167. Thus EV-miRNAs which are negative regulators of osteogenesis could be the potential targets for enhancing bone regeneration as they can by directly delivered as anti-mir oligonucleotides into bone injury sites. The approach of using EVs as vehicles to deliver miRNA mimics or anti-mirs is certainly a potential tool in future bone regeneration applications.
IsomiRs, which are functionally different from their canonical microRNA are most probably generated by variation in processing by Drosha and/or Dicer and differ by variations of a few bases at 5′ and 3′ end of miRNA68,69. They affect the half-life of miRNA, their sub-cellular localization and also their target specification70,71,72. IsomiRs with variations at the 3′ end were the predominant isomiR category identified in our data in both stem cell types (Fig. 8), and the ratio of isomiR reads to canonical miRNA reads was similar to earlier studies33,73. Exact function of isomiRs with 3′ modifications are not clearly known but are suggested to be related to a disease state74. In hPSC for example, we found that miR-302a, an important miRNA for hESCs self-renewal, showed predominant expression of the isomiR species with 5′ –end variations over its canonical miRNA. mir-302a isomiRs with unique seed sequences have indicated specificity in their target selection and therefore likely to be significant regulators of cellular differentiation75. Importance of the isomiRs in different stem cell types with variations at 3′ and 5′ end were reported by Tan et al.69, indicating that isomiRs are active in vivo and thereby have functionally importance as they co-immunoprecipitate with Ago proteins and are active in in vitro luciferace and cleavage assays.
Apart from miRNAs, 16 snRNA and 67 snoRNAs were the most highly-expressed non-coding RNA in hPSC-EVs. These non-coding RNAs play important roles in mRNA biogenesis and maturation8,76 and are also known to be involved in human cancers77. ncRNAs such as y and tRNA fragments displayed selectivity for AT-MSC which was consistent with recent reports12. Recent studies have reported that Y RNA fragments may play role in histone mRNA processing and cell damage78,79 and DNA replication80. Fragments derived from tRNA play important roles in regulating many biological processes, such as reverse transcription and guidance of other RNAs81. Functions of these ncRNAs in stem cell biology needs to be further elucidated.
Fetal bovine serum (FBS) used in cell culture experiments contains bovine EVs which interferes with the EVs produced by the cells. In our previous work we have reported that FBS used in cell culture experiments contains abundant amounts of EVs15. Ultracentrifugation (100,000 g for 19 hours), the most commonly used protocol for removal of FBS derived EVs, does not completely remove the EVs15. The current study support our previous findings, confirming presence of FBS derived miRNAs and small ncRNA in the EV depleted media (Fig. 7). Specifically, mir-122-5p, mir-423-5p and mir-148a-3p were the abundant miRNAs present in the media. mir-122 has also been previously reported to be highly enriched in FBS82. Other miRNA and small ncRNA present in EV depleted FBS are shown in the Supplemental 3. This issue should be taken into consideration when drawing final conclusions about cell culture results as the interference and the biological effects associated with FBS derived RNA is not clearly known. hPSC medium used for culturing hPSCs, as expected, showed insignificant amounts of miRNAs and ncRNAs, assumed to be derived from medium components.
Conclusion
Overall, in our small scale analysis, we found that both AT-MSCs and hPSCs secrete a selective pattern of small ncRNA in there cell-free secretome. Their transfer to the target cells could be a mechanism of action for maintaining the stem cell specific characteristics, orchestrating gene expression, and to mediating communication between neighboring cells. Understanding of paracrine regulation of pluripotency and differentiation commitment of stem cells, could have implications on the improvement of stem cell cultures and differentiation protocols. Thus, in-depth understanding of EV-derived ncRNA regulatory mechanisms could provide strategies for developing engineered EVs with therapeutic RNA.
Material and Methods
Culture and characterization of stem cells
hPSC lines
Human ESC line hPSC-1 was derived from surplus, blastocyst stage human embryo as described previously83. Human iPSC line hPSC-2 was generated at Prof. Katriina Aalto-Setälä’s laboratory at University of Tampere from dermal fibroblasts (Table 3). For this study, hPSCs were cultured in xeno- and serum-free Essential 8™ Flex Medium (E8 flex, Thermo Fisher Scientific), supplemented with 50 U/ml Penicillin-Streptomycin (Gibco, Thermo Fisher Scientific) on Corning® CellBIND® -well plates coated with 0.55 µg/cm2 human recombinant laminin-521 (LN-521, Biolamina, Sweden). Single cell passaging with TrypLE™ Select Enzyme (Gibco, Thermo Fisher Scientific) was carried out twice a week using split ratio of 40 000-50 000 cells/cm2 as described by Hongisto et al.84.
hPSC characterization
Human PSCs were characterized as described previously84. Briefly, the hPSCs were monitored regularly microscopically, and characterized by immunofluorescence staining (IHC) for expression of pluripotency markers using the following primary antibodies; Nanog (1:200, R&D Systems, AF1997), OCT-3/4 (1:200, R&D Systems, AF1759), SSEA-3 (1:300, Novus Biologicals NB100–1832), SSEA-4 (1:200, R&D Systems, MAB1435), TRA-1-60 (1:200, Millipore, MAB4360), TRA-1-81 (1:200, Santa Cruz Biotechnology SC-21706), and early marker for differentiation SSEA-1 (1:200, Santa Cruz Biotechnology, SC-21702). Alexa Fluor-conjugated (1:400, ThermoFisher Scientific A-11055, A-21042, A-10037), and FITC-conjugated (1:400, Novus Biologicals, NB7102) secondary antibodies were used. Nuclei were counterstained with 4′,6′diamidino-2-phenylidole (DAPI) (Vector Laboratories Inc., Burlingame, CA).
Pluripotency was verified with in vitro pluripotency assay by spontaneous differentiation as embryonic bodies84, followed by immunofluorescence analysis for alpha-smooth muscle actin (SMA, 1:400, R&D Systems, MAB1420) for mesoderm, alpha-fetoprotein (AFP, 1:200, R&D Systems MAB1369) for endoderm, and OTX2 (1:200, R&D Systems, AF1979) for ectoderm. Karyotyping was performed at Finnish Microarray and Sequencing Centre (FMSC), Turku Centre for Biotechnology with the KaryoLite BoBs assay (Perkin Elmer).
AT-MSC lines
Human AT-MSC were obtained from water jet-assisted liposuction aspirates85 from three donors using mechanical and enzymatic isolation as described previously86. All donors were female, age range of 32–53 (average 45) and BMI range of 22.7–31.2 (average 25.8) (Table 3). Cells were cultured in AT-MSC media consisting of Dulbecco’s modified Eagle’s medium/Ham’s Nutrient Mixture F-12 with 1% L-alanyl-L-glutamine (DMEM/F-12 1:1 GlutaMAX; Gibco ref. 31331–028, lot. 1765999), 1% antibiotics (100 U/ml penicillin, 0.1 mg/ml streptomycin; Lonza ref. DE 17–602 E, lot. 5MB 068) and 10% FBS (Fetal Bovine Serum, South American ref. 10270106, lot. 42F8554K) at 37 °C and 5% CO215. Once AT-MSCs had adhered to the culture flask, non-adherent populations were gently washed away with PBS and fresh culture media was added.
AT-MSC characterization
Cells cultured in AT-MSC media (n = 3) were characterized using BD Accuri C6 flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) to confirm the mesenchymal origin of the cells. Allophycocyanin (APC)-conjugated monoclonal antibodies against CD14 (clone: M5E2), CD19 (clone: HIB19), CD34 (clone: 581), CD45RO (clone: UCHL1), CD54 (clone: HA58), CD73 (clone: AD2), CD90 (clone: 5E10), CD105 (clone: 266) and HLA-DR (clone: G46-6) (BD Pharmingen, Becton Dickinson) were used. Analysis was performed on 100.000 cells per sample, and the positive expression was defined as the level of fluorescence greater than 99% of the corresponding unstained cell sample15. Flow cytometric data is shown as average with standard deviation. Cells from three donors were plated at 2.000 cells/cm2 in four replicates per condition in 24-multiwell plate to analyse the osteogenic potential of AT-MSCs. Cells were first plated in AT-MSC media. After 24 h, osteogenic differentiation was induced using osteogenic media (OM; AT-MSC media supplemented with 50 µM L-ascorbic acid 2-phosphate, 10 mM β-glycerophosphate disodium salt hydrate and 5 nM dexamethasone (all Sigma-Aldrich). At 7, 14 and 21days time points, osteogenesis was assessed using quantitative alkaline phosphatase activity (qALP) analysis which was normalized with total DNA quantification and quantitative Alizarin red staining (qARS) as reported previously15,87.
EV isolation and basic characterization
hPSC media collection for EV extraction was performed from four consecutive passages for both cell lines. Media were collected from 80–100% confluent cells cultures. The day after passaging, the hPSCs were rinsed twice with DPBS (Lonza), and fresh hPSC medium was added to the cells. 48 hours later, the hPSC conditioned media were collected to 50 ml Falcon tube, and immediately centrifuged 2,000 × g for 10 minutes at 4 °C to remove cell debris. Supernatant was transferred to a new Falcon tube and frozen to -80 °C for storage. Non-conditioned hPSC medium was collected as control. Prior to EV isolation, AT-MSCs were grown in EV-depleted medium for 72 hours. EV-depletion was performed as described previously15. Briefly, EV depleted FBS (dFBS) was prepared by 19 h ultracentrifugation of regular FBS at 26 000 rpm (121 896 gmax) using an SW28 rotor (k-factor 284.7, Beckmann-Coulter). Only the light coloured top layers of the supernatant (approx. 9/10) were retained and used in the subsequent analyses. The dFBS was filtered with a 0.22 μm filter (Millipore Stericup-GP, 0.22 μm, polyethersulfone filter) before addition to cell-culture medium. For characterization, EVs were isolated from conditioned culture medium using ultracentrifugation as described previously15. Briefly, the conditioned medium was depleted of cell debris by centrifuging for 10 min at 2,500 x g and only the AT-MSC supernatant was filtered through a 0.45 μm sterile filter (Merck Millipore). EVs were extracted using ultracentrifuge at 26,000 rpm (121 896 gmax) for 2 hours at 4 °C with SW28 rotor to collect the EV pellet, which was washed by filtered PBS and stored in Protein LoBind microcentrifuge tubes (Eppendorf) at −80 °C. For RNA sequencing, EVs were isolated from the conditioned medium by precipitation using the miRCURYTM Exosome Isolation Kit (Exiqon A/S, Vedbaek, Denmark) according to the manufacturer’s instructions.
Nanoparticle tracking analysis (NTA)
The number and size distribution of particles from isolated EV samples were analysed using Nanosight model LM14 (NanoSight Technology, Salisbury, U.K., http://www.malvern.com) equipped with blue (404 nm, 70 mW) laser and CMOS camera (Hamamatsu Photonics K.K., Hamamatsu City, Japan). For the analyses samples were diluted in filtered (0.1 µm) DPBS to obtain the optimal detection concentration of 106–109 particles/ml, and triplicate 60 s videos were recorded using camera level 13. The data was analysed using NTA software 3.0 with the detection threshold 588.
Transmission electron microscopy
Particle morphology was examined using transmission electron microscopy (TEM) Tecnai 12 (FEI Company, Eindhoven, the Netherlands) operating at 80 kV as described previously89. Briefly, after loading to 200 mesh copper grids and fixation with 2% PFA in 0.1 M NaPO4 buffer (pH 7.0), samples were washed with the 0.1 M NaPO4 buffer and deionized water, negatively stained with 2% neutral uranyl acetate and embedded in methyl cellulose uranyl acetate mixture (1.8/0.4%). Images were taken with Gatan Orius SC 1000B CCD-camera (Gatan Inc., USA) with 4008 × 2672 px image size and no binning. Samples from 2–3 biological replicates were viewed.
Western blotting
Western blotting was performed as described previously89 using primary antibodies against Hsp70 (#554243, BD Biosciences), and CD63 (#556019, BD Biosciences) at 1:1000 dilution, as well as TSG101 (#SAB2702167, Sigma-Aldrich), and CD90 (#WH0007070M1, Sigma-Aldrich) at 1:500 dilution. The lack of calnexin served as an indicator for the purity of EVs samples, calnexin is an endoplasmic reticulum protein; probing of such protein involved the use of Calnexin (C5C9) Rabbit mAb (#2679, Cell Signaling Technology) at 1:800 dilution. EVs isolated by ultracentrifugation from equal volumes (20 mL) of each sample were loaded to gels. As controls, 30 µg of protein from AT-MSC and hPSC lysates measured by BCA assay (Pierce BCA Protein Assay Kit) were used, growth media for both AT-MSC and hPSC served as negative control samples. Probing for CD63 and CD90 was done on individual samples, however, due to limited amount of EV samples, Hsp70 and TSG101 protein detection was detected from pooled samples. Samples were denatured at 95 °C for 5 min in reducing Laemmli sample buffer; except for CD63 detection which was run in non-reducing conditions, separated using Mini-PROTEAN® TGX™ 12% gradient SDS-PAGE gel (Bio-Rad, Hercules, CA, USA) with prestained protein ladder (BlueSTAR Prestained Protein Marker, # MWP03, Nippon Genetics Europe GmbH) as a standard, running conditions were 150 V for 60 minutes. Blotting involved semi-dry transfer of proteins on nitrocellulose membranes, 0.2 µm (#162–0112, BIORAD), using 40 mA per gel for 60 minutes. Blocking and antibody incubations were performed in Odyssey blocking buffer (LI-COR) with and without 0.1% Tween-20. After primary antibody overnight incubation at +4 °C, membranes were washed 4 × 5 minutes in TBS-T, and probed with secondary IRDye® 800CW Goat (Li-COR) at 1: 15,000 for 1 hour at RT. After incubation, membranes were washed 4 × 5 minutes in TBS-T at RT and briefly rinsed with PBS 1×, then imaged on an Odyssey FC Imager (Li-COR). All original WB images are shown in Supplemental 4.
Small RNA sequencing
Sequencing experiments for three AT-MSC samples and two hPSC samples and the corresponding pure media were conducted at Exiqon Services, Denmark. EVs were isolated from 3 ml conditioned cell culture media using miRCURY™ Exosome Isolation kit (Exiqon A/S). Before precipitation, two centrifugation steps were used to remove cellular components and two supernantant removal steps to get rid of the un-precipitated material, followed by EV lysis and RNA isolation using miRCURY™ RNA Isolation Kits - Cell & Plant (Exiqon A/S). Good performance of Exiqons EV isolation kit has been reported22. NGS libraries were prepared using the NEBNext® Small RNA Library preparation kit (New England Biolabs), consisting of adapter ligation, cDNA conversion, PCR amplification (18 cycles) and purification. From a total of 50 µl isolated RNA, 6 μl was converted into microRNA NGS libraries. Library preparation QC was performed using Bioanalyzer 2100 (Agilent). Based on the quality of the inserts and the concentration measurements the libraries were pooled in equimolar ratios. The pool was then size selected using the LabChip XT (PerkinElmer) aiming to select the fraction with the size corresponding to microRNA libraries (~145 nt). The library pools were quantified using the qPCR KAPA Library Quantification Kit (KAPA Biosystems). Single-ended sequencing with 50 cycles was then performed on the library pools using a NextSeq500 sequencing instrument according to the manufacturer instructions. Raw data was de-multiplexed and FASTQ files for each sample were generated using the bcl2fastq software (Illumina inc.).
Reads were mapped using Bowtie 2 software90 according to the Exiqon NGS Service microRNA/smallRNA pipeline. First, reads mapped to spike-ins or outmapped (mapped to adapter sequences, ribosomal RNA, ΦX174-genome, mitochondrial RNA, or polyA and polyC homopolymers) with the following parameters: ‘-N 0 -L 32–no-1mm-upfront -R 10 -D 15–n-ceil C,0–score-min C,0’, were discarded. Second, reads were mapped to mature miRNAs from miRBase 20 with the same parameters. Third, unaligned reads were mapped to GRCh37 genome with the parameters ‘-N 1 -L 32 -R 4 -D 20–n-ceil L,0,0.15–rdg 200,51–rfg 200,51–score-min C,-250’. The reads were then quantified (including isomiR quantification) using custom Ruby scripts. IsomiRs were defined as sequences with a single base substitution or having 5′ and 3′ insertion/deletion compared to the mature canonical miRNA sequence.
Only RNAs with more than 1 CPM in at least two samples were included in the analysis. Unsupervised clustering was performed using Euclidean distance metric and average linkage. Differential expression was calculated using the exact test for negative-binomially distributed counts with tagwise dispersion in the edgeR package 3.20.191. For a large proportion of the identified miRNAs and smRNAs, similar quantities were also detected in the corresponding unconditioned media, especially for AT-MSC derived EVs (Fig. 8C,D), but also for hPSC EVs (Fig. 8A,B). Therefore, to filter out the RNAs possibly originating from the media, the following procedure was employed: first, raw read counts were normalized to external spike-ins (UniSp100-UniSp151), since assuming same levels of RNA for unconditioned media and EV samples is not appropriate. Second, RNAs overexpressed in either sample group (|Fold change| > 2) were required to have at least 2-fold higher spike-in normalized counts in all samples than in the corresponding media to be included in the analyses. RNAs with similar expression levels between samples (|Fold change| < 2) were required to have at least 2-fold normalized counts in both sample groups. Multiple hypothesis correction was performed using the Benjamini-Hochberg procedure92 with a FDR limit of 0.01.
We have submitted all relevant data of our EV characterization to the EV-TRACK knowledgebase (EV-TRACK ID: EV180022)93.
Ethical approval and informed consent
For the hPSCs, the research groups at the University of Tampere have approval of the National Authority for Medicolegal Affairs Finland (Dnro 1426/32/300/05) to use human embryos for research purposes, and supportive statements of the Ethical Committee of the Pirkanmaa Hospital District to derive, culture, and differentiate hESC lines (Skottman/R05116) and to use hiPSC lines in ophthalmic research (Skottman/R14023). No new hPSC lines were derived for this study.
For the AT-MSCs, the study was carried out with supportive statements of the ethical committee of Helsinki and Uusimaa Hospital District for the use of adipose tissue and derivatives (DNro 217/13/03/02/2015) and with informed consent from the donors. All methods were carried out in accordance with the relevant guidelines and regulations.
Data Availability
The RNA sequencing data has been deposited to the GEO (accession number GSE113868). We have submitted all relevant data of our EV characterization to the EV-TRACK knowledgebase (EV-TRACK ID: EV180022).
References
Yunjoon, J., Gerhard, B. & Nolta, J. A. Concise Review: Induced Pluripotent Stem Cell‐Derived Mesenchymal Stem Cells: Progress Toward Safe Clinical Products. Stem Cells 30, 42–47 (2011).
Santos, A. R. Jr., Nascimento, V. A., Genari, S. C. & Lombello, C. B. Mechanisms of Cell Regeneration — From Differentiation to Maintenance of Cell Phenotype, Cells, and Biomaterials in Regenerative Medicine. InTech (2014).
Riazifar, M., Pone, E. J., Lötvall, J. & Zhao, W. Stem Cell Extracellular Vesicles: Extended Messages of Regeneration. Annu. Rev. Pharmacol. Toxicol. 57, 125–154 (2017).
Nawaz, M. et al. Extracellular Vesicles: Evolving Factors in Stem Cell Biology. Stem Cells Int 2016, 1073140 (2016).
Judson, R. L., Babiarz, J. E., Venere, M. & Blelloch, R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 27, 459–461 (2009).
Zhang, W. et al. Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Res Ther 8, 30 (2017).
Fatima, F. et al. Non-coding RNAs in Mesenchymal Stem Cell-Derived Extracellular Vesicles: Deciphering Regulatory Roles in Stem Cell Potency, Inflammatory Resolve, and Tissue Regeneration. Front Genet 8, 161 (2017).
Wei, Z. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells.. Nat Commun. 26, 8(1), 1145 (2017).
Fatima, F. & Nawaz, M. Vesiculated Long Non-Coding RNAs: Offshore Packages Deciphering Trans-Regulation between Cells, Cancer Progression and Resistance to Therapies. Noncoding RNA. 23, 3(1) (2017).
Mens, M. M. J. & Ghanbari, M. Cell Cycle Regulation of Stem Cells by MicroRNAs. Stem Cell Rev (2018).
De Luca, L. et al. MiRNAs and piRNAs from bone marrow mesenchymal stem cell extracellular vesicles induce cell survival and inhibit cell differentiation of cord blood hematopoietic stem cells: a new insight in transplantation. Oncotarget 7, 6676–6692 (2016).
Baglio, S. R. et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res Ther 6, 127 (2015).
Nawaz, M. Extracellular vesicle-mediated transport of non-coding RNAs between stem cells and cancer cells: implications in tumor progression and therapeutic resistance. Stem Cell Investig. 24, 4, 83 (2017).
Patrikoski, M. et al. Development of fully defined xeno-free culture system for the preparation and propagation of cell therapy-compliant human adipose stem cells. Stem Cell Res Ther 4, 27 (2013).
Kornilov, R. et al. Efficient ultrafiltration-based protocol to deplete extracellular vesicles from fetal bovine serum. J Extracell Vesicles 7, 1422674 (2018).
Dominici, M. et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8, 315–317 (2006).
Sidney, L. E., Branch, M. J., Dunphy, S. E., Dua, H. S. & Hopkinson, A. Concise Review: Evidence for CD34 as a Common Marker for Diverse Progenitors. STEM CELLS 32, 1380–1389 (2014).
Lindroos, B. et al. Differential gene expression in adipose stem cells cultured in allogeneic human serum versus fetal bovine serum. Tissue Eng Part A 16, 2281–2294 (2010).
Maas, S. L. N. et al. Possibilities and limitations of current technologies for quantification of biological extracellular vesicles and synthetic mimics. Journal of Controlled Release 200, 87–96 (2015).
Mateescu, B. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J Extracell Vesicles. 7, 6(1), 1286095 (2017).
Li, P., Kaslan, M., Lee, S. H., Yao, J. & Gao, Z. Progress in Exosome Isolation Techniques. Theranostics. 7, 789–804 (2017).
Helwa, I. et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS One. 23, e0170628 (2017).
Gebraad, A. et al. Monocyte-derived extracellular vesicles stimulate cytokine secretion and gene expression of matrix metalloproteinases by mesenchymal stem/stromal cells. FEBS J. 285, 2337–2359 (2018).
Buschmann, D. et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J Extracell Vesicles. 4, 1481321 (2018).
Billing, A. M. et al. Comprehensive transcriptomic and proteomic characterization of human mesenchymal stem cells reveals source specific cellular markers. Sci Rep 6, 21507 (2016).
Shao, L. et al. MiRNA-Sequence Indicates That Mesenchymal Stem Cells and Exosomes Have Similar Mechanism to Enhance Cardiac Repair. Biomed Res Int 2017, 4150705 (2017).
Camussi, G., Deregibus, M. C. & Cantaluppi, V. Role of stem-cell-derived microvesicles in the paracrine action of stem cells. Biochem. Soc. Trans. 41, 283–287 (2013).
Tosar, J. P. et al. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 23, 5601–16 (2015).
Sork, H. et al. Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Sci Rep. 17, 10813 (2018).
Nolte-‘t Hoen, E. N. et al. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 40, 9272-85 (2012).
Fiskaa, T. et al. Distinct Small RNA Signatures in Extracellular Vesicles Derived from Breast Cancer Cell Lines. PLoS One. 31, e0161824 (2016).
Lunavat, T. R. et al. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells–Evidence of unique microRNA cargos. RNA Biol. 12, 810–23 (2015).
Berardocco, M. et al. RNA-seq reveals distinctive RNA profiles of small extracellular vesicles from different human liver cancer cell lines. Oncotarget. 8, 82920–82939 (2017).
Qi, J. et al. microRNAs regulate human embryonic stem cell division. Cell Cycle 8, 3729–3741 (2009).
Wang, Y. et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 40, 1478–1483 (2008).
Card, D. A. G. et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol. 28, 6426–6438 (2008).
Siti Razila, A. R. et al. Profiling of MicroRNA in Human and Mouse ES and iPS Cells Reveals Overlapping but Distinct MicroRNA Expression Patterns. PLoS One 8, e73532 (2013).
Wilson, K. D. et al. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 18, 749–758 (2009).
Lipchina, I. et al. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 25, 2173–2186 (2011).
Laurent, L. C. et al. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells 26, 1506–1516 (2008).
Anokye-Danso, F. et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8, 376–388 (2011).
Subramanyam, D. et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 29, 443–448 (2011).
Bar, M. et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 26, 2496–2505 (2008).
Morin, R. D. et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 18, 610–621 (2008).
Liang, Y., Ridzon, D., Wong, L. & Chen, C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8, 166 (2007).
Virant-Klun, I., Ståhlberg, A., Kubista, M. & Skutella, T. MicroRNAs: From Female Fertility, Germ Cells, and Stem Cells to Cancer in Humans. Stem Cells Int 2016, 3984937 (2016).
Ratajczak, J. et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20, 847–856 (2006).
Katsman, D., Stackpole, E. J., Domin, D. R. & Farber, D. B. Embryonic stem cell-derived microvesicles induce gene expression changes in Müller cells of the retina. PLoS ONE 7, e50417 (2012).
Clark, E. A., Kalomoiris, S., Nolta, J. A. & Fierro, F. A. Concise review: MicroRNA function in multipotent mesenchymal stromal cells. Stem Cells 32, 1074–1082 (2014).
Ferguson, S. W. et al. The microRNA regulatory landscape of MSC-derived exosomes: a systems view. Sci Rep 8, 1419 (2018).
Kapinas, K. & Delany, A. M. MicroRNA biogenesis and regulation of bone remodeling. Arthritis Res. Ther. 13, 220 (2011).
Papaioannou, G., Mirzamohammadi, F. & Kobayashi, T. MicroRNAs involved in bone formation. Cell. Mol. Life Sci. 71, 4747–4761 (2014).
Peter, M. E. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle 8, 843–852 (2009).
Wei, J. et al. let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2. Stem Cells Dev. 23, 1452–1463 (2014).
Xu, J. et al. Altered microRNA expression profile in exosomes during osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. PLoS ONE 9, e114627 (2014).
Zhao, R. et al. miR-199b-5p modulates BMSC osteogenesis via suppressing GSK-3β/β-catenin signaling pathway. Biochem. Biophys. Res. Commun. 477, 749–754 (2016).
Chen, X. et al. Nanoparticle delivery of stable miR-199a-5p agomir improves the osteogenesis of human mesenchymal stem cells via the HIF1a pathway. Biomaterials 53, 239–250 (2015).
Huang, S. et al. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 21, 2531–2540 (2012).
Qin, Y., Wang, L., Gao, Z., Chen, G. & Zhang, C. Bone marrow stromal/stem cell-derived extracellular vesicles regulate osteoblast activity and differentiation in vitro and promote bone regeneration in vivo. Sci Rep 6, 21961 (2016).
Kim, Y. J., Bae, S. W., Yu, S. S., Bae, Y. C. & Jung, J. S. miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J. Bone Miner. Res. 24, 816–825 (2009).
Peng, Y. et al. Transcriptional characterization of bone morphogenetic proteins (BMPs)-mediated osteogenic signaling. J. Cell. Biochem. 90, 1149–1165 (2003).
Zeng, Y. et al. MicroRNA-100 regulates osteogenic differentiation of human adipose-derived mesenchymal stem cells by targeting BMPR2. FEBS Lett. 586, 2375–2381 (2012).
Wang, H. et al. MiR-125b Regulates the Osteogenic Differentiation of Human Mesenchymal Stem Cells by Targeting BMPR1b. Cell. Physiol. Biochem. 41, 530–542 (2017).
Zhang, G. et al. MicroRNA-98 regulates osteogenic differentiation of human bone mesenchymal stromal cells by targeting BMP2. J. Cell. Mol. Med. 21, 254–264 (2017).
Almeida, M. I. et al. miR-195 in human primary mesenchymal stromal/stem cells regulates proliferation, osteogenesis and paracrine effect on angiogenesis. Oncotarget 7, 7–22 (2016).
Hassan, M. Q. et al. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc. Natl. Acad. Sci. USA 107, 19879–19884 (2010).
Yin, J. et al. MiR-615-3p inhibits the osteogenic differentiation of human lumbar ligamentum flavum cells via suppression of osteogenic regulators GDF5 and FOXO1. Cell Biol. Int. 41, 779–786 (2017).
Neilsen, C. T., Goodall, G. J. & Bracken, C. P. IsomiRs–the overlooked repertoire in the dynamic microRNAome. Trends Genet. 28, 544–549 (2012).
Tan, G. C. et al. 5′ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 42, 9424–9435 (2014).
Ameres, S. L. & Zamore, P. D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 14, 475–488 (2013).
Li, S. et al. miRNA arm selection and isomiR distribution in gastric cancer. BMC Genomics 13(Suppl 1), S13 (2012).
Cloonan, N. et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 12, R126 (2011).
Lee, L. W. et al. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA. 16, 2170–80 (2010).
Telonis, A. G., Loher, P., Jing, Y., Londin, E. & Rigoutsos, I. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 43, 9158–9175 (2015).
Hinton, A. et al. sRNA-seq analysis of human embryonic stem cells and definitive endoderm reveals differentially expressed microRNAs and novel IsomiRs with distinct targets. Stem Cells 32, 2360–2372 (2014).
Jorjani, H. et al. An updated human snoRNAome. Nucleic Acids Res. 44, 5068–5082 (2016).
Ronchetti, D. et al. Small nucleolar RNAs as new biomarkers in chronic lymphocytic leukemia. BMC Med Genomics 6, 27 (2013).
Kohn, M. et al. The Y3** ncRNA promotes the 3′ end processing of histone mRNAs. Genes Dev. 29, 1998–2003 (2015).
Chakrabortty, S. K. et al. Extracellular vesicle-mediated transfer of processed and functional RNY5 RNA. RNA 21, 1966–1979 (2015).
Zhang, A. T. et al. Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J Cell Sci. 124, 2058–69 (2011).
Anderson, P. & Ivanov, P. tRNA fragments in human health and disease. FEBS Lett 588, 4297–304 (2014).
Wei, Z., Batagov, A. O., Carter, D. R. & Krichevsky, A. M. Fetal Bovine Serum RNA Interferes with the Cell Culture derived Extracellular RNA. Sci Rep. 9(6), 31175 (2016).
Skottman, H. Derivation and characterization of three new human embryonic stem cell lines in Finland. In Vitro Cell. Dev. Biol. Anim. 46, 206–209 (2010).
Hongisto, H., Ilmarinen, T., Vattulainen, M., Mikhailova, A. & Skottman, H. Xeno- and feeder-free differentiation of human pluripotent stem cells to two distinct ocular epithelial cell types using simple modifications of one method. Stem Cell Res Ther 8, 291 (2017).
Peltoniemi, H. H. et al. Stem cell enrichment does not warrant a higher graft survival in lipofilling of the breast: a prospective comparative study. J Plast Reconstr Aesthet Surg 66, 1494–1503 (2013).
Gimble, J. M., Katz, A. J. & Bunnell, B. A. Adipose-derived stem cells for regenerative medicine. Circ. Res. 100, 1249–1260 (2007).
Kyllönen, L. et al. Exogenously added BMP-6, BMP-7 and VEGF may not enhance the osteogenic differentiation of human adipose stem cells. Growth Factors 31, 141–153 (2013).
Saeed, K. et al. Comprehensive Drug Testing of Patient-derived Conditionally Reprogrammed Cells from Castration-resistant Prostate Cancer. Eur. Urol. 71, 319–327 (2017).
Puhka, M. et al. KeepEX, a simple dilution protocol for improving extracellular vesicle yields from urine. Eur J Pharm Sci 98, 30–39 (2017).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society. Series B (Methodological), 289–300 (1995).
Van Deun, J. et al. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods. 14(3), 228–232 (2017).
Foshay, K. M. & Gallicano, G. I. miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev. Biol. 326, 431–443 (2009).
Balzano, F. et al. MiR200 and miR302: Two Big Families Influencing Stem Cell Behavior. Molecules 23 (2018).
Nguyen, P. N. N., Huang, C., Sugii, S., Cheong, S. K. & Choo, K. B. Selective activation of miRNAs of the primate-specific chromosome 19 miRNA cluster (C19MC) in cancer and stem cells and possible contribution to regulation of apoptosis. J. Biomed. Sci. 24, 20 (2017).
Mizuno, Y. et al. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem. Biophys. Res. Commun. 368, 267–272 (2008).
Laine, S. K., Alm, J. J., Virtanen, S. P., Aro, H. T. & Laitala-Leinonen, T. K. MicroRNAs miR-96, miR-124, and miR-199a regulate gene expression in human bone marrow-derived mesenchymal stem cells. J. Cell. Biochem. 113, 2687–2695 (2012).
Schoolmeesters, A. et al. Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. PLoS ONE 4, e5605 (2009).
Hou, C. et al. MiR-193b regulates early chondrogenesis by inhibiting the TGF-beta2 signaling pathway. FEBS Lett. 589, 1040–1047 (2015).
Acknowledgements
Professor Katriina Aalto-Setälä from University of Tampere is cordially thanked for providing the hiPSC line used in the study. The authors thank Flow Cytometry Core Facility, EV Core Facility and Electron Microscopy Unit of the Institute of Biotechnology (all University of Helsinki) as well as Tampere Imaging Facility (TIF) for providing equipment for fluorescence imaging and Finnish Microarray and Sequencing Centre (FMSC), Turku Centre for Biotechnology for the karyotyping. Biomedical laboratory technicians Heidi Husu, Outi Melin, Hanna Pekkanen, and Emma Vikstedt are thanked for excellent technical assistance, and DDS Roman Kornilov for scientific support. This research was supported by University of Helsinki project funding (WBS490302, WBS73714112), Helsinki University Hospital State funding for university-level health research (Y1014SUL05, TYH2016130), Paulo foundation, the Finnish Dental Society Apollonia, Business Finland grant (CraMaxS 5773/31/16), Selma and Maja-Lisa Selander foundation, Egyptian Ministry of Higher Education (MoHE), Jenny and Antti Wihuri Foundation, Biomedicum Helsinki Foundation and Nissi Foundation and Academy of Finland (297886).
Author information
Authors and Affiliations
Contributions
Sippy Kaur the author contributed to the planning and designing the study, analysis of data, writing of manuscript. Ahmed G. Abu-Shahba the author contributed by performing analyses (Western blot, MSC characterization), analysis of the data and proof-reading of the manuscript. Riku O. Paananen The author contributed by analyzing the data (RNAseq), participating in the writing of the manuscript and proofreading of the manuscript. Heidi Hongisto the author contributed by performing analyses (hPSC characterization, hPSC EV collection), participated in the writing of the manuscript, proof-reading of the manuscript. Hanna Hiidenmaa the author contributed by performing analyses (EV characterization), proof-reading the manuscript. Heli Skottman the author contributed by reviewing the results and proof-reading the manuscript. Riitta Seppänen-Kaijansinkko the author contributed by proving the facilities and financial support to the study, proof-reading of the manuscript. Bettina Mannerström the author contributed to the planning and designing the study, analysis of data, performing administrative tasks, participating writing of manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kaur, S., Abu-Shahba, A.G., Paananen, R.O. et al. Small non-coding RNA landscape of extracellular vesicles from human stem cells. Sci Rep 8, 15503 (2018). https://doi.org/10.1038/s41598-018-33899-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-33899-6
Keywords
This article is cited by
-
Retroviral infection of human neurospheres and use of stem Cell EVs to repair cellular damage
Scientific Reports (2022)
-
Extracellular Vesicles from Human Adipose-Derived Mesenchymal Stem Cells: A Review of Common Cargos
Stem Cell Reviews and Reports (2022)
-
Extracellular vesicle-derived miRNA as a novel regulatory system for bi-directional communication in gut-brain-microbiota axis
Journal of Translational Medicine (2021)
-
Mesenchymal Stromal Cells in Viral Infections: Implications for COVID-19
Stem Cell Reviews and Reports (2021)
-
Stem Cell Extracellular Vesicles and their Potential to Contribute to the Repair of Damaged CNS Cells
Journal of Neuroimmune Pharmacology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.