Abstract
New therapeutic targets for advanced colorectal cancer (CRC) are critically needed. Our laboratory recently performed an insertional mutagenesis screen in mice to identify novel CRC driver genes and, thus, potential drug targets. Here, we define Transmembrane 9 Superfamily 2 (TM9SF2) as a novel CRC oncogene. TM9SF2 is an understudied protein, belonging to a well conserved protein family characterized by their nine putative transmembrane domains. Based on our transposon screen we found that TM9SF2 is a candidate progression driver in digestive tract tumors. Analysis of The Cancer Genome Atlas (TCGA) data revealed that approximately 35% of CRC patients have elevated levels of TM9SF2 mRNA, data we validated using an independent set of CRC samples. RNAi silencing of TM9SF2 reduced CRC cell growth in an anchorage-independent manner, a hallmark of cancer. Furthermore, CRISPR/Cas9 knockout of TM9SF2 substantially diminished CRC tumor fitness in vitro and in vivo. Transcriptome analysis of TM9SF2 knockout cells revealed a potential role for TM9SF2 in cell cycle progression, oxidative phosphorylation, and ceramide signaling. Lastly, we report that increased TM9SF2 expression correlates with disease stage and low TM9SF2 expression correlate with a more favorable relapse-free survival. Taken together, this study provides evidence that TM9SF2 is a novel CRC oncogene.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) arises from a stepwise accumulation of mutations that transform normal epithelia into cancerous tissue1,2. Decades of research analyzing the genetic basis of CRC has resulted in the identification of several important driver genes including APC, KRAS, SMAD4, and TP53. In addition, recent large scale genomic analyses, such as The Cancer Genome Atlas (TCGA), have identified numerous additional recurrent somatic mutations, focal copy number alterations and gene expression changes3,4. From these studies it is clear that CRC has a complex genetic etiology. It is important that we understand the functional significance of these genetic changes so that we can develop better therapies, especially for advanced disease.
To functionally define genetic drivers of CRC, we and others have used Sleeping Beauty (SB) transposon mutagenesis screens in mice, an unbiased method of finding genetic drivers of CRC. These studies have produced multiple lists of genes suspected of contributing to CRC when altered by transposon mutagenesis5,6,7,8. With the goal of finding potential therapeutic targets we are using cross-species bioinformatics approaches to select genes from these lists for further study. This approach has resulted in the identification of potential actionable targets including KCNQ1, CFTR, and RSPO2/39,10,11. In this study, we report our findings on TM9SF2, a transmembrane protein belonging to the transmembrane-9 superfamily (TM9SF), which includes TM9SF1-4. Although it is well conserved evolutionarily, very little is known about the function of TM9SF proteins in mammalian cells, nor their role in cancer12,13. TM9SF1 has been implicated in autophagosome formation and has been linked to bladder cancer14,15. It has been reported that TM9SF3 is upregulated in chemoresistant breast cancer cells after combination treatment with paclitaxel and an HDAC inhibitor and may also play a role in gastric cancer16,17. The most well studied member, TM9SF4, is reportedly overexpressed in human melanoma cells and has also been described as a proton pump associated protein18,19.
In this study, we identify TM9SF2 as a novel oncogene in CRC. We found that TM9SF2 is potentially regulated by the Ets-family transcription factor ELF1, and TM9SF2 is upregulated in approximately one-third of human CRC samples. We used RNAi and CRISPR/Cas9 to either reduce or knockout the expression of TM9SF2, which had the effect of reducing tumor fitness in both the in vitro and in vivo settings. Finally, we performed transcriptome analysis to gain insight into the potential role of TM9SF2 as a cell cycle regulating protein.
Results
Insertional mutagenesis screens identify TM9SF2 as candidate cancer gene
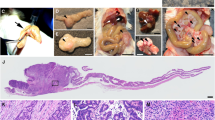
Our laboratory previously performed an insertional mutagenesis screen in mice to identify novel gastrointestinal (GI) tract cancer driver genes5. In this study we used the Sleeping Beauty (SB) DNA system consisting of an oncogenic DNA transposon (T2/Onc) capable of disrupting tumor suppressor genes and activating oncogenes, which is activated by tissue-specific expression of the SB transposase20,21,22. We identified 77 candidate cancer genes whose activity was potentially altered by T2/Onc transposition based on common insertion site (CIS) analysis23. Of these 77 candidate cancer genes, we chose to focus on TM9SF2 for further study because we found this gene to be overexpressed in a large percentage of human CRC samples, suggesting a potential oncogenic function. TM9SF2 is a member of a highly conserved family of proteins that span the lipid bilayer nine times. The predicted function of the TM9SF2 protein product is to act as a small molecule transporter or ion channel. In our screen the T2/Onc transposon insertions were mapped to the murine Tm9sf2 gene in nine tumor samples (Fig. 1A).
SB screen identifies TM9SF2 as candidate CRC driver gene. Tm9sf2 is a CIS gene in SB transposon screens. (A) schematic representation of gastrointestinal tract tumor-T2/onc insertion sites within the murine Tm9sf2 gene. Triangles depict the location of insertion as well as the orientation of the promoter-splice donor within the transposon. (B) The frequency of tumors with SB insertions in Tm9sf2 in digestive tract, solid tumor, liquid tumors, and all tumors analyzed in the SBCD database. Gray bars represented instances where Tm9sf2 is a progression diver gene. White bars are not significantly altered cases. (C) The frequency of Tm9sf2 insertions in intestinal-specific mutagenesis screens in mice with predisposing mutations in Trp53 (R172H allele) or Kras (G12D allele). Tm9sf2 insertions are predicted to act as a progression driver gene in both studies.
To further explore the role of TM9SF2 as a cancer gene, we used two publicly available databases that catalog cancer genes discovered using DNA transposon insertional mutagenesis. The Candidate Cancer Gene Database (CCGD, http://ccgd-starrlab.oit.umn.edu/about.php) catalogs cancer genes identified in 69 insertional mutagenesis studies covering 12 tumor types8. Mining the CCGD database revealed that Tm9sf2 was a transposon-targeted mutation in an additional eight forward genetic screens, including screens for liver, pancreatic, breast, and gastric cancer (see Supplementary Table S2). The Sleeping Beauty Cancer Driver Database (SBCDDB: http://sbcddb.moffitt.org/index.html) catalogs over 1.5 million transposon insertions from 2 354 tumors taken from approximately 1 000 mice from 19 tumor types24. Mining of the SBCDDB revealed that Tm9sf2 was a common insertion site in 7.2% (121/1674 tumors) of all digestive tumors, which includes liver, pancreas, intestine, and stomach tumors, but was not identified as a driver in hematopoietic tumors (Fig. 1B). Several insertional mutagenesis studies were conducted using mice that are predisposed to GI tract cancer by manipulating known genetic drivers, including Apc, Kras, Smad4, and Trp536,7,25. Interestingly, TM9SF2 was not identified as a driver gene in mice with Apc or Smad4 mutations but was identified as a driver in mice with Kras or Trp53 mutations. Mice harboring the activating Kras G12D allele had Tm9Sf2 transposon insertions in 13 out of 173 tumors (7.5%; p = 1.68e-05) and mice harboring the dominant negative R172H Trp53 mutation had insertions in 7 out of 55 tumors (12.7%; p < 0.005) (Fig. 1C and Supplementary Table S3). This analysis indicates TM9SF2 has a contributing role in the formation of murine intestinal tumors.
TM9SF2 is overexpressed in human CRC
To determine if TM9SF2 is altered in human disease we evaluated TM9SF2 mutation data from the Catalogue of Somatic Mutations in Cancer (COSMIC) database (see: http://cancer.sanger.ac.uk/cosmic/publications)26. Interestingly, data from COSMIC revealed that mutations in TM9SF2 are rarely catalogued in human tumors. We found that tumors derived from the endometrium contained the highest rate of TM9SF2 mutation at a tumor frequency of approximately 2% (13/656 total tested). Tumors from the small and large intestine were the next most likely to contain mutations in TM9SF2 but the mutation frequency remained very low at 1.92% and 1.71% respectively. These data suggest that point mutations and small in/dels in TM9SF2 are unlikely to play a significant role in CRC development and progression.
We used several approaches to measure TM9SF2 gene expression in CRC. First, we used cBioportal to analyze gene expression in CRC tumors from The Cancer Genome Atlas27,28. Analyses of gene expression levels using microarray or RNA sequencing reveal that, when compared to the mean expression distribution of tumor samples that are diploid for TM9SF2, TM9SF2 is overexpressed in approximately one-third (194/601 tumors) of large intestine tumors (Fig. 2A). These data suggest that TM9SF2 may function as a proto-oncogene in CRC. For our second approach, we evaluated the mRNA levels of TM9SF2 in a panel of nine commonly used colorectal cancer cell lines using qRT-PCR. For comparison we used the Human Colonic Epithelial Cell (HCEC) line, which is a non-oncogenic immortalized cell line, as a normal (non-tumor) control sample29. All cell lines tested had significantly upregulated levels of TM9SF2 transcript when compared to HCEC. DLD1, HCT-8, and HT-29 cells expressed the highest levels of TM9SF2 ranging from approximately 4 to 5-fold that of HCEC cells (Fig. 2B). Finally, we performed RNA-sequencing on a set of 44 CRC tumor and matched normal samples collected at the University of Minnesota. Expression levels were significantly increased in tumors compared to matched normal samples (P = 4.461 × 10−6; Fig. 2C). In all but eight samples the levels of TM9SF2 mRNA were increased in the patient’s tumor sample compared to the matched normal sample (Fig. 2C). These data support the hypothesis that TM9SF2 is overexpressed in CRC and may play a role as an oncogene.
TM9SF2 is overexpressed in CRC cell lines and patient samples. qRT-PCR and transcriptomics demonstrate TM9SF2 overexpression in CRC cell lines and patient derived samples. (A) (top) Oncoprint of genomic alterations in 379 TCGA patient colorectal adenocarcinomas subjected to RNA sequencing. TM9SF2 copy number alterations and mRNA expression changes are shown in addition to a heatmap depicting the intensity of TM9SF2 mRNA changes. (A) (bottom) Oncoprint showing alterations in TM9SF2 in 222 TCGA patients subjected to Agilent microarray analysis. (B) qRT-PCR analysis for TM9SF2 expression in human CRC cell lines. HCEC, an immortalized but non-transformed colon epithelial cell line, was used as the control. (C) RNA sequencing results showing TM9SF2 expression levels in a set of CRC tumor and matched normal tissue samples from the University of Minnesota. Green lines highlight samples with an increase in TM9SF2 mRNA in tumor versus normal and red lines indicate a decrease. Expression differences between healthy and tumor tissue were tested for significance using non-parametric Mann-Whitney U test (paired). T-test *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001.
TM9SF2 functions as an oncogene in CRC cell lines
To determine if TM9SF2 plays an oncogenic role in CRC, we generated stable TM9SF2 knockdowns in DLD1 cells using lentiviral shRNA vectors (Fig. S1A,B). Reducing the level of TM9SF2 in DLD1 cells resulted in reduced anchorage-independent growth compared to empty-vector control cells based on their reduced ability to form colonies in soft agar. We observed a significant reduction in colony numbers in two independent knockdown DLD1 cell lines (shRNA3: 25% and shRNA7: 37% (Figs 3A and S1C).
CRISPR/Cas9 knockout of TM9SF2 reduces cellular proliferation rate and colony formation in vitro. CRISPR/Cas9 mediated deletion of TM9SF2 reduces cellular proliferation and anchorage-independent cell growth. (A) Quantification of colonies indicating that TM9SF2 knockdown reduces DLD1 growth in soft agar. Two independent knockdown clones, sh3 and sh7, were used in this experiment. (B) Representative quantification of colonies indicating that TM9SF2 knockout reduces cell growth in soft agar. Two independent single cell clones, A3 and C6, were used in this experiment. (B) Images of colonies stained with crystal violet ten days post plating. (C) proliferation assay measuring trypan blue exclusion in parental (black line) and TM9SF2 HT-29 knockout clones (gray lines). (D) Representative quantification of colonies indicating TM9SF2 overexpression increases cell growth in soft agar. T-test *P < 0.05. T-test ****P < 0.0001.
To further validate the oncogenic role of TM9SF2, we used CRISPR/Cas9 editing to knockout the TM9SF2 gene in HT-29 and HCT116 CRC cell lines (Supplementary Fig. 2). We generated multiple independent clones that did not express TM9SF2 based on sequencing and Western blot analysis (Supplementary Fig. S2D). Similar to the DLD1 knockdown cell line, loss of TM9SF2 in HT-29 resulted in reduced anchorage-independent growth based on the soft agar assay (Fig. 3B). In addition, the proliferation rate was decreased in HT-29 TM9SF2 KO cells (Fig. 3C). Anchorage-independent growth and proliferation were not affected in TM9SF2 knockout HCT116 cells (data not shown), which is likely due to the already low level of expression in the parental HCT116 cells (Fig. 2B). In fact, overexpression of TM9SF2 in HCT116 cells resulted in increased colony growth in soft agar, which further supports the finding that TM9SF2 functions as a CRC driving oncogene (Figs 3D and S1E).
TM9SF2 functions as an oncogene in vivo
To determine if TM9SF2 knockout has an effect on tumor growth in vivo we performed a xenograft experiment using HT-29 TM9SF2 KO cells compared to parental cells. By day 20, mean tumor volume for parental control cells exceeded 1 000 mm3 compared to 600 mm3 and 325 mm3 for knockout clones A3 and C6 respectively (Fig. 4A). This reduced tumor burden corresponded to a significantly increased overall survival for mice bearing TM9SF2 knockout tumor cells. Mice grafted with clones A3 and C6 had a median survival of 39 and 43 days, while mice grafted with control cells survived on average only 25 days (Fig. 4B). Taken together, these data support the hypothesis that TM9SF2 functions as an oncogene in CRC.
TM9SF2 knockout reduces tumor growth and prolongs survival in a xenograft model. TM9SF2 knockout reduces tumor growth and extends survival in an in vivo xenograft model. (A) HT-29 TM9SF2 knockout cells were injected subcutaneously in the rear flank of athymic nude mice and tumor volume was measured with calipers approximately every other day. TM9SF2 knockout cells have a significantly reduced ability to grow versus control cells. (B) Kaplan-Meier survival curves for control animals and two groups bearing tumors from two independent TM9SF2 knockout clones. *P < 0.05. **P < 0.01. T-test ****P < 0.0001.
Cell cycle and metabolic pathways upregulated by TM9SF2
To uncover the molecular pathways affected by TM9SF2 we performed RNASeq and quantified transcript levels in the parental and TM9SF2 knockout HT-29 cells. We identified 835 genes that were differentially expressed, with 596 being increased and 239 being decreased in the knockout cells (see Supplementary Table S5). The expression of several notable cell cycle checkpoint genes including CCNA2, CCNB2, and AURKA, as well as the Myc target proliferation-related genes PCNA and NPM1, were all significantly lower in the TM9SF2 knockout cells. The reduced proliferative phenotype observed in TM9SF2 knockout cells can likely be explained by the downregulation of these genes.
To identify gene sets that are affected by TM9SF2 loss we performed Gene Set Enrichment analysis (GSEA) on the set of altered genes30. Gene sets that were significantly enriched in parental controls included the G2-M cell cycle checkpoint genes, Myc target genes, and genes in the reactive oxygen species pathway (Fig. 5 and Table 1). The highest-ranking member of the reactive oxygen species pathways was the glucose-6-phosphate dehydrogenase gene (G6PD). Loss or downregulation of this gene has been known to cause embryonic lethality in mice and lead to cellular senescence in various cell types31,32,33. TM9SF2 knockout cells have more than a two-fold decrease in G6PD, which likely contributes to the reduced oncogenic behavior of these cells.
We used Ingenuity Pathway Analysis to identify canonical pathways altered in TM9SF2 knockout cells. This analysis found significant alterations in the expression of genes associated with Ceramide signaling, Mitotic Roles of Polo-like Kinase, and Protein Kinase A signaling pathways (Supplementary Table S6). Notable Ceramide signaling genes altered in the HT-29 knockout cells included PIK3R3 (−2.1-fold), S1PR2 (+5.7-fold), SMPD3 (−8.9-fold), and SPHK1 (+5.2-fold). These genes, and the majority of the other molecules associated with Ceramide signaling, are altered in the direction consistent with pathway activation. These genes are known to play an integral part in the metabolism of Sphingolipids and they have been associated with various neoplasms34,35,36,37,38.
ELF1 transcription factor regulates TM9SF2 expression
To identify regulatory factors responsible for upregulation of TM9SF2 in CRC we analyzed HCT116 CHIP-Seq data from the ENCODE project39. Based on ENCODE data, there was a strong ELF1 binding motif (chr13: 100,153,732–100,153,742) in the 5′UTR of TM9SF2. The consensus ELF1 binding motif is 5′-CGGAAGT, which is a near perfect sequence match to the observed ELF1 DNA binding site in the TM9SF2 promoter region (5′-CGGAACT). Based on ENCODE Chip-seq data, the location of the ELF1 binding site overlaps with elevated levels of H3K4 trimethylation, an epigenetic mark commonly associated with promoters (Figs 6A and S3).
The Ets family transcription factor ELF1 regulates TM9SF2 expression. ELF1 is a potential regulator of TM9SF2 gene expression. (A) Position weight matrix from ENCODE CHIP-seq experiments showing a potential ELF1 transcription factor binding motif in the 5′ UTR of the TM9SF2 gene. (B) ELF1 and TM9SF2 mRNA co-expression scatter plot using TCGA RNAseq data from CRC patients. (C) qRT-PCR analysis for ELF1 expression in human CRC cell lines. HCEC, an immortalized but non-transformed colon epithelial cell line, was used as the control. (D) Data from ELF1 ChIP normalized against those from IgG mock ChIP. Schematic shows binding location of two qPCR primer sets (black and gray arrows) used to quantify TM9SF2 promoter DNA after ELF1 ChIP. Translation start site is indicated with bent arrow. t-test, ns = not significant, *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001.
If ELF1 is required for elevated expression of TM9SF2, we predicted that ELF1 mRNA levels would correlate with TM9SF2 expression. To test this prediction, we analyzed ELF1 and TM9SF2 mRNA expression levels in 382 TCGA CRC patient samples and found a strong positive correlation (Pearson: 0.781; Spearman 0.744) between the expression of TM9SF2 and ELF1 (Fig. 6B). For further validation we performed qRT-PCR for ELF1 in our panel of CRC cell lines. ELF1 expression remains generally unchanged in the five cell lines with lower TM9SF2 expression but is highly upregulated in the three cell lines with the highest TM9SF2 expression (HT-29, HCT-8 and DLD1) (Figs 2B and 6C).
To verify that ELF1 binds to the TM9SF2 promoter we performed CHIP-qPCR using DLD1 cells. At steady state we observed a 6-15-fold enrichment in TM9SF2 promoter DNA after chromatin pull-down with an ELF1 antibody (Fig. 6D). These data are in support of ELF1 as a TM9SF2 regulating transcription factor.
High levels of TM9SF2 correlate with higher stage cancer and decreased disease-free survival
Based on our data supporting a role for TM9SF2 as an oncogene in CRC we tested the hypothesis that TM9SF2 expression correlates with worse patient prognosis. We analyzed data from Staub et al., (Staub, 2009, GSE12945) where they measured gene expression in 62 patient samples spanning all clinical stages of CRC (I-IV). We found that TM9SF2 expression correlates with disease severity with late stage (III & IV) cancers having a significantly higher level of expression compared to stage I (Fig. 7A).
TM9S2 expression correlates with stage and predicts relapse free survival. TM9SF2 expression correlates with disease stage and predicts relapse-free survival. (A) Microarray data (Staub, 2009, GSE12945) depicting TM9SF2 mRNA expression level in patient samples from stages I through IV. (B,C) Kaplan-Meier curves depicting relapse-free survival with data stratified by intensity of TM9SF2 mRNA measured in CRC samples via microarray (Left, Marissa, 2013, GSE39582, Right, Sieber Smith, 2010, GSE14333 plus GSE17538 minus identical samples).
In two additional studies with 566 patients (Marisa et al.) and 355 patients (Sieber et al.; Smith et al.) we tested the association of TM9SF2 mRNA levels with disease-free survival40,41,42. In both studies, we observed a significantly favorable disease-free survival probability in patients with the lowest levels of TM9SF2 expression (Fig. 7B,C).
Discussion
Our studies support the hypothesis that TM9SF2 functions as an oncogene in CRC. Analysis of data from our previous SB transposon forward genetic screen identified Tm9sf2 as a top candidate cancer gene. Mining of other publicly available SB transposon databases revealed further support for Tm9sf2 as a candidate cancer gene. Transposon insertions in the murine Tm9sf2 gene occurred in over 7% of the 1 674 analyzed digestive tract tumors and is predicted to be a progression driver gene24. We demonstrated using both TCGA data and our own independent data set that TM9SF2 mRNA is overexpressed in up to one-third of CRC patients. Finally, using RNAi and CRISPR/Cas9 gene editing we demonstrated that reduction or complete knockout of TM9SF2 resulted in reduced tumor fitness in the in vitro and in vivo settings.
Our data also support TM9SF2 expression as a potential prognostic indicator as we found that mRNA expression levels correlate with both disease stage and relapse-free survival probability. We observed a significant increase in TM9SF2 expression in patients with stage III/IV disease versus those with stage I, suggesting that TM9SF2 expression may promote either the migration of cancerous cells or their seeding and growth in distant organs.
The molecular mechanism of TM9SF2’s oncogenic function in CRC carcinogenesis is currently unknown. Others have demonstrated that TM9SF proteins are responsible for controlling surface expression of adhesion molecules43,44. In our RNA-seq data we observed alterations in several cell adhesion related genes including ITGA1 and three members of the CEACAM family (CEACAM5, CEACAM6, CEACAM19). The mRNA levels of CEACAM5 and CEACAM6 were reduced by 4.9-fold and 12.3-fold respectively in knockout cells. Both CEACAM5 and CEACAM6 are known to play a critical role in facilitating tumorigeneis and metastasis by inhibiting anoikis45,46. The reduced ability of TM9SF2 knockout cells to grow in soft agar could be explained by an increased sensitivity to anoikis due to the accompanying decrease in CEACAM expression.
It was recently reported that TM9SF4 plays a role in the assembly of the V-ATPase proton pump in CRC cells19. Silencing of TM9SF4 expression led to a more acidic cytoplasmic pH with an accompanying alkalization of intracellular vesicles. Ingenuity pathway analysis revealed that the top canonical pathway altered in TM9SF2 knockout cells was the Ceramide Signaling pathway. Ceramides are bioactive lipids that are critical for modulating multiple cellular processes including cell cycle, apoptosis, senescence, and stemness47. Ceramide levels within cells are tightly controlled by pH sensitive phospholipases called sphingomyelinases48. With a close similarity in amino acid sequence and structure to TM9SF4, we hypothesize that TM9SF2 may function as a membrane-bound vesicular protein involved in the regulation of intracellular acidity. Future studies are required to determine the role of TM9SF2 in intracellular ion homeostasis and regulation of ceramide signaling molecules.
An increased understanding of the genetic basis of CRC will be useful for designing new targeted therapies. Current efforts to target the known drivers of CRC, including APC, SMAD4, TP53, and KRAS, have not yet resulted in significant improvements in survival for advanced CRC. In the current study we identify TM9SF2 as an oncogene that is most likely regulated by the transcription factor ELF1. Our findings provide a rationale for exploring the efficacy of drugs targeting either TM9SF2 or ELF1 for treating advanced-stage CRC.
Materials and Methods
All experiments were performed in accordance with relevant guidelines and regulations.
Cells
All cell lines (DLD1, LoVo, HCT116, HT-29, HCT-8, SW480, SW620, and T84) except HCEC were obtained from the ATCC and were minimally passaged. Human colonic epithelial cells (HCEC) were immortalized by expression of cyclin dependent kinase 4 (Cdk4) and the active components of human telomerase (hTERT) and were kindly provided by Dr. Jerry Shay (UT Southwestern, Dallas TX). HCEC cells were maintained in DMEM media with 2% calf serum, 25 ng/ml EGF (Sigma Aldrich, St. Louis, MO), 2 μg/ml transferrin (Sigma Aldrich, St. Louis, MO), 10 μg/ml insulin (Sigma Aldrich), 5 nM sodium selenite (Sigma Aldrich, St. Louis, MO), 1 μg/ml hydrocortisone (Sigma Aldrich), and 50 μg/ml gentamicin (Gemini Bio-products, West Sacramento, CA). HCEC cells were grown at 37 °C on Primaria flasks (Corning Inc., Conring NY) placed in chambers purged with 93% nitrogen 5% carbon dioxide and 2% oxygen gas. All other cell lines were maintained in DMEM with 4 mM L-glutamine, 10% FBS, and 1x penicillin/streptomycin at 37 °C and 5% carbon dioxide.
Vectors
RNAi experiments were performed using pLKO.1 vectors obtained through the University of Minnesota Genomics Center (UMGC) partnership with the Open Access Program from Open Biosystems. The pLKO.1 vector with the oligo ID (TRCN0000059768) was used to generate DLD1 shRNA3 cells and the vector with oligo ID (TRCN0000059772) was used to generate DLD1 shRNA7 cells. Non-silencing control vectors were obtained in the same manner. The lentiviral packaging vector psPAX2 was a gift from Didier Trono (Addgene plasmid #12260) and viral envelope encoding vector pCMV-VSV-G was a gift from Bob Weinberg (Addgene plasmid #8454). The lentiCRISPR v2 vector used to generate TM9SF2 knockout cells was a gift from Feng Zhang (Addgene plasmid # 52961).
ShRNA knockdown cells
DLD1 stable TM9SF2 knockdown cells were created by transduction with lentiviral particles encoding TM9SF2 shRNAs followed by puromycin selection. Virus production was performed using standard 293 T packaging protocols and the pLKO.1 vectors.
CRISPR/Cas9 knockout cells
CRISPR lentiviral constructs targeting TM9SF2 were generated by annealing together primers for sgTM9SF2 CRISPR1, sgTM9SF2 CRISPR2, and sgTM9SF2 CRISPR6 and cloning the annealed product into lentiCRISPR v2 as described previously49. The sgRNA primer sequences are listed in Supplementary Table S1. The sgRNAs were designed to specifically target coding regions of the TM9SF2 gene using the CRISPR design tool from Dr. Feng Zhang’s laboratory (http://crisper.mit.edu/). After transducing cells with TM9SF2 targeting CRISPR/Cas9 lentiviral particles, single cell puromycin-resistant clones were isolated by limiting dilution. DNA was extracted from single cell clones for PCR amplification of the genomic loci targeted by CRISPR/Cas9. After PCR, the amplicons were purified and subsequently sequenced. Sequencing data from these clones was then used for Tracking of Indels by Decomposition (TIDE) analysis and mutants were confirmed by immunoblot with an anti-TM9SF2 antibody (Sigma Aldrich, St. Louis, MO, Catalog # HPA005657) using a standard Western Blotting protocol.
Soft Agar assay and cell proliferation by trypan blue exclusion
For soft agar assays, cells were grown in between layers of 0.5% Sea Plaque Low Melt Agarose (Lonza Cat. # 50101) in DMEM supplemented with 10% FBS and 1x penicillin/streptomycin for 10 days. Colonies were fixed, stained with crystal violet, and imaged. Colony counts were analyzed using ImageJ software. Equal numbers of HT-29 control and TM9SF2 knockout cells were plated in triplicate wells of 24-well culture plates and allowed to adhere for 24 hours. Cells were detached using trypsin and, after adding trypan blue, counted with an automated cell counter (Thermo Fisher Scientific, Waltham MA). The total number of live cells was determined for each well and the average number of viable cells for each triplicate was plotted for days 1 through 4.
RNA extraction and qRT-PCR
Total RNA was extracted using the RNeasy Plus mini kit (Qiagen, Germantown, MD) according to the manufacturer’s protocol. Total RNA concentration and quality were measured using the BioTek Epoch microplate spectrophotometer (BioTek, Winooski, VT). One microgram of total RNA was reverse transcribed using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). RT-qPCR was then performed in triplicate using diluted cDNA, FastStart Essential DNA Green Master mix (Roche, Basel, Switzerland), and either ELF1 or TM9SF2 specific primers. Samples were run in the LightCycler 96 (Roche, Basel, Switzerland). Data were normalized to human beta actin and fold change was calculated using the delta-delta Ct method. Primer sequences are listed in Supplementary Table S1.
Xenograft model
Athymic nude mice were obtained from The Jackson Laboratory (Stock No. 002019) and protocols were approved by the University of Minnesota’s Institutional Animal Care and Use committee. Five million HT-29 control and TM9SF2 knockout cells were resuspended in 200 µL of DMEM media and subcutaneously injected into the rear flank. Each mouse carried duplicate tumors on the left and right flank. Tumor volume was measured with digital calipers every other day and tumor volume was calculated using the formula: volume = (width)2 × (length)/2, where the width is the smaller of the measurements. All animal experimental protocols were approved by the University of Minnesota’s Institutional Animal Care and Use Committee (IACUC Protocol ID: 1702-34600A). All experiments were performed in accordance with the American Association for Accreditation of Laboratory Animal Care (AAALAC) relevant guidelines and regulations for performing live vertebrate experiments.
Gene expression analysis
Oncoprint data showing the genomic alterations in TM9SF2 were generated using cBioPortal (version 1.12.1; http://www.cbioportal.org/index.do) using both RNA-seq and Microarray data from Colorectal Adenomacarcinoma (TCGA, Provisional)27,28. cBioPortal enrichment analysis revealed that TM9SF2 and ELF1 were highly positively correlated as measured by Pearson’s or Spearman’s correlation.
RNA sequencing
Matched frozen tumor and normal samples were acquired from 44 patients (88 samples total) from the University of Minnesota’s Tissue Repository. All samples were de-identified. For RNA-seq of patient samples, total RNA was extracted using a previously established protocol50,51. Purified RNA was submitted to the University of Minnesota Genomics Center for library preparation and sequencing. A quality check of raw sequence data (FASTQ files) was performed using FastQC software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to assess overall sequence quality, GC content, adaptor content, etc. Quality trimming was performed to remove sequence adaptors and low-quality bases using Trimmomatic with a 3 bp sliding window trimming from 3′ end requiring minimum Q16 (phred33)52. FastQC was run on the resulting trimmed FASTQ files to ensure good quality of sequences. The paired-end reads were mapped to NCBI v38 H. sapiens reference genome using HISAT2, resulting in average alignment rate of 87.11% overall for 88 samples53. SAMtools was used for sorting and indexing the aligned bam files. After alignment, featureCounts was used to generate gene abundance, and Cuffnorm was used to generate an FPKM expression table54. We used ensembl web-service from biomaRt package in R to map gene transcripts (ensemble gene id) to HGNC gene symbols55.
RNA-seq of HT-29 knockout cells: 50 bp FastQ paired-end reads (n = 8.8 million per sample) were trimmed using Trimmomatic (v 0.33) enabled with the optional “-q” option; 3 bp sliding-window trimming from 3′ end requiring minimum Q30. Quality control checks on raw sequence data for each sample were performed with FastQC. Read mapping was performed via HISAT2 (v2.0.2) using the UCSC human genome (hg38) as reference. Gene quantification was done via Cuffquant for FPKM values and Feature Counts for raw read counts. Differentially expressed genes were identified using the edgeR (negative binomial) feature in CLCGWB (Qiagen, Valencia, CA) using raw read counts. We filtered the generated list based on a minimum 2X Absolute Fold Change and Bonferroni corrected p < 0.05. These filtered genes were then imported into Ingenuity Pathway Analysis Software (Qiagen, Valencia, CA) for pathway identification.
Chromatin Immunoprecipitation and qPCR
DLD1 cells were plated at 3 × 106 cells per 10 cm plate in standard fully supplemented DMEM overnight. ChIP was performed using an ELF-1 specific antibody (Santa Cruz, SC-631) or a non-specific IgG rabbit isotype control using Protein G magnetic beads (Active Motif). Analysis was performed using previously described methods56. qPCR data are presented as Fold enrichment relative to the negative (IgG) sample. Primers are listed in Supplemental Table S1.
Patient Prognoses and Survival
The Staub cohort of 62 CRC patient samples used for transcriptome analysis by microarray (GSE12945) was previously described57. Patients were grouped by disease stage and the mean expression value of TM9SF2 in each stage was plotted using box plots. Statistical significance between groups were determined using one-way ANOVA with a Tukey’s post hoc test. Kaplan Meier curves depicting relapse-free survival (RFS) probability were generated using the R2 genomics analysis and visualization platform (http://hgserver1.amc.nl/). The Marisa et al. cohort consisting of 566 samples was used for RFS analysis with two groups (high vs. low) TM9SF2 mRNA expression divided based on optimal expression separation. RFS groups were compared using the log-rank test. The same method was used when analyzing the 355-patient cohort provided by Sieber and Smith (GSE1433 and GSE175)41,42.
References
Vogelstein, B. et al. Genetic alterations during colorectal-tumor development. The New England journal of medicine 319, 525–532, https://doi.org/10.1056/NEJM198809013190901 (1988).
Tanaka, T. Colorectal carcinogenesis: Review of human and experimental animal studies. J Carcinog 8, 5 (2009).
Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337, https://doi.org/10.1038/nature11252 (2012).
Haan, J. C. et al. Genomic landscape of metastatic colorectal cancer. Nat Commun 5, 5457, https://doi.org/10.1038/ncomms6457 (2014).
Starr, T. K. et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 323, 1747–1750, https://doi.org/10.1126/science.1163040 (2009).
Starr, T. K. et al. A Sleeping Beauty transposon-mediated screen identifies murine susceptibility genes for adenomatous polyposis coli (Apc)-dependent intestinal tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America 108, 5765–5770, https://doi.org/10.1073/pnas.1018012108 (2011).
Takeda, H. et al. Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nature genetics 47, 142–150, https://doi.org/10.1038/ng.3175 (2015).
Abbott, K. L. et al. The Candidate Cancer Gene Database: a database of cancer driver genes from forward genetic screens in mice. Nucleic Acids Res 43, D844–848, https://doi.org/10.1093/nar/gku770 (2015).
Than, B. L. et al. The role of KCNQ1 in mouse and human gastrointestinal cancers. Oncogene 33, 3861–3868, https://doi.org/10.1038/onc.2013.350 (2014).
Than, B. L. N. et al. CFTR is a tumor suppressor gene in murine and human intestinal cancer. Oncogene 36, 3504, https://doi.org/10.1038/onc.2017.3 (2017).
Seshagiri, S. et al. Recurrent R-spondin fusions in colon cancer. Nature 488, 660–664, https://doi.org/10.1038/nature11282 (2012).
Pruvot, B. et al. Comparative analysis of nonaspanin protein sequences and expression studies in zebrafish. Immunogenetics 62, 681–699, https://doi.org/10.1007/s00251-010-0472-x (2010).
Chluba-de Tapia, J., de Tapia, M., Jaggin, V. & Eberle, A. N. Cloning of a human multispanning membrane protein cDNA: evidence for a new protein family. Gene 197, 195–204 (1997).
He, P. et al. High-throughput functional screening for autophagy-related genes and identification of TM9SF1 as an autophagosome-inducing gene. Autophagy 5, 52–60 (2009).
Zaravinos, A., Lambrou, G. I., Boulalas, I., Delakas, D. & Spandidos, D. A. Identification of common differentially expressed genes in urinary bladder cancer. PloS one 6, e18135, https://doi.org/10.1371/journal.pone.0018135 (2011).
Chang, H. et al. Identification of genes associated with chemosensitivity to SAHA/taxane combination treatment in taxane-resistant breast cancer cells. Breast cancer research and treatment 125, 55–63, https://doi.org/10.1007/s10549-010-0825-z (2011).
Oo, H. Z. et al. Identification of novel transmembrane proteins in scirrhous-type gastric cancer by the Escherichia coli ampicillin secretion trap (CAST) method: TM9SF3 participates in tumor invasion and serves as a prognostic factor. Pathobiology 81, 138–148, https://doi.org/10.1159/000357821 (2014).
Lozupone, F. et al. The human homologue of Dictyostelium discoideum phg1A is expressed by human metastatic melanoma cells. EMBO reports 10, 1348–1354, https://doi.org/10.1038/embor.2009.236 (2009).
Lozupone, F. et al. TM9SF4 is a novel V-ATPase-interacting protein that modulates tumor pH alterations associated with drug resistance and invasiveness of colon cancer cells. Oncogene 34, 5163–5174, https://doi.org/10.1038/onc.2014.437 (2015).
Starr, T. K. & Largaespada, D. A. Cancer gene discovery using the Sleeping Beauty transposon. Cell Cycle 4, 1744–1748, https://doi.org/10.4161/cc.4.12.2223 (2005).
Dupuy, A. J., Akagi, K., Largaespada, D. A., Copeland, N. G. & Jenkins, N. A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436, 221–226, https://doi.org/10.1038/nature03691 (2005).
Collier, L. S., Carlson, C. M., Ravimohan, S., Dupuy, A. J. & Largaespada, D. A. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 436, 272–276, https://doi.org/10.1038/nature03681 (2005).
Copeland, N. G. & Jenkins, N. A. Harnessing transposons for cancer gene discovery. Nature reviews. Cancer 10, 696–706, https://doi.org/10.1038/nrc2916 (2010).
Newberg, J. Y., Mann, K. M., Mann, M. B., Jenkins, N. A. & Copeland, N. G. SBCDDB: Sleeping Beauty Cancer Driver Database for gene discovery in mouse models of human cancers. Nucleic Acids Res 46, D1011–D1017, https://doi.org/10.1093/nar/gkx956 (2018).
March, H. N. et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nature genetics 43, 1202–1209, https://doi.org/10.1038/ng.990 (2011).
Forbes, S. A. et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45, D777–D783, https://doi.org/10.1093/nar/gkw1121 (2017).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2, 401–404, https://doi.org/10.1158/2159-8290.CD-12-0095 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling 6, pl1, https://doi.org/10.1126/scisignal.2004088 (2013).
Roig, A. I. et al. Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro. Gastroenterology 138(1012–1021), e1011–1015, https://doi.org/10.1053/j.gastro.2009.11.052 (2010).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550, https://doi.org/10.1073/pnas.0506580102 (2005).
Stanton, R. C. Glucose-6-phosphate dehydrogenase, NADPH, and cell survival. IUBMB Life 64, 362–369, https://doi.org/10.1002/iub.1017 (2012).
Ho, H. Y. et al. Enhanced oxidative stress and accelerated cellular senescence in glucose-6-phosphate dehydrogenase (G6PD)-deficient human fibroblasts. Free Radic Biol Med 29, 156–169 (2000).
Longo, L. et al. Maternally transmitted severe glucose 6-phosphate dehydrogenase deficiency is an embryonic lethal. EMBO J 21, 4229–4239 (2002).
Widau, R. C., Jin, Y., Dixon, S. A., Wadzinski, B. E. & Gallagher, P. J. Protein phosphatase 2A (PP2A) holoenzymes regulate death-associated protein kinase (DAPK) in ceramide-induced anoikis. The Journal of biological chemistry 285, 13827–13838, https://doi.org/10.1074/jbc.M109.085076 (2010).
Powell, J. A. et al. Targeting sphingosine kinase 1 induces MCL1-dependent cell death in acute myeloid leukemia. Blood 129, 771–782, https://doi.org/10.1182/blood-2016-06-720433 (2017).
Adada, M. M. et al. Intracellular sphingosine kinase 2-derived sphingosine-1-phosphate mediates epidermal growth factor-induced ezrin-radixin-moesin phosphorylation and cancer cell invasion. FASEB J 29, 4654–4669, https://doi.org/10.1096/fj.15-274340 (2015).
Shida, D., Takabe, K., Kapitonov, D., Milstien, S. & Spiegel, S. Targeting SphK1 as a new strategy against cancer. Curr Drug Targets 9, 662–673 (2008).
Kim, W. J. et al. Mutations in the neutral sphingomyelinase gene SMPD3 implicate the ceramide pathway in human leukemias. Blood 111, 4716–4722, https://doi.org/10.1182/blood-2007-10-113068 (2008).
Consortium, E. P. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74, https://doi.org/10.1038/nature11247 (2012).
Marisa, L. et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 10, e1001453, https://doi.org/10.1371/journal.pmed.1001453 (2013).
Jorissen, R. N. et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research 15, 7642–7651, https://doi.org/10.1158/1078-0432.CCR-09-1431 (2009).
Smith, J. J. et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 138, 958–968, https://doi.org/10.1053/j.gastro.2009.11.005 (2010).
Froquet, R. et al. TM9/Phg1 and SadA proteins control surface expression and stability of SibA adhesion molecules in Dictyostelium. Molecular biology of the cell 23, 679–686, https://doi.org/10.1091/mbc.E11-04-0338 (2012).
Perrin, J. et al. TM9 family proteins control surface targeting of glycine-rich transmembrane domains. Journal of cell science 128, 2269–2277, https://doi.org/10.1242/jcs.164848 (2015).
Ordonez, C., Screaton, R. A., Ilantzis, C. & Stanners, C. P. Human carcinoembryonic antigen functions as a general inhibitor of anoikis. Cancer research 60, 3419–3424 (2000).
Ilantzis, C., DeMarte, L., Screaton, R. A. & Stanners, C. P. Deregulated expression of the human tumor marker CEA and CEA family member CEACAM6 disrupts tissue architecture and blocks colonocyte differentiation. Neoplasia 4, 151–163, https://doi.org/10.1038/sj/neo/7900201 (2002).
Lai, M. et al. Complete Acid Ceramidase ablation prevents cancer-initiating cell formation in melanoma cells. Scientific reports 7, 7411, https://doi.org/10.1038/s41598-017-07606-w (2017).
Goni, F. M. & Alonso, A. Sphingomyelinases: enzymology and membrane activity. FEBS letters 531, 38–46 (2002).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784, https://doi.org/10.1038/nmeth.3047 (2014).
Burns, M. B. et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494, 366–370, https://doi.org/10.1038/nature11881 (2013).
Burns, M. B., Lynch, J., Starr, T. K., Knights, D. & Blekhman, R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med 7, 55, https://doi.org/10.1186/s13073-015-0177-8 (2015).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120, https://doi.org/10.1093/bioinformatics/btu170 (2014).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360, https://doi.org/10.1038/nmeth.3317 (2015).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930, https://doi.org/10.1093/bioinformatics/btt656 (2014).
Durinck, S., Spellman, P. T., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 4, 1184–1191, https://doi.org/10.1038/nprot.2009.97 (2009).
Chan, S. C. et al. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res 43, 5880–5897, https://doi.org/10.1093/nar/gkv262 (2015).
Staub, E. et al. An expression module of WIPF1-coexpressed genes identifies patients with favorable prognosis in three tumor types. J Mol Med (Berl) 87, 633–644, https://doi.org/10.1007/s00109-009-0467-y (2009).
Acknowledgements
This work was supported by a 3M Science and Technology Fellowship award and the Norman Wells Memorial Fellowship from the Minnesota Colorectal Cancer Research Foundation (Clark CR). Additional financial support came from the National Cancer Institute of the National Institutes of Health, No. 5R00CA1516723-03, the Mezin-Koats Colorectal Cancer Research Fund and the University of Minnesota Masonic Cancer Center (Starr, TK). Other support came from the Minnesota Supercomputing Institute and the University of MN Genomics Center. The authors thank Robert T. Cormier for critically reviewing and editing the manuscript.
Author information
Authors and Affiliations
Contributions
Data acquisition: C.R.C., M.M., P.B., S.R.H., A.S., W.D., S.P.; Data analysis and interpretation: C.R.C., S.P., M.B., R.B., J.E.A., T.K.S.; Wrote the manuscript: C.R.C.; Conception, design, and supervision of study: C.R.C., T.K.S. All authors contributed to manuscript revision.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Clark, C.R., Maile, M., Blaney, P. et al. Transposon mutagenesis screen in mice identifies TM9SF2 as a novel colorectal cancer oncogene. Sci Rep 8, 15327 (2018). https://doi.org/10.1038/s41598-018-33527-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-33527-3
Keywords
This article is cited by
-
Identification of sulfur metabolism-related gene signature in osteoarthritis and TM9SF2’s sustenance effect on M2 macrophages' phagocytic activity
Journal of Orthopaedic Surgery and Research (2024)
-
Novel biomarkers to predict treatment response and prognosis in locally advanced rectal cancer undergoing neoadjuvant chemoradiotherapy
BMC Cancer (2023)
-
Retroviral integrations contribute to elevated host cancer rates during germline invasion
Nature Communications (2021)
-
LINC01232 exerts oncogenic activities in pancreatic adenocarcinoma via regulation of TM9SF2
Cell Death & Disease (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.