Abstract
A particular challenge in genome engineering has been the simultaneous introduction of mutations into linked (located on the same chromosome) loci. Although CRISPR/Cas9 has been widely used to mutate individual sites, its application in simultaneously targeting of linked loci is limited as multiple nearby double-stranded DNA breaks created by Cas9 routinely result in the deletion of sequences between the cleavage sites. Base editing is a newer form of genome editing that directly converts C∙G-to-T∙A, or A∙T-to-G∙C, base pairs without introducing double-stranded breaks, thus opening the possibility to generate linked mutations without disrupting the entire locus. Through the co-injection of two base editors and two sgRNAs into mouse zygotes, we introduced C∙G-to-T∙A transitions into two cytokine-sensing transcription factor binding sites separated by 9 kb. We determined that one enhancer activates the two flanking genes in mammary tissue during pregnancy and lactation. The ability to introduce linked mutations simultaneously in one step into the mammalian germline has implications for a wide range of applications, including the functional analysis of linked cis-elements creating disease models and correcting pathogenic mutations.
Similar content being viewed by others
Introduction
Clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 genome editing1,2 has been widely used to disrupt individual and multiple targets3,4,5,6. However, co-targeting two or more sites on the same chromosome usually results in the excision of the DNA between targets3,7,8,9,10, especially for sites in close proximity (up to 1 Mb deletions have been reported)8,9,10. To tackle this particular technical challenge, we turned to base editing (BE)11,12,13 with its advantage of changing specific nucleotides in the genome without inducing double-strand breaks, which should make it less likely to cause undesired mutations, such as deletions or insertions. The deaminases currently in use facilitate the conversion of cytosine to uracil or adenosine to inosine and subsequent DNA repair results in a C∙G-to-T∙A or A∙T-to-G∙C substitution11,12,13. Thus, this approach should enable the simultaneous mutation of linked sites without causing deletions.
The introduction of mutations into linked loci is essential for experimental approaches to understand complex loci with multiple haplotypes of single-nucleotide polymorphisms (SNPs) related to diseases14,15, roles of individual enhancers within super-enhancers7,16, and tumorigenesis17,18. This approach is also needed to examine the possibility of correcting co-occurring somatic mutations14,15. Therefore, it is paramount to establish a reliable tool that permits the efficient and faithful introduction of two or more linked mutations in one step. In this study we have investigated the ability of cytosine base editors to simultaneously introduce point mutations in two sites separated by 9 kb. To ensure a sensitive and reliable readout, we chose to mutate enhancers that possibly activate genes in mammary tissue during pregnancy.
Results
Simultaneous targeting of linked loci by cytosine-deaminase-mediated base editing
First, we aimed to determine the extent of deletions introduced upon simultaneously targeting linked sites by CRISPR/Cas9. We co-microinjected two or more sgRNAs together with Cas9 mRNA into mouse zygotes. Although it is known that co-targeting two or more linked loci can result in the deletion of the entire sequence between the targets, we were surprised by the prevalence of big deletions spanning both sites compared to small deletions at each cutting site. Targeting two sites separated by 18 kb resulted in the complete deletion of the locus in 11 out of 14 founders (Supplementary Fig. 1a). In a second experiment, 15 out of 23 founder mice carried a complete deletion between target sites that were 9 kb apart (Supplementary Fig. 1a). Other studies have reported deletion sizes between 10 kb and 0.5 Mb at frequencies between 10% and 90% (Supplementary Fig. 1a and b). These results emphasize the extraordinary efficiency of CRISPR/Cas9 technology, but also suggest that Cas9 proteins may not function independent of each other. Deleting sequences between two sites requires that both Cas9/sgRNA complexes cut the same allele at exactly the same moment. Therefore, deletions should not occur if a cut introduced by Cas9 is repaired through non-homologous end joining (NHEJ) before the second cut is introduced. The surprisingly high efficiency of big deletions suggests that Cas9-created double strand breaks are not immediately repaired by NHEJ, or multiple Cas9 molecules may communicate with each other and cut DNA simultaneously.
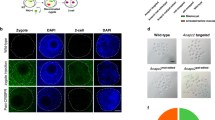
Next, we investigated the ability of base editors to introduce mutations into linked loci without disrupting sequences between target sites. We focused on a 75 kb region in the mouse casein locus, which contains at least eight putative enhancers (A-H) (Fig. 1a). These enhancers were identified using ChIP-seq experiments and are characterized by the binding of transcription factors STAT5, GR, ELF5, MED1 and the presence of the active histone marker H3K27ac (Supplementary Fig. 2). The location of these putative enhancers infers a regulatory role in controlling expression of the associated Csn2 and Csn1s2a genes during pregnancy and lactation. We used VQR-BE3, which recognizes a NGA PAM19, and BE4, which recognizes a NGG PAM13, to mutate transcription factor binding motifs in sites C and E, respectively. We co-injected VQR-BE3 and BE4 mRNAs and their corresponding guide RNAs, targeting the STAT5 motif (TTCNNNGAA) in site C and an ELF5 motif (GGAA/T) in site E (Fig. 1a and Supplementary Fig. 2), into mouse zygotes and transferred injected embryos into oviducts of pseudo-pregnant recipients. Out of the 32 founder mice, 9% carried target mutations exclusively in site C, 19% only in site E, and 47% carried target mutations in both sites (Figs 1b, 1c and 2a). Twenty-five percent of the founders did not carry any mutation (Fig. 1c). Homozygosity was prevalent with 28% of the founders at site C and 24% at site E (Figs 1b and 2b). In nine out of the 15 co-targeted founders, the mutations in sites C and E were linked, i.e. they co-located on the same homologous chromosome (Fig. 1d). Mutations were passed on through the germline (Figs 3a and b). Unlike conventional CRISPR/Cas9 genome editing, which results in the deletion of sequences between sites targeted by sgRNAs3,7,8,9,10 (Supplementary Fig. 1), we did not detect such deletions in any of the 32 founders and their offspring. However, we found indels around target sites and out of the 32 founder mice, one carried a 93 bp deletion at site C and five mice carried deletions between 2 to 11 bp at site E (Fig. 3c). Presumably, those deletions are the results of the nickase activity of BE on a single strand and cell’s endogenous DNA repair machinery20. Our results demonstrate that base editing, in contrast to CRISPR/Cas9 genome editing, can be used to simultaneously and efficiently introduce linked mutations in the mouse germline without disrupting the targeted locus.
Simultaneously targeting of two linked genomic loci by cytosine-deaminase-mediated base editing. (a) Schematic diagram of target sites in the casein locus. The eight putative enhancers (A-H) and the two promoters in the Csn2 and Csn1s2a locus were identified by ChIP-seq analysis for enhancer marks. Sites C and E are 9 kb apart and were targeted simultaneously with two sgRNAs and VQR-BE3 and BE4. (b) Summary of data obtained from mouse zygotes co-injected with VQR-BE3 and BE4 mRNA, and two sgRNAs. Experiments were conducted with founder mice and established mouse lines. (c) C-to-T mutation frequency observed at the two sites. (d) Distribution of linked (on the same chromosome) and non-linked mutations.
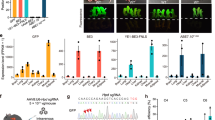
Alignment of sequences from founder mice carrying mutations in sites C and E. (a) sgRNA sequences are underlined and the PAM sites are shown in brown. The C-to-T on-target substitutions are shown in green. Mutant mice carrying homozygous mutations are marked in bold purple. Unintended nucleotide substitutions are shown in red. Deletions are shown as underlines. WT, wild-type. (b) Sanger sequencing chromatograms of DNA from WT and mutant mice carrying homozygous mutations (founder 166, 169, 175, and 186). The sgRNA sequences are underlined. C-to-T transitions are seen at target sites C and E and marked with a red asterisk.
Inheritance of intended mutations. (a) Female founder F883 was mated with a WT male and mutations in site C were analyzed in its offspring. F883 was homozygous for the intended C-to-T conversion on site C and all offspring were heterozygous for this mutation. The C-to-T editing is shown in green. (b) F165 was mated with male founder M183, and intended mutations and deletions are co-segregated. (c) The indel frequency and deletion length were analyzed in founder mice.
Enhancer mutations selectively impact neighboring genes
Given the complexity of the enhancer landscape associated with the Csn2 and Csn1s2a genes and the possibility of compensatory functions among the eight constituent enhancers (Fig. 1a, Supplementary Fig. 2), the biological consequences of the mutations were far from clear. Csn2 and Csn1s2a mRNA levels increase 60- and 130-fold, respectively, during pregnancy21. To investigate whether the mutations in sites C and E curb the induction of these two genes, we measured their mRNA levels at day six of pregnancy (p6), and days one (L1) and ten (L10) of lactation. While Csn2 expression at p6 was not affected by the mutation in site C, expression at L1 was reduced by 30% (Fig. 4a), revealing a contribution of this enhancer in gene activation during pregnancy. Expression of Csn1s2a, which is located 50 kb from site C, was affected to a lesser extent (Fig. 4b). Csn2 expression at L10 was not curtailed by the mutation in site C suggesting a high degree of compensation between enhancers during lactation, similar to that observed in another mammary super-enhancer7. Mutations of an ELF5 site in enhancer E did not affect Csn2 or Csn1s2a mRNA levels during pregnancy and lactation (Fig. 4c). Our findings lend biological meaning to an enhancer in the activation of two casein genes during pregnancy. The moderate activity of enhancer C within the complex casein super-enhancer is in line with other studies addressing genes under the control of multiple enhancers and super-enhancers, including Wap in mammary tissue7, Il2ra in T cells16, and α-globin in erythroid cells22.
Biological consequence of enhancer mutations during pregnancy. (a–c) Expression of the Csn2 and Csn1s2a genes in mammary tissue from WT and mutant mice carrying nucleotide substitution at site C or E, respectively, by qRT-PCR. mRNA levels were normalized to Gapdh levels. Results are shown as the means ± s.e.m. of independent biological replicates (WT and mutants, n = 3). T-test was used to evaluate the statistical significance of differences between WT and mutants. ****P < 0.00001. n.s., not significant.
Off-target analysis by WGS
Finally, to assess off-target effects, we initially used computational prediction and identified potential off-target sites for each sgRNA, with up to 4-nucleotide mis-matches in the mouse genome (Supplementary Fig. 3a). A total of 434 potential off-target sites were identified for the sgRNA used with VQR-BE3 and 143 for the sgRNA used with BE4. To evaluate targeting at predicted off-target sites, we performed whole genome sequencing (WGS) of two founder mice carrying linked mutations in sites C and E and a cohort of mice23 from the same genetic background as controls. We did not detect any SNPs and indels at the predicted off-target sites (Supplementary Fig. 3b and Supplementary Table 1).
Discussion
Our study addressed a major unresolved technical challenge and conclusively demonstrates that cytosine base-editing can be used to simultaneously and efficiently introduce linked mutations in mouse embryos without deleting sequences between the target sites. This experimental approach opens opportunities, both in basic and translational research, to address the biology of complex loci carrying several haplotypes. Our findings also provide biological significance to a constituent enhancer within a complex super-enhancer, which contributes to the activation of the Csn2 gene during mammary differentiation24. Our data also suggest limited enhancer redundancy within this locus and a more complete understanding of its regulation will require the introduction of mutations into most, if not all of the eight constituent enhancers. Base-editing could be the preferred option and, depending on the editing window and the sequences of potential protospacer adjacent motif (PAM), the use of several different base editors recognizing different PAMs will be required.
Methods
Mice
All animals were housed and handled according to the guidelines of the Animal Care and Use Committee (ACUC) of the NIH (https://oacu.oir.nih.gov) and all animal experiments were approved by the ACUC of National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK, MD) and performed under the NIDDK animal protocol K089-LGP-17. Base editing- and CRISPR/Cas9-targeted founder mice were generated using C57BL/6 N mice (Charles River Laboratories, MD) by the Transgenic Core of the National Heart, Lung, and Blood Institute (NHLBI, MD).
Targeted loci
Using CRISPR/Cas9 genome editing, two sites in the casein locus that are 9 kb apart and two sites in Wap-Ramp3 locus that are 18 kb apart, were co-targeted by Cas9 and two sgRNAs, respectively. In the 75 kb locus with 8 enhancers, transcription binding sites on sites C and E that were 9 kb apart were targeted simultaneously using VQR-BE3 and BE4 base editors, and two sgRNAs to introduce C-to-T transitions.
mRNA preparation and microinjection into mouse zygotes
The sgRNAs were designed based on the nearest PAM of the target sequence. Each sgRNA was cloned into the pDR274 plasmid vector (Addgene #42250, MA), and in vitro transcribed using the MEGAshortscript T7 kit (ThermoFisher Scientific, MA). pCMV-VQR-BE3 and BE4 mRNAs was synthesized in vitro using the mMESSAGE mMACHINE T7 kit (ThermoFisher Scientific). Cas9 (100 ng/μl) and deaminase fused-Cas9 mRNA (50 ng/μl for each base editor) and sgRNAs (20 ng/μl for each sgRNA) were mixed and co-microinjected into the cytoplasm of fertilized eggs collected from superovulated C57BL/6 N female mice (Charles River Laboratories) and implanted into oviducts of pseudopregnant fosters (Swiss Webster, NY).
Generation of V5-tagged ELF5 mouse
V5-tagged Elf5 mutant mouse by injecting the oligo donor with V5-tag and sgRNA into zygotes.
Genotyping
Genomic DNA of all mice was isolated from the tip of the tail, amplified by PCR, and followed by Sanger sequencing. Large deletions were identified by serial PCR genotyping using primers that were designed to amplify 400~500 bp encompassing the target sequence or long-range PCR.
RNA isolation and quantitative real-time PCR (qRT–PCR)
Total RNA was extracted from frozen mammary tissue of wild-type and mutant mice using a homogenizer and the PureLink RNA Mini kit according to the manufacturer’s instructions (Invitrogen, MA). Total RNA (1 μg) was reverse transcribed for 50 min at 50 °C using 50 μM oligo dT and 2 μl of SuperScript III (Invitrogen) in a 20 μl reaction. Quantitative real-time PCR (qRT-PCR) was performed using TaqMan probes (Csn2, Mm04207885_m1; Csn1s2a, Mm00839343_m1; mouse Gapdh, Mm99999915_g1, ThermoFisher scientific) on the CFX384 Real-Time PCR Detection System (Bio-Rad, CA) according to the manufacturer’s instructions. PCR conditions were 95 °C for 30 s, 95 °C for 15 s, and 60 °C for 30 s for 40 cycles. All reactions were done in triplicate and normalized to the housekeeping gene Gapdh. Relative differences in PCR results were calculated using the comparative cycle threshold (CT) method.
Statistical analyses
Shapiro-Wilk normality test returned for all groups a p-value above 0.05. Thus, the hypothesis that the samples come from a population with normal distribution was not rejected. For comparison of samples, data were presented as standard deviation in each group and were evaluated with a t-test using PRISM GraphPad. Statistical significance was obtained by comparing the measures from wild-type or control group, and each mutant group. A value of *P < 0.05, **P < 0.001, ***P < 0.0001, ****P < 0.00001 was considered statistically significant.
Chromatin immunoprecipitation sequencing (ChIP-seq) and data analysis
Mammary tissue was harvested at day one of lactation and stored at −80 °C. Frozen tissues were ground into powder in liquid nitrogen. Chromatin was fixed with formaldehyde (1% final concentration) for 15 min at room temperature, and then quenched with glycine (0.125 M final concentration). Samples were processed as previously described25. The following antibodies were used for ChIP-seq: V5 tag antibody (ThermoFisher Scientific, R960-25). Libraries for next-generation sequencing were prepared and sequenced with a HiSeq 2500 instrument (Illumina). Quality filtering and alignment of the raw reads was done using Trimmomatic26 (version 0.36) and Bowtie27 (version 1.1.2), with the parameter -m to keep only uniquely mapped reads, using the reference genome mm10. Picard tools (Broad Institute. Picard, http://broadinstitute.github.io/picard/. 2016) was used to remove duplicates and subsequently, Homer28 (version 4.8.2) software was applied to generate bedGraph files. Integrative Genomics Viewer29 (version 2.3.81) was used for visualization.
Off-target analysis
Off-target sites were predicted using http://crispor.tefor.net/30. The resulting off-target sites were filtered using the same criteria as for SNPs and indels, to consider only those areas of the genome which do not coincide with black regions31 (ENCODE31; http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/mm10-mouse) or repetitive elements32 (UCSC’s masked repeats plus simple repeats; http://hgdownload.soe.ucsc.edu/goldenPath/mm10/database). Mutations, which were present in the population and not only in base-edited mice, but identified at predicted off-target sites, were not considered as a consequence of base editing.
GATK analysis
WGS (60×) was performed of two founder mice carrying a base substitution at site C and E using three guide RNAs and two base editors. In addition, we analyzed 30 mice as control, 24 wild-types mice (males and females) and six of their non-injected progeny. The analysis was done accordingly to the GATK best practices guidelines33,34,35 for germline mutations (version 3.8-0). Thus, BBmap36 (version 37.36) was applied for quality control, followed by BWA MEM37 (version 0.7.15) for the alignment step (reference genome mm10). The aligned BAM files were subsequently split up to a chromosome level (for runtime optimization) and reads aligned to different chromosomes were filtered using SAMtools38 (version 1.5). Additionally, Picard tools39 (version 2.9.2) was applied to mark duplicates. The subsequent GATK analysis workflow comprised: (i) base recalibration - GATK BaseRecalibrator, AnalyzeCovariates, and PrintReads - using the databases of known polymorphic sites, dbSNP142 and MGPv5 (provided by the high-performance computing team of the NIH (Biowulf)); (ii) variant calling - GATK HaplotypeCaller - with the genotyping mode “discovery”, the “ERC” parameter for creating gvcf and a minimum phred-scaled confidence threshold of 30. The final step included merging the VCF files of each chromosome (GenomeAnalysisTK, GATK).
GATK SNP analysis
Joint genotyping was applied on each of the three groups independently (wild-type, non-injected controls and base-edited mice). On each of the groups hard filtering with the following parameters was applied: “QD < 2.0||FS > 60.0||MQ < 40.0||MQRankSum < −12.5||ReadPosRankSum < −8.0||SOR > 3”. The resulting SNPs were filtered by removing those overlapping with repetitive elements32 (UCSC’s masked repeats plus simple repeats; http://hgdownload.soe.ucsc.edu/goldenPath/mm10/database) and black regions (ENCODE31; http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/mm10-mouse/). On an individual level those SNPs with a genotype of 0/1 or 1/1 were kept. Further filtering included the removal of SNPs with a read depth smaller than 10, an excessive read depth40 (d + 3√d, d = average read depth), an allele frequency less than 10% and a quality smaller than 130 using a variety of tools41,42,43. All SNPs within +/−5 bp of an indel border were removed as likely false-positives.
Simple GATK indel analysis
Indels were extracted from the GATK joint genotyping file and hard filters were applied based on the GATK recommendations: “QD < 2.0||FS > 200.0||ReadPosRankSum < −20.0||SOR > 10.0”. Indels overlapping with repetitive elements32 (UCSC’s masked repeats plus simple repeats; http://hgdownload.soe.ucsc.edu/goldenPath/mm10/database) or black regions (ENCODE31; http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/mm10-mouse/) were removed. Subsequently, each file was filtered keeping only indels with the genotypes of 0/1 and 1/1, removing those with a read depth smaller than 10 as well as sites with an excessive number of reads40 (d + 3√d, d = average read depth). The last step comprised the removal of all simple indels that overlap with complex indels identified using LUMPY. For all those steps a variety of tools41,42,43 was applied.
Complex indel analysis using LUMPY
Indel analysis was done on the same samples as described above using Lumpy44 according to the guidelines. Thus, BWA MEM37, with the parameters “–excludeDups–addMateTags–maxSplitCount 2–minNonOverlap 20” was applied for mapping (reference genome mm10), followed by Lumpy44 using the discordant and split reads as input. Post-processing was carried out using SVTyper45 to identify genotypes. The filtering step comprised the selection of indels with a genotype of 0/1 and 1/1 and the removal of indels with a quality smaller than 100 and an excessive read coverage (d + 3√d40, where d is the average read depth). Indels overlapping with repetitive elements32 or black regions31 were excluded.
Data Availability
ChIP-seq data of wild-type mammary tissue at L1 were obtained from GSE74826 and GSE115370 in the Gene Expression Omnibus (GEO). ChIP-seq for V5 tag have been deposited under GSE119657. The WGS data of the wild-type mice are available at SRA PRJNA470569. The WGS data of the base-edited mice are deposited at SRA PRJNA489707.
References
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823, https://doi.org/10.1126/science.1231143 (2013).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821, https://doi.org/10.1126/science.1225829 (2012).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918, https://doi.org/10.1016/j.cell.2013.04.025 (2013).
Yang, H. et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379, https://doi.org/10.1016/j.cell.2013.08.022 (2013).
Fujii, W., Kawasaki, K., Sugiura, K. & Naito, K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic acids research 41, e187, https://doi.org/10.1093/nar/gkt772 (2013).
Seruggia, D., Fernandez, A., Cantero, M., Pelczar, P. & Montoliu, L. Functional validation of mouse tyrosinase non-coding regulatory DNA elements by CRISPR-Cas9-mediated mutagenesis. Nucleic acids research 43, 4855–4867, https://doi.org/10.1093/nar/gkv375 (2015).
Shin, H. Y. et al. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat Genet 48, 904–911, https://doi.org/10.1038/ng.3606 (2016).
Hara, S. et al. Microinjection-based generation of mutant mice with a double mutation and a 0.5 Mb deletion in their genome by the CRISPR/Cas9 system. J Reprod Dev 62, 531–536, https://doi.org/10.1262/jrd.2016-058 (2016).
Wang, L. et al. Large genomic fragment deletion and functional gene cassette knock-in via Cas9 protein mediated genome editing in one-cell rodent embryos. Sci Rep 5, 17517, https://doi.org/10.1038/srep17517 (2015).
Shin, H. Y. et al. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat Commun 8, 15464, https://doi.org/10.1038/ncomms15464 (2017).
Komor, A. C. et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T: Abase editors with higher efficiency and product purity. Sci Adv 3, eaao4774, https://doi.org/10.1126/sciadv.aao4774 (2017).
Kim, Y. B. et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol 35, 371–376, https://doi.org/10.1038/nbt.3803 (2017).
Gaudelli, N. M. et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471, https://doi.org/10.1038/nature24644 (2017).
Nairismagi, M. L. et al. JAK-STAT and G-protein-coupled receptor signaling pathways are frequently altered in epitheliotropic intestinal T-cell lymphoma. Leukemia 30, 1311–1319, https://doi.org/10.1038/leu.2016.13 (2016).
Crescenzo, R. et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell 27, 516–532, https://doi.org/10.1016/j.ccell.2015.03.006 (2015).
Li, P. et al. STAT5-mediated chromatin interactions in superenhancers activate IL-2 highly inducible genes: Functional dissection of the Il2ra gene locus. Proc Natl Acad Sci USA 114, 12111–12119, https://doi.org/10.1073/pnas.1714019114 (2017).
Vahedi, G. et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520, 558–562, https://doi.org/10.1038/nature14154 (2015).
Bahr, C. et al. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 553, 515–520, https://doi.org/10.1038/nature25193 (2018).
Kleinstiver, B. P. et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485, https://doi.org/10.1038/nature14592 (2015).
Alexis C. Komor, Ahmed H. Badran, David R. Liu. Editing the Genome Without Double-Stranded DNA Breaks. ACS Chemical Biology 13(2), 383–388 (2017).
Yamaji, D., Kang, K., Robinson, G. W. & Hennighausen, L. Sequential activation of genetic programs in mouse mammary epithelium during pregnancy depends on STAT5A/B concentration. Nucleic Acids Res 41, 1622–1636, https://doi.org/10.1093/nar/gks1310 (2013).
Hay, D. et al. Genetic dissection of the alpha-globin super-enhancer in vivo. Nat Genet 48, 895–903, https://doi.org/10.1038/ng.3605 (2016).
Willi, M., Smith, H. E., Wang, C., Liu, C. & Hennighausen, L. Mutation frequency is not increased in CRISPR-Cas9-edited mice. Nat Methods 15, 756–758, https://doi.org/10.1038/s41592-018-0148-2 (2018).
Kabotyanski, E. B. et al. Lactogenic hormonal induction of long distance interactions between beta-casein gene regulatory elements. The Journal of biological chemistry 284, 22815–22824, https://doi.org/10.1074/jbc.M109.032490 (2009).
Metser, G. et al. An autoregulatory enhancer controls mammary-specific STAT5 functions. Nucleic Acids Res 44, 1052–1063, https://doi.org/10.1093/nar/gkv999 (2016).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120, https://doi.org/10.1093/bioinformatics/btu170 (2014).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25, https://doi.org/10.1186/gb-2009-10-3-r25 (2009).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589, https://doi.org/10.1016/j.molcel.2010.05.004 (2010).
Thorvaldsdottir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14, 178–192, https://doi.org/10.1093/bib/bbs017 (2013).
Haeussler, M. et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17, 148, https://doi.org/10.1186/s13059-016-1012-2 (2016).
Consortium, E. P. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74, https://doi.org/10.1038/nature11247 (2012).
Casper, J. et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res 46, D762–D769, https://doi.org/10.1093/nar/gkx1020 (2018).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303 (2010).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43, 491–498 (2011).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43, 11 10 11–33 (2013).
Bushnell, B. BBMap short-read aligner, and other bioinformatics tools, http://sourceforge.net/projects/bbmap/ (2016).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Broad Institute. Picard http://broadinstitute.github.io/picard/ (2016).
Li, H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30, 2843–2851 (2014).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Neph, S. et al. BEDOPS: high-performance genomic feature operations. Bioinformatics 28, 1919–1920, https://doi.org/10.1093/bioinformatics/bts277 (2012).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Layer, R. M., Chiang, C., Quinlan, A. R. & Hall, I. M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15, R84, https://doi.org/10.1186/gb-2014-15-6-r84 (2014).
Chiang, C. et al. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Methods 12, 966–968, https://doi.org/10.1038/nmeth.3505 (2015).
Acknowledgements
We thank Chul Min Yang for help with establishing the ELF5-V5 mutant mouse line. This study was supported by the IRPs of NHLBI and NIDDK. Research support was received from the NIH Director’s Challenge Innovation Award program. S.M.M. was supported by an NSF graduate fellowship. S.M.M. and D.R.L. acknowledge support from DARPA HR0011-17-2-0049, U.S. NIH RM1 HG009490, U01 AI142756, and R35 GM118062, and HHMI. We also thank the team of the NIH HPC Biowulf cluster.
Author information
Authors and Affiliations
Contributions
Study design: H.K.L., C.L., L.H. Design of protospacers and providing materials: S.M. and D.R.L. Generation of mutant mice: C.L. Experimental mouse work and data analysis: H.K.L. Computational analysis: M.W. and H.S. All authors contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
D.R.L. is a co-founder and consultant for Beam Therapeutics, Editas Medicine, and Pairwise Plants, companies that use genome editing. The other authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, H.K., Willi, M., Smith, H.E. et al. Simultaneous targeting of linked loci in mouse embryos using base editing. Sci Rep 9, 1662 (2019). https://doi.org/10.1038/s41598-018-33533-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-33533-5
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.