Abstract
Results from previous experiments indicated that the Gram-negative α-proteobacterium Serratia liquefaciens strain ATCC 27592 was capable of growth under low temperature (0 °C), low pressure (0.7 kPa), and anoxic, CO2-dominated atmosphere–conditions intended to simulate the near-subsurface environment of Mars. To probe the response of its transcriptome to this extreme environment, S. liquefaciens ATCC 27592 was cultivated under 4 different environmental simulations: 0 °C, 0.7 kPa, CO2 atmosphere (Condition A); 0 °C, ~101.3 kPa, CO2 atmosphere (Condition B); 0 °C, ~101.3 kPa, ambient N2/O2 atmosphere (Condition C); and 30 °C, ~101.3 kPa, N2/O2 atmosphere (Condition D; ambient laboratory conditions). RNA-seq was performed on ribosomal RNA-depleted total RNA isolated from triplicate cultures grown under Conditions A-D and the datasets generated were subjected to transcriptome analyses. The data from Conditions A, B, or C were compared to laboratory Condition D. Significantly differentially expressed transcripts were identified belonging to a number of KEGG pathway categories. Up-regulated genes under all Conditions A, B, and C included those encoding transporters (ABC and PTS transporters); genes involved in translation (ribosomes and their biogenesis, biosynthesis of both tRNAs and aminoacyl-tRNAs); DNA repair and recombination; and non-coding RNAs. Genes down-regulated under all Conditions A, B, and C included: transporters (mostly ABC transporters); flagellar and motility proteins; genes involved in phenylalanine metabolism; transcription factors; and two-component systems. The results are discussed in the context of Mars astrobiology and planetary protection.
Similar content being viewed by others
Introduction
A central goal of Astrobiology is to understand the potential for habitability in the universe, including determination of the physical limits at which life can exist and the mechanisms used by living organisms to survive and grow in extreme environments1,2. Of particular interest have been investigations of whether Earth life could inhabit the environments of Mars, our closest potentially habitable neighbor. These studies are relevant to two related areas: (i) the potential for transport of life between Earth and Mars by natural impact processes (i.e., lithopanspermia), and (ii) the potential forward contamination of Mars as a consequence of human exploration activities (i.e., planetary protection)3,4,5,6,7. Due to their well-known resistance properties and ubiquitous distribution in extreme terrestrial environments, prokaryotes are considered the most likely candidates for interplanetary transfer by natural processes or human spaceflight activities, and much attention has logically focused on highly resistant or extremophilic microbes such as spores of Bacillus subtilis or the extremely radiation-resistant Deinococcus radiodurans4,8. Relatively less attention has been paid to more common environmental or human-associated bacteria which may actually be more likely to find themselves inadvertently transported to the martian surface. It is therefore important to understand the potential for terrestrial microbes not normally considered as extremophiles9 to proliferate in the martian environment, especially considering the recent increase of interest in travel to Mars by non-governmental entities not necessarily subject to the planetary protection requirements agreed to by the United Nations and administered by the Committee on Space Research (COSPAR)10.
Early studies confirmed that the UV radiation environment was a potent factor limiting the survival of Earth microbes on the martian surface, but that burial of cells at even minimal depths in the regolith provided effective UV shielding3,11,12. Subsequent studies testing the ability of various bacteria to grow under increasingly Mars-like laboratory simulations revealed that growth of most of the microbes tested, mostly strains obtained from laboratory collections or spacecraft assembly facilities, was inhibited by low temperature, low pressure, and anoxic, predominantly CO2 atmospheres, applied either singly or in pairwise combination13,14. Although most bacteria were unable to grow under the conditions applied in these early experiments, recently a small but growing subset of bacterial species have been discovered that can grow under simultaneously applied conditions of low temperature (0 °C), low pressure (0.7 kPa) and atmospheric gas composition (anoxic CO2) intended to simulate the physical conditions in the near subsurface of Mars. The origin of these bacteria were mostly Arctic soils or Siberian permafrost, and they were identified by 16S rDNA sequencing as hardy environmental species belonging to the genera Bacillus, Carnobacterium, Clostridium, Cryobacterium, Exiguobacterium, Paenibacillus, Rhodococcus, Streptomyces, and Trichococcus15,16.
Recently a screen of laboratory strains of bacteria for growth under the above “Mars-like” environmental conditions resulted in the identification of several species of the genus Serratia able to grow, including the type strain of S. liquefaciens, strain ATCC 2759217. Although in early publications the interest in S. liquefaciens stemmed from its being an opportunistic human pathogen18,19,20,21,22, recent reports point towards its physiological versatility allowing it to occupy ecologically diverse environments such as cold raw milk23, thawed cryoprecipitate24 or pulp mill effluent25. Because Serratia spp. have also been found on and within spacecraft and their assembly facilities26,27, S. liquefaciens is considered a potential forward contaminant of Mars-bound missions. The discovery that S. liquefaciens is capable of growth under Mars-like environmental conditions naturally leads to the question: what are the cellular and molecular mechanisms responsible? As a first step towards addressing this question, here we investigated how the global transcription profile (i.e., the transcriptome) of this organism responds when cultivated under environmental conditions mimicking those found in the martian near-subsurface. This study represents the first transcriptome profiling of a microorganism exposed to a simulation of the physical conditions prevailing on Mars.
Results and Discussion
Characterization of the S. liquefaciens transcriptomic response to different physical environments
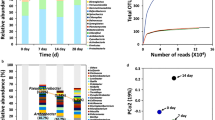
RNA-seq was utilized to analyze transcriptional changes in S. liquefaciens strain ATCC 27592 in response to four environmental conditions of temperature, pressure, and atmospheric gas composition simulating: the physical environment of Mars (Condition A), Earth-ambient laboratory conditions (Condition D), or a mixture of the two environments (Conditions B and C) (Table 1). Total RNA was isolated from cells grown under each condition as described in Materials and Methods, and determination of RNA integrity number (RIN) values (Table 1) demonstrated that all RNA samples were of the high quality required for further processing. RNA-seq analysis was performed on three replicates from each condition, resulting in 12 libraries which were sequenced on an Illumina HiSeq2500 instrument and subjected to the bioinformatic and statistical workflow described in detail in Materials and Methods and summarized in Fig. 1. Transcripts were defined as significantly differentially expressed if they exhibited a >4-fold difference with an adjusted P value < 0.01.
Overview of transcriptome analysis
In order to elucidate differences in gene expression of S. liquefaciens under the four conditions tested, each of the datasets obtained from Conditions A, B, or C were subjected to pairwise comparison with dataset D serving as the “Earth-like” control. The comparative analysis identified 493 differentially expressed genes in Condition A (193 up- and 300 down-regulated); 708 genes in condition B (209 up- and 499 down-regulated) and 429 genes in Condition C (153 up- and 276 down-regulated) with respect to the reference Condition D. The results of this analysis are summarized in Table 2 and represented graphically as Venn diagrams (Fig. 2). Inspection of the data revealed that exposure of S. liquefaciens to Conditions A, B, or C each produced its own set of transcriptome responses. These sets were partially unique and partially overlapping in any combination examined (Fig. 2). For example, a common set of 34 up-regulated and 156 down-regulated transcripts were found in cells exposed to all three Conditions A, B, and C; on the other hand, exposure to Condition A produced differential expression of 112 genes unique to Condition A (75 up- and 37 down-regulated) (Fig. 2). An Excel file listing the differential transcripts in common with Conditions A, B, and C vs. D is presented in Supplemental Table S1.
Venn diagram depicting the number of differentially up-regulated genes (left) and down-regulated genes (right) identified from Conditions A, B, and C compared to Condition D. Numbers in overlapping regions denote genes shared between the different comparisons and numbers in non-overlapping regions denote genes unique to each comparison. See Table 1 for description of the conditions.
Expression differences due to temperature
The transcriptome of S. liquefaciens ATCC 27592 consists of 5,003 total genes, of which 4,894 are protein-coding genes28,29. Comparison of Conditions C vs. D, in which only the temperature of incubation was different (0 °C vs. 30 °C), resulted in significant alteration of transcript levels of a total of 429 genes, or approximately 8.7% of the protein-coding transcriptome, with 153 and 276 transcripts being up- and down-regulated, respectively (Table 2). An Excel file listing the differentially expressed transcripts found in comparison of Condition C vs. D is presented in Supplemental Table S2.
Expression differences due to temperature and atmospheric composition
Comparison of Conditions B vs. D, which differed both in temperature (0 °C vs. 30 °C) and atmospheric gas composition (anoxic CO2 vs. N2/O2), revealed differential expression of 708 total transcripts, or ~14.5% of the protein-coding transcriptome, with 209 and 499 transcripts being up- and down-regulated, respectively (Table 2). An Excel file listing the differentially expressed transcripts found in Condition B vs. D is presented in Supplemental Table S3. Clearly, exposure of S. liquefaciens to the combined conditions of low temperature and anoxic atmosphere provoked a massive restructuring of its transcriptome to accommodate growth under this new environmental regime.
Expression differences in simulated Mars conditions
Cultures of S. liquefaciens grown under Conditions A vs. D differed by three parameters: the temperature of cultivation (0 °C vs. 30 °C), atmospheric gas composition (anoxic CO2 vs. N2/O2), and pressure (0.7 kPa vs. ~101 kPa); these represent a comparison of simulated Mars conditions vs. Earth (laboratory) conditions. Comparison of Condition A vs. D revealed a total of 493 transcripts altered, ~10.1% of the protein-coding transcriptome, with 193 and 300 transcripts being up- or down-regulated, respectively (Table 2). An Excel file listing the differentially expressed transcripts found in Condition A vs. D is presented in Supplemental Table S4.
We were surprised to find that Condition A (Mars simulation), which we presumed to be the most drastic deviation from Condition D (Earth control), resulted in a less dramatic restructuring of the S. liquefaciens transcriptome (10.1%) than was observed with the 0 °C, CO2-enriched anoxic atmosphere in Condition B (14.5%), and which was comparable in magnitude to the 0 °C only environment in Condition C (8.7%). It appeared as if the lowered pressure of Condition A was somehow ameliorating the dramatic transcriptomic response seen in Condition B. Similar effects among these conditions have been observed for the growth of S. liquefaciens and Carnobacterium spp.14,15 exposed to identical Mars-like conditions as were tested here. The observation that the low-pressure environment in Condition A seemed to mitigate some of the stresses in S. liquefaciens in Condition B prompted us to more closely examine gene expression differences due exclusively to pressure in Conditions A vs. B.
Expression differences due to pressure alone (A vs. B)
Comparison of the transcriptome data from Condition A (0.7 kPa) vs. B (~101 kPa) revealed significant differential expression of 184 genes, or ~3.8% of the S. liquefaciens protein-coding genome, of which 178 were up-regulated and 6 were down-regulated under low pressure (Supplemental Table S5). Examination of the data sets failed to identify specific genes which might be reasoned to facilitate growth at low pressure. Indeed, the analysis revealed that among the most strongly up-regulated genes were those involved in transport and utilization of various sugars (lactose, arabinose, maltose, galactose, or general α-glucosides) or the polyol m-inositol. It should be noted that while TSA medium contains glucose at 0.25% final concentration, none of these other sugars are present in the medium. Of all the environmental parameters tested in our experiments, only low pressure is not encountered in nature on Earth, suggesting that up-regulation of this plethora of carbohydrate utilization systems at 0.7 kPa is likely maladaptive in S. liquefaciens. This may partly explain why S. liquefaciens grew so slowly and to such a low cell density when cultivated under Condition A (Table 1). Examination of the down-regulated genes showed that the most strongly affected were involved in transport of sulfate or the sulfur-containing amino acid cystine (Supplemental Table S5). Again, the possible relevance of the lowered expression of these genes to growth at low pressure would be speculative at best.
Principal component analysis
Because the datasets generated from our RNA-seq experiments consisted of hundreds of genes, we turned to Principal Component Analysis (PCA) to assist us in assessing (i) the reproducibility of the replicates within a Condition, and (ii) whether different treatments result in different groupings. PCA was performed on the 12 datasets, and 4 distinct population clusters were identified which coincided with the 4 environmental conditions tested (Fig. 3). In the PCA, the first and second principal components explained 37% and 24% of the variance, respectively. In each population cluster, the three replicate samples grouped rather tightly, indicating good agreement among replicates for each condition. From inspection of the PCA plot it could be seen that Conditions A, B, and C differed markedly from Condition D, the laboratory control. However, Conditions A and B, which differed in pressure (0.7 kPa vs. ~101 kPa, respectively) clustered rather close to one another, indicating a relatively low degree of gene expression difference (Fig. 3).
PCA plot of the data. Depicted are triplicate samples from Condition (A) (red) (B) (blue), (C) (green), and (D) (black). See Table 1 for description of the conditions.
Assignment of KEGG categories
Genes whose transcripts were identified as significantly up- or down-regulated were further categorized and assigned to pathways as defined by the Kyoto Encyclopedia of Genes and Genomes (KEGG)30. Up- and down-regulated transcripts are presented in Figs 4 and 5 respectively. Visual inspection of the data revealed that genes up-regulated under all Conditions A, B, and C included: transporters (ABC and PTS transporters); genes involved in translation (ribosomes and their biogenesis, biosynthesis of both tRNAs and aminoacyl-tRNAs); DNA repair and recombination; and non-coding RNAs (Fig. 4). Genes down-regulated under all Conditions A, B, and C included: transporters (mostly ABC transporters); flagellar and motility proteins; genes involved in phenylalanine metabolism; transcription factors; and two-component systems (Fig. 5). Visual examination of the KEGG pathway data failed to identify specific functions that might be postulated to be responsible for the ability of S. liquefaciens to grow, or not, under a particular Condition.
Up-regulated genes sorted by KEGG orthology. The number of genes of a particular KEGG pathway are depicted as a fraction of the total number of genes classified in that pathway in the genome of S. liquefaciens. See Table 1 for description of the conditions.
Down-regulated genes sorted by KEGG orthology. The number of genes of a particular KEGG pathway are depicted as a fraction of the total number of genes classified in that pathway in the genome of S. liquefaciens. See Table 1 for description of the conditions.
Adaptation of microbes to pressure changes
Of the environmental conditions used in this study, low temperature and anaerobic atmosphere can be found on Earth; only low pressure is a parameter unique to Mars, and the present study is the first to measure the transcriptomic response of a bacterium to low pressure comparable to that prevailing at the martian surface (0.7 kPa). To date, only one prior study has been published in which the transcriptomic response of a microorganism to low pressure exposure has been measured, that of the Gram-positive bacterium Bacillus subtilis grown at 5 kPa and 27 °C in Earth-normal 80% nitrogen/20% oxygen atmosphere31. In that study, exposure to low pressure led to up-regulation of transcripts from several global regulons (AbrB, CcpA, CodY, Fur, IolR, ResD, Rok, SigH, and Spo0A). Notably, the highest number of up-regulated genes, 86, belonged to the SigB-mediated General Stress Response (GSR) regulon31. In Gram-negative bacteria, the GSR is controlled by RNA polymerase containing the sigma factor RpoS32. In the present study, we did not observe significant up-regulation of expression of rpoS or RpoS-dependent GSR genes by S. liquefaciens exposed to simulated Mars conditions.
On the other end of the pressure spectrum, recent studies have reported the global transcriptional responses of piezophilic microorganisms to changes in pressure33,34,35. The closely related piezophiles Desulfovibrio hydrothermalis and D. piezophilus each grow at an optimum pressure of 10 MPa. Growth of the organisms at 0.1, 10, and 26 MPa resulted in alterations in expression of genes involved in lactate oxidation, energy metabolism, and amino acid biosynthesis33,35, but very few homologous genes were found to be differentially expressed in both organisms. Likewise, a study comparing the transcriptomic response of two closely related species, Thermococcus barophilus and T. kodakarensis, to changes in hydrostatic pressure failed to identify transcriptional responses in common34.
The previous studies cited, as well as the present study, were conducted with the intent of searching for new insights into the molecular mechanisms responsible for microbial adaptation to alterations in their pressure environment. In all cases, the sets of genes observed to be altered by pressure appeared to be organism-specific; no shared “pressure-responsive” genes or pathways have been discovered in the studies cited above. This finding parallels numerous studies which have tested explicitly for effects on the transcriptome of 2 or more environmental stressors acting simultaneously. Such studies have uncovered complex effects on gene expression that would not have been predicted from single stressor treatments. Furthermore, exposure to extreme environments can provoke maladaptive responses36. Because S. liquefaciens, like all other Earth organisms, has never been exposed to a low pressure of 0.7 kPa during its entire evolutionary history, it is not surprising that exposure to low pressure (Condition A vs. B), resulted in the up-regulation of a number of genes for utilization of carbohydrates not present in the medium, suggesting a maladaptive response to low pressure. What was surprising was that growth of S. liquefaciens at low pressure was not the result of a major rearrangement in its transcriptome, as visualized by the PCA plot (Fig. 3). This observation highlights the point that multiple interacting variables affect the transcriptome in ways that are neither intuitive nor predictable.
Relevance to the potential growth of S. liquefaciens on Mars
The Martian surface is bathed in numerous biocidal factors among which UV irradiation, low water activity, low pressure, low temperature, and oxidizing conditions predominate5,17. Despite these harsh conditions, a few studies have demonstrated that as many as 31 bacterial species, including S. liquefaciens, are capable of active metabolism and growth under 0.7 kPa, 0 °C, and CO2-enriched anoxic conditions if cells are incubated in stable UV-protected, hydrated, and nutrient-rich environments15,16,17. The current study is the first to characterize up- and down-regulated transcripts at 0.7 kPa in a bacterium that may be plausibly transported to Mars (Serratia spp. have been recovered from spacecraft surfaces prior to launch26,27); thus our results begin to reveal how at least one terrestrial hypobarophile might be able to grow on Mars. The key for its success will be whether relevant metabolic pathways can remain active under 0.7 kPa, 0 °C, and CO2-dominated anoxic atmosphere, and whether cells can be dispersed into hydrated niches containing usable nutrients.
Although organic compounds have been directly detected37 or inferred38 in Martian regolith, the exact composition of the in situ organics remains unsolved because of the degradation of the organics by perchlorate salts during thermal pyrolysis of surface fines38,39. However, it is now accepted that in situ organics persist in the shallow subsurface on Mars. Furthermore, approx. 2.4 × 105 kg of meteoritic carbon is accreted annually to Mars in the form of small carbonaceous chondrites and interplanetary dust particles (IDPs)40. The composition of carbonaceous chondrites41 and IDPs42 have been extensively studied, and a comparison between accreted organics to Mars and the up-regulated pathways for S. liquefaciens described here for 0.7 kPa Mars simulations suggest that at least some common metabolic pathways may remain active on the surface of Mars in which accreted organics might be accessible. For example, KEGG analyses indicated that several amino acid, purine, and sugar carbohydrate pathways were up-regulated at 0.7 kPa (Fig. 4), suggesting that these metabolic pathways might remain active on Mars, and that cells of S. liquefaciens might be able to utilize similar classes of organics (i.e., aldehydes, amino acids, ketones, and purines) found in IDPs and carbonaceous chondrites41,42,43. Our results are consistent with the hypothesis that some terrestrial bacteria are capable of metabolism and growth in stable UV-protected and hydrated niches on Mars, and that accreted or in situ organics may provide the required nutrients for heterotrophic metabolism. However, more research is required to examine the malleability of the transcriptome of S. liquefaciens and other hypobarophiles to fluctuating conditions on the Martian surface.
Finally, the habitability of any martian landing site will be a key consideration for future robotic and crewed spacecraft in which either life-detection experiments will be conducted or human exploration traverses completed. If: (i) hypobarophilic microorganisms are present on spacecraft or payload surfaces prior to launch; (ii) landing sites are near terrains that might harbor an extant Mars microbiota or support the growth of terrestrial microbes (i.e., defined as Special Regions5); and (iii) metabolic pathways remain active at 0.7 kPa that can utilize in situ or accreted organics on Mars, then the risk of the forward contamination of such sites will remain high.
Materials and Methods
Bacterial strain and medium
The bacterial strain used in this study was Serratia liquefaciens strain ATCC 27592 obtained from the American Type Culture Collection, Manassas, VA USA. Its complete genome sequence has been determined28 and is deposited in the National Center for Biotechnology Information (NCBI) GenBank database under accession number CP006252.1. Medium used throughout was Tryptic Soy Agar (TSA) (BD Difco, Franklin Lakes, NJ USA).
Growth conditions
The experimental conditions used are described in Table 1. In brief, simulated Mars conditions (Condition A) were chosen based on the near-ubiquitous extent of low pressure between 0.7 and 1.0 kPa at the surface44,45, the nominal CO2-dominated (96%) atmosphere46, and the requirement to maintain a stable liquid environment near the triple-point of water at 0 °C and 0.7 kPa47. The control conditions B, C, and D were chosen in order to discriminate the effects of pressure, gas composition, and temperature, respectively, compared to the Martian conditions tested. Although numerous other environmental and geochemical conditions might be present at the surface of Mars (e.g., diel temperature swings, low water activity, UV irradiation, high salt or perchlorate concentrations, oligotrophic nutrient regimes in the regolith) the first-order Mars simulations used here were chosen to create a stable hydrated and nutrient-rich environment in order to characterize transcriptomic changes in S. liquefaciens under a core set of three common environmental conditions on the Martian surface. The simulated Martian conditions used are consistent with models that suggests stable liquid water may occur on present-day Mars47,48.
Cultures were prepared in triplicate. For each replicate at each growth condition, the number of TSA plates indicated in Table 1 was inoculated with cells from a freshly-prepared culture grown overnight in a laboratory incubator at 30 °C. Cultures were incubated in a desiccator/low-pressure control system as described previously16,17. Briefly, TSA plates seeded with S. liquefaciens cells were placed into a 4-L polycarbonate desiccator (model 08-642-7, Fisher Scientific, Pittsburg, PA, USA), surrounded by four anaerobic pouches (Remel Anaerobic Pack sachets, Fisher Scientific), the desiccator top attached to the desiccator body, and the lab atmosphere flushed with filter-sterilized, ultra-high purity CO2 gas for 3–4 min, resulting in an atmosphere of essentially 100% CO2. The desiccator was sealed, connected to an externally mounted pump and low-pressure control system (model PU-842, KNF Neuberger, Trenton, NJ, USA), and placed within a microbial incubator set at 0 °C.
The equilibration process to stabilize the cultures at low-pressure conditions without boiling the media required approximately 60 min. The desiccators were pumped down to 10, 5, 2.5, and 0.7 kPa sequentially for approx. 15 min per time-step. The cumulative 60-min equilibration time allowed the cultures to outgas laboratory air or dissolved CO2 while also simultaneously cooling the media slowly to 0 °C. At 7-day intervals, the desiccators were vented to lab air and exhausted anaerobic pouches were replaced with fresh ones. Anaerobic pouches similar to those described above have been shown to remove O2 to a concentration of ≤0.1% within 1 h in small closed containers49; this concentration closely matches the concentration of O2 in the martian atmosphere (0.145%)46.
After incubation, cells were removed from agar surfaces by rubbing with a sterile glass rod and suspended in sterile, ice-cold PBS buffer50. Cell suspensions were immediately centrifuged (7,000 × g, 0 °C, 20 min) and the cell pellets stored at −70 °C until use.
Isolation of total RNA
Total RNA was extracted from cell pellets and treated with RNase-free DNase I using the RiboPureTM RNA Purification Kit (Thermo Fisher Scientific, Waltham, MA) following the manufacture’s protocol. Total RNA content in samples was determined using a Qubit fluorometer (Invitrogen, Thermo Fisher Scientific Inc, Waltham, MA). Sample quality was measured using the RNA 6000 Nano Chip and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The RNA Integrity Number (RIN)51 values of the samples are presented in Table 1.
RNA-seq and data analysis
RNA samples were shipped on dry ice to the Hudson Alpha Institute for Biotechnology, (Huntsville, AL) where they were subjected to ribosomal RNA reduction, library preparation, and multiplex 50-nucleotide single-end sequencing on an Illumina HiSeq2500 instrument. The raw Illumina sequences were imported into the University of Florida’s High Performance Research Computer HiPerGator 2 platform as.fastq files for further processing. Low-quality base calls were trimmed from the sequences using fastx-trimmer version 0.0.14 with a quality threshold of 33. Trimmed reads were mapped to the S. liquefaciens strain ATCC 27592 genome (NCBI RefSeq NC_021741.1)28 using bowtie2 version 2.3.252, and the aligned sequence files were imported into HTSeq version 0.6.153 for read counting. All differential expression analyses were performed in R version 3.4.2 using the package limma54,55,56. Per limma recommendation, transcripts with less than 10 total counts across all samples were removed. Gene counts were normalized using the trimmed mean of M values (TMM) method, and the normalized counts were transformed using the built-in voom method. The log2-fold change values for each transcript were determined using limma’s default eBayes method and the P values were adjusted for multiple testing using the Benjamini-Hochberg method57. Genes were considered to be differentially expressed if they exhibited at least a 4-fold (i.e., a log2 value >2) difference in transcript levels between conditions with a Benjamini-Hochberg adjusted P value less than 0.01. Principal Component Analysis (PCA) was performed on the normalized gene counts using the built-in R package stats, and the loading scores for the first two principal components were plotted in R. The processed read data are available in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database under accession number GSE120390.
Functional enrichment analysis was performed using BLAST2GO version 4.1.958. FASTA sequences for differentially expressed genes were retrieved from the PATRIC database59 using the KEGG locus tags associated with each gene. A BlastX-fast program was run for all FASTA sequences against the non-redundant database with an α-proteobacterial (taxa: 28211, Alphaproteobacteria) filter. Default mapping and annotation parameters were used, and Blast expectation values were set at a threshold of 1.0E-3. Annotation configurations were run with the default parameters and successfully annotated hits were mapped to Gene Ontology (GO) terms and pathways in the KEGG database30,60,61.
References
Chyba, C. F. & Hand, K. P. In Annu. Rev. Astron. Astrophys. Vol. 43 31–74. (Annual Reviews, 2005).
Cockell, C. S. et al. Habitability: A Review. Astrobiology 16, 89–117, https://doi.org/10.1089/ast.2015.1295 (2016).
Fajardo-Cavazos, P., Schuerger, A. C. & Nicholson, W. L. Testing interplanetary transfer of bacteria between Earth and Mars as a result of natural impact phenomena and human spaceflight activities. Acta Astronaut. 60, 534–540 (2007).
Nicholson, W. L., Munakata, N., Horneck, G., Melosh, H. J. & Setlow, P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64, 548–572, https://doi.org/10.1128/mmbr.64.3.548-572.2000 (2000).
Rummel, J. D. et al. A new analysis of Mars “Special Regions”: findings of the second MEPAG Special Regions Science Analysis Group (SR-SAG2). Astrobiology 14, 887–968, https://doi.org/10.1089/ast.2014.1227 (2014).
Nicholson, W. L. Ancient micronauts: interplanetary transport of microbes by cosmic impacts. Trends Microbiol. 17, 243–250, https://doi.org/10.1016/j.tim.2009.03.004 (2009).
Nicholson, W. L., Schuerger, A. C. & Race, M. S. Migrating microbes and planetary protection. Trends Microbiol. 17, 389–392, https://doi.org/10.1016/j.tim.2009.07.001 (2009).
Munteanu, A., Uivarosi, V. & Andries, A. Recent progress in understanding the molecular mechanisms of radioresistance in Deinococcus bacteria. Extremophiles 19, 707–719, https://doi.org/10.1007/s00792-015-0759-9 (2015).
Horikoshi, K. et al. Extremophiles Handbook. 608 pp. (Springer, 2011).
Meltzer, M. When Biospheres Collide: A History of NASA’s Planetary Protection Programs. 521 pp. (NASA, Washington DC, 2011).
Nicholson, W. L., Schuerger, A. C. & Setlow, P. The solar UV environment and bacterial spore UV resistance: considerations for Earth-to-Mars transport by natural processes and human spaceflight. Mutat. Res. 571, 249–264, https://doi.org/10.1016/j.mrfmmm.2004.10.012 (2005).
Moores, J. E., Smith, P. H., Tanner, R., Schuerger, A. C. & Venkateswaran, K. J. The shielding effect of small-scale martian surface geometry on ultraviolet flux. Icarus 192, 417–433 (2007).
Schuerger, A. C. & Nicholson, W. L. Interactive effects of hypobaria, low temperature, and CO2 atmospheres inhibit the growth of mesophilic Bacillus spp. under simulated martian conditions. Icarus 185, 143–152, https://doi.org/10.1016/j.icarus.2006.06.014 (2006).
Berry, B. J., Jenkins, D. G. & Schuerger, A. C. Effects of simulated Mars conditions on the survival and growth of Escherichia coli and Serratia liquefaciens. Appl Environ Microbiol 76, 2377–2386, https://doi.org/10.1128/AEM.02147-09 (2010).
Nicholson, W. L., Krivushin, K., Gilichinsky, D. & Schuerger, A. C. Growth of Carnobacterium spp. from permafrost under low pressure, temperature, and anoxic atmosphere has implications for Earth microbes on Mars. Proc Natl Acad Sci USA 110, 666–671, https://doi.org/10.1073/pnas.1209793110 (2013).
Schuerger, A. C. & Nicholson, W. L. Twenty species of hypobarophilic bacteria recovered from diverse soils exhibit growth under simulated martian conditions at 0.7 kPa. Astrobiology 16, 964–976, https://doi.org/10.1089/ast.2016.1587 (2016).
Schuerger, A. C., Ulrich, R., Berry, B. J. & Nicholson, W. L. Growth of Serratia liquefaciens under 7 mbar, 0 °C, and CO2-enriched anoxic atmospheres. Astrobiology 13, 115–131, https://doi.org/10.1089/ast.2011.0811 (2013).
Grohskopf, L. A. et al. Serratia liquefaciens bloodstream infections from contamination of epoetin alfa at a hemodialysis center. N Engl J Med 344, 1491–1497, https://doi.org/10.1056/NEJM200105173442001 (2001).
Engelhart, S. et al. Severe Serratia liquefaciens sepsis following vitamin C infusion treatment by a naturopathic practitioner. J Clin Microbiol 41, 3986–3988 (2003).
Mossad, S. B. The world’s first case of Serratia liquefaciens intravascular catheter-related suppurative thrombophlebitis and native valve endocarditis. Clin Microbiol Infect 6, 559–560 (2000).
Pinna, A., Usai, D., Sechi, L. A., Carta, A. & Zanetti, S. Detection of virulence factors in Serratia strains isolated from contact lens-associated corneal ulcers. Acta Ophthalmol 89, 382–387, https://doi.org/10.1111/j.1755-3768.2009.01689.x (2011).
Roth, V. R. et al. Transfusion-related sepsis due to Serratia liquefaciens in the United States. Transfusion 40, 931–935 (2000).
Machado, S. G. et al. Pseudomonas spp. and Serratia liquefaciens as predominant spoilers in cold raw milk. J Food Sci 80, M1842–1849, https://doi.org/10.1111/1750-3841.12957 (2015).
Ramirez-Arcos, S., Jenkins, C. & Sheffield, W. P. Bacteria can proliferate in thawed cryoprecipitate stored at room temperature for longer than 4 h. Vox Sang 112, 477–479, https://doi.org/10.1111/vox.12517 (2017).
Haq, I., Kumar, S., Kumari, V., Singh, S. K. & Raj, A. Evaluation of bioremediation potentiality of ligninolytic Serratia liquefaciens for detoxification of pulp and paper mill effluent. J Hazard Mater 305, 190–199, https://doi.org/10.1016/j.jhazmat.2015.11.046 (2016).
Ghosh, S., Osman, S., Vaishampayan, P. & Venkateswaran, K. Recurrent isolation of extremotolerant bacteria from the clean room where Phoenix spacecraft components were assembled. Astrobiology 10, 325–335, https://doi.org/10.1089/ast.2009.0396 (2010).
Moissl, C. et al. Molecular bacterial community analysis of clean rooms where spacecraft are assembled. FEMS Microbiol Ecol 61, 509–521, https://doi.org/10.1111/j.1574-6941.2007.00360.x (2007).
Nicholson, W. L. et al. Complete Genome Sequence of Serratia liquefaciens Strain ATCC 27592. Genome Announc 1, e00548–00513, https://doi.org/10.1128/genomeA.00548-13 (2013).
Markowitz, V. M. et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Research 40, D115–D122, https://doi.org/10.1093/nar/gkr1044 (2012).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 (2000).
Waters, S. M., Robles-Martínez, J. A. & Nicholson, W. L. Exposure of Bacillus subtilis to low pressure (5 kPa) induces several global regulons including the sigB-mediated General Stress Response. Appl Environ Microbiol 80, 4788–4794, https://doi.org/10.1128/AEM.00885-14 (2014).
Landini, P., Egli, T., Wolf, J. & Lacour, S. sigmaS, a major player in the response to environmental stresses in Escherichia coli: role, regulation and mechanisms of promoter recognition. Environmental Microbiology Reports 6, 1–13, https://doi.org/10.1111/1758-2229.12112 (2014).
Amrani, A. et al. Deciphering the adaptation strategies of Desulfovibrio piezophilus to hydrostatic pressure through metabolic and transcriptional analyses. Environ Microbiol Rep 8, 520–526, https://doi.org/10.1111/1758-2229.12427 (2016).
Vannier, P., Michoud, G., Oger, P., Marteinsson, V. & Jebbar, M. Genome expression of Thermococcus barophilus and Thermococcus kodakarensis in response to different hydrostatic pressure conditions. Res Microbiol 166, 717–725, https://doi.org/10.1016/j.resmic.2015.07.006 (2015).
Amrani, A. et al. Transcriptomics reveal several gene expression patterns in the piezophile Desulfovibrio hydrothermalis in response to hydrostatic pressure. PLoS One 9, e106831, https://doi.org/10.1371/journal.pone.0106831 (2014).
DeBiasse, M. B. & Kelly, M. W. Plastic and evolved responses to global change: what can we learn from comparative transcriptomics? J Hered 107, 71–81, https://doi.org/10.1093/jhered/esv073 (2016).
Ming, D. W. et al. Volatile and organic compositions of sedimentary rocks in Yellowknife Bay, Gale crater, Mars. Science 343, 1245267, https://doi.org/10.1126/science.1245267 (2014).
Navarro-Gonzalez, R., Vargas, E., de la Rosa, J., Raga, A. C. & McKay, C. P. Reanalysis of the Viking results suggests perchlorate and organics at midlatitudes on Mars. Journal of Geophysical Research-Planets 115, 11, https://doi.org/10.1029/2010je003599 (2010).
Glavin, D. P. et al. Evidence for perchlorates and the origin of chlorinated hydrocarbons detected by SAM at the Rocknest aeolian deposit in Gale Crater. Journal of Geophysical Research-Planets 118, 1955–1973, https://doi.org/10.1002/jgre.20144 (2013).
Flynn, G. J. The delivery of organic matter from asteroids and comets to the early surface of Mars. Earth, Moon & Planets 72, 469–474, https://doi.org/10.1007/bf00117551 (1996).
Sephton, M. A. Organic compounds in carbonaceous meteorites. Nat. Prod. Rep. 19, 292–311, https://doi.org/10.1039/b103775g (2002).
Sephton, M. A. & Botta, O. Recognizing life in the Solar System: guidance from meteoritic organic matter. Int. J. Astrobiol. 4, 269–276, https://doi.org/10.1017/s1473550405002806 (2005).
Clemett, S. J., Maechling, C. R., Zare, R. N., Swan, P. D. & Walker, R. M. Identification of complex aromatic molecules in individual interplanetary dust particles. Science 262, 721–725, https://doi.org/10.1126/science.262.5134.721 (1993).
Gómez-Elvira, J. et al. Curiosity’s rover environmental monitoring station: Overview of the first 100 sols. J. Geophys. Res. Planets 119, 1680–1688 (2014).
Hess, S. L., Ryan, J. A., Tillman, J. E., Henry, R. M. & Leovy, C. B. The annual cycle of pressure on Mars measured by Viking Landers 1 and 2. Geophys. Res. Letters 7, 197–200 (1980).
Mahaffy, P. R. et al. Abundance and isotopic composition of gases in the martian atmosphere from the Curiosity rover. Science 341, 263–266, https://doi.org/10.1126/science.1237966 (2013).
Haberle, R. M. et al. On the possibility of liquid water on present-day Mars. J. Geophys. Res. 106, 23317–23326 (2001).
Lobitz, B., Wood, B. L., Averner, M. M. & McKay, C. P. Use of spacecraft data to derive regions on Mars where liquid water would be stable. Proc Natl Acad Sci USA 98, 2132–2137, https://doi.org/10.1073/pnas.031581098 (2001).
Van Horn, K. G., Warren, K. & Baccaglini, E. J. Evaluation of the AnaeroPack system for growth of anaerobic bacteria. J. Clin. Microbiol. 35, 2170–2173 (1997).
Nicholson, W. L. & Setlow, P. In Molecular biological methods forBacillus. (eds C. R. Harwood & S. M. Cutting) 391–450 (J. Wiley & Sons, 1990).
Schroeder, A. et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7, 3, https://doi.org/10.1186/1471-2199-7-3 (2006).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25, https://doi.org/10.1186/gb-2009-10-3-r25 (2009).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, https://doi.org/10.1093/bioinformatics/btu638 (2015).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47, https://doi.org/10.1093/nar/gkv007 (2015).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29, https://doi.org/10.1186/gb-2014-15-2-r29 (2014).
Liu, R. et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res 43, e97, https://doi.org/10.1093/nar/gkv412 (2015).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Statist. Soc. B 57, 289–300 (1995).
Götz, S. et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36, 3420–3435, https://doi.org/10.1093/nar/gkn176 (2008).
Wattam, A. R. et al. Assembly, annotation, and comparative genomics in PATRIC, the All Bacterial Bioinformatics Resource Center. Methods Mol Biol 1704, 79–101, https://doi.org/10.1007/978-1-4939-7463-4_4 (2018).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44, D457–462, https://doi.org/10.1093/nar/gkv1070 (2016).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45, D353–D361, https://doi.org/10.1093/nar/gkw1092 (2017).
Acknowledgements
This work was funded by the National Aeronautics and Space Administration (NASA) Planetary Protection grant NNX12AJ84G.
Author information
Authors and Affiliations
Contributions
P.F.-C., W.L.N. and A.C.S. conceived and performed the experiments. K.M.M. and M.D.M. performed the bioinformatics data processing. W.L.N. performed the biological analysis. All authors participated in writing the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fajardo-Cavazos, P., Morrison, M.D., Miller, K.M. et al. Transcriptomic responses of Serratia liquefaciens cells grown under simulated Martian conditions of low temperature, low pressure, and CO2-enriched anoxic atmosphere. Sci Rep 8, 14938 (2018). https://doi.org/10.1038/s41598-018-33140-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-33140-4
Keywords
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.