Abstract
DNA oxidative damage repair is strongly involved in the pathogenesis of age-related cataract (ARC). The sequence variants of in coding region of DNA repair genes have been shown to be associated with ARC. It is known that single nucleotide polymorphisms (SNPs) in the 3′-terminal untranslated region (3′-UTR) can alter the gene expression by binding with microRNAs (miRNAs). We hypothesize that SNP(s) in miRNA binding site of certain DNA oxidative damage repair genes might associate with ARC risk. We examined 10 miRNA binding SNPs in 3′-UTR of 7 oxidative damage genes and revealed the XPC- rs2229090 C allele was associated with nuclear type of ARC (ARNC) risk in Chinese population. The individuals with the variant G allele (CG and GG) of XPC- rs2229090 had higher XPC mRNA expression compared to individuals carrying CC genotype. The in vitro assay showed that luciferase reporter gene expression can be down regulated by hsa-miR-589-5p in cells transfected with rs2229090 C allele compared to G allele. These results suggested that the C allele of XPC-2229090 increase the risk with ARNC. The mechanism underlying might be due to the stronger interation of the C allele with hsa-miR-589-5p, resulting in lower XPC expression and DNA repair capability than the individuals carring G allele in lens.
Similar content being viewed by others
Introduction
Age-related cataract (ARC) is a multifactorial disease that is the major cause of blindness worldwide1. Studies demonstrated that genes and environmental factors including aging, gender, ultraviolet rays, ionizing radiation and chemicals attribute to ARC2,3. One of the mechanisms for the above interaction is that the process triggers DNA oxidative damage and further leads to age-related diseases including ARC4.
DNA oxidative damage and inefficient of DNA repair capacity in lens epithelial cells (LECs) have been shown to be associated with ARC pathogenesis4,5,6,7. The repair of DNA damage is a pivotal mechanism to keep the homeostasis in mammalian cells. Nucleotide excision repair (NER), base excision repair (BER), double-strand break repair (DSBR) and mismatch repair (MMR)8 repair the various types DNA damage. Most oxidative DNA damage are rapidly repaired by NER and DSBR pathways9,10. NER mainly clears bulky adducts caused by chemical agents. DSBR may be rectified by either homologous or no homologous recombination pathways10. There is strong association between oxidative damage repair gene disrupted by gene variation in coding region and ARC5,11,12. However, a few studies focus on the 3′-untranslated region (3′-UTR) variation of gene.
Gene variation include copy number variations (CNVs) and single nucleotide polymorphisms (SNPs)13. In human genome, the most abundant form of DNA variation are SNPs13,14. SNP exist in any regions of DNA including intron, coding and untranslated region15.
MicroRNAs (miRNAs) are a group of noncoding RNAs, mature miRNAs contain approximately 22 nucleotides. They mainly interact with the 3′-UTR of mRNAs and restraint the gene transcript or lead to the mRNA degradation to regulate gene mRNA level16,17,18. SNPs located at miRNA binding sites (miRSNPs) can affect the base pairing between miRNA and target mRNA19, then further regulate miRNA-mediated genes mRNA level. It has been reported that SNPs in miRSNPs can adjust the expression of target genes of age-related disease, including cancer20,21, hypertension22,23, Parkinson’s disease24, Alzheimer disease25.
In our previous study, we have reported some miRSNPs in DNA repair genes such as ZNF350 is related to ARC26. In this study, we select genes involved in the two pathways for ARC association, including XPA and XPC in NER pathway and ATR, XRCC5, XRCC4, RAD55 and RAD54 in DSBR pathway10,19. In current case-control study, we selected 10 SNPs located in the 3′-UTR of these genes to testify the relationship between ARC and these SNPs. Subsequently, in vitro assays were used to reveal the function of the SNPs.
Materials and Methods
Study Participants
The study was approved by the Ethics Committee of Affiliated Hospital of Nantong University and conducted in compliance with the Declaration of Helsinki. All participants were told the purpose and obtained the Informed consent.
This nested case-control study included cases and controls from a population based epidemiologic cohort of the Jiangsu Eye Study located in Funing and Qidong counties. All participants were followed up with a series of ophthalmic evaluation including vision acuity, lens examination by a slit lamp biomicroscope after mydriasis, and ophthalmoscopic examination.
According to the opacity region of lens, the type of ARC was classified into four subtypes: cortical cataract (C), nuclear cataract (N), posterior sub capsular cataract (PSC) and mixed cataract (M)27. The Lens Opacities Classification System III (LOCSIII) was used to diagnose and grade lens opacities28. The age- and sex-matched controls who have transparent lens were also included from same communities. The criteria of our epidemiological investigation for the ARC group was LOCSIII > C2; >N2; >P2, while the control group was LOCSIII ≤ C1; ≤N1; ≤P1. The covered area of the study has a relatively stable and ethnically homogenous population. The details in the inclusion/exclusion of the case-control design were described in our previous study11. Consequently, 993 ARC patients (C = 453, N = 276, PSC = 52, M = 212) and 993 controls were included (Table 1).
To collect ocular tissue and matched veinal blood, additional 20 ARNC patients (LOCSIII > N2) and 20 age-, sex- and ethnically-matched controls from inpatients/outpatients of our hospital were recruited (Table S1). The average age is 65.8 ± 6.7 years in ARNC patients and 65.3 ± 6.4 years in controls. The ratio of sex is 0.55 in ARNC patients and 0.5 in controls. There is no difference between age and sex. The lens capsule samples were collected for measuring mRNA level and the rate of oxidative damage of LECs. The veinal blood was drawn for DNA genotyping and oxidative damage assessment of lymphocytes. All those ARNC patients’ capsule samples was harvested by phacoemulsification. The controls’ LECs from transparent lens were obtained from patients who had lens extraction during vitrectomy. We excluded the patients (both cases and controls) who had lens trauma, diabetes, uveitis glaucoma and high myopia (>6D) according our previous study4.
DNA, RNA and cDNA preparation
Genomic DNA extraction from Veinal blood was used by Qiagen Blood DNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions.
Total RNA was isolated by Trizol reagent from lens capsule samples (Invitrogen, Carlsbad, CA). Then cDNAs were performed by PrimeScript RT reagent Kit (TaKaRa, Dalian, China).
Selection of SNPs and genotyping
Haplotype-tagging SNPs located in the 3′-UTR regions of some DNA repair genes were selected by searching Han Chinese data in NCBI dbSNP (https://www.ncbi.nlm.nih.gov/snp). The SNPs with a MAF ≥ 10% were included while excluding those having strong linkage disequilibrium (LD) between adjacent variants with r2 threshold ≤0.80 (Table 2).
SNP Genotyping was performed with the TaqMan genotyping assay (Thermos fisher, Foster City, CA, USA) according to the manufacturer’s instructions, as described in our previous publications29,30.
In silico analysis
The PolymiRTS database 3.0 (http://compbio.uthsc.edu/miRSNP) and miRNA Target Detection (http://www.microrna.org/microrna/getGeneForm.do) were used to predict the candidate miRNAs which bind the selected 3′-UTR sequences. LD analysis was analyzed based on the 1000 Genomes data for the CEU population using the SNP Annotation and Proxy (SNAP) tool (http://www.broadinstitute.org/mpg/snap)31. An online software, the RNAhybrid program (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid), was employed to calculate the minimum free energy (MFE) of hybridization between miRNAs and their potential target sequences with MFE < −20 kcal/mol as the threshold were selected according previous study29,31.
Comet Assay
Comet assay, the single cell gel electrophoresis assay, is a sensitive technique to detect the DNA breaks. We measured DNA damage of LECs from the capsule samples and lymphocytes from peripheral venous blood using comet Assay kit (Trevigen, Gaithersburg, Maryland, USA) according to the manufacturer’s protocol. LECs and lymphocytes were isolated and then suspended at 1 × 104 cells/ml in PBS. The data analysis was performed with measuring the percentage of DNA in the tail of comets (%Tail DNA) and the olive tail moment (OTM) according to the method described by our previous study32,33.
Quantification of XPC mRNA expression
TaqMan gene expression assay probes (Thermos fisher) were used for XPC mRNA quantification (assay ID: Hs01104205_g1). Human GAPDH (Hs02786624_g1) was used as housekeeping gene control. Real-time PCR analysis was performed by ABI StepOne plus real-time PCR system (Applied Biosystems, Foster City, CA, USA). The fold change of genes mRNA level was calculated using 2(−ΔΔCt) algorithm.
Plasmids construct
The 3′-UTR of the XPC 2229090-C or XPC 2229090-G was cloned to pmiR-RB-REPORT™ vector (Ribobio, Guangzhou, China). The designed and synthesized primer of reporter construct (XPC-rs2229090) were as follows:
XPC -WT-f TCGAGCATGCCCAGCCCCTGGTGGTGGGGGCTTCTCTGCTGAGAAGGCAAACTGAGGC;
XPC -WT-r GGCCGCCTCAGTTTGCCTTCTCAGCAGAGAAGCCCCCACCACCAGGGGCTGGGCATGC;
XPC -MUT-f TCGAGCATGCCCAGCCCCTGGTGGTGGGGGGTTCTCTGCTGAGAAGGCAAACTGAGGC;
XPC -MUT-r GGCCGCCTCAGTTTGCCTTCTCAGCAGAGAACCCCCCACCACCAGGGGCTGGGCATGC.
The synthetic sequences of reporter construct (XPC-rs2229090) were as follows:
XPC-WT:
GGCGATCGCTCGAGCATGCCCAGCCCCTGGTGGTGGGGGCTTCTCTGCTGAGAAGGCAAACTG;
XPC-MUT:
GAGCATGCCCAGCCCCTGGTGGTGGGGGGTTCTCTGCTGAGAAGGCAAACTCGAGGCGGCCGCTGGCCGCAAT.
DNA sequencing was used to confirm the recombinant constructs.
Cells culture and transfection
SRA01/04 cell line originated from human lens epithelium was bought from Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco’s modified eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Lonza, Basel, Switzerland), 1% Penicillin-Streptomycin Solution (100 U/ml of penicillin and 0.1 mg/ml of Streptomycin) according our previous study18, in a humidified atmosphere with 5% CO2 at 37 °C. The cells were plated into 96-well plates at a density of 2 × 105 cells/well. Transfection was conducted when cells reached 60–70% confluence.
The cells only transfected with miRNA mimics or inhibitors (Ribobio) were for qRT-PCR assays and co-transfected with miRNA mimics or inhibitors reporter and plasmids were for Luciferase reporter assay. We used riboFECTTM Transfection kit (Ribobio) to transfect 100 ng/well reporter plasmids (XPC 2229090-C, XPC 2229090-G or Blank controls), 50 nmol/L hsa-miR-589-5p mimics, 100 nmol/L hsa-miR-589-5p inhibitors, or 50 nmol/L miRNA mimics controls, 100 nmol/L inhibitor controls into the cells, respectively.
Luciferase reporter assay
The cells were prepared 48 h after transfection, and 100 μl of supernatants was removed from each well for luminescence assay. A luciferase assay kit (Promega, Madison, Wisconsin) was used to measure luciferase activity. Experiments were repeated at least three times.
Statistical analysis
Statistical analyses were used by Stata software (Stata Corp, College Station, TX). The χ2 test was performed to test the association between the alleles frequencies of ARC groups and controls, and to calculate odds ratios (OR) and 95% confidence interval (CI). Hardy- Weinberg Equilibrium (HWE) of genotype distributions were also tested by the χ2 test. Bonferroni correction was conducted when positive association exist in the initial allele analysis. Various genetic model analyses were performed to characterize the association as dominant (mutant type homozygote versus wild type homozygotes and heterozygote), recessive model (heterozygotes and mutant type homozygotes versus wild type homozygotes). We only present the most significant model in the results. P < 0.05 was considered as statistically significant. The values of the Comet assay were expressed as mean ± SD. The ANOVA was used to compare the differences of the Comet assay parameters between the genotypes. P < 0.05 was considered as statistically significant. The qRT-PCR and Luciferase assay in this study were repeated at least 3 times independently. Data were presented as means ± SD. The t test was used to compare the average values of two groups.
Results
Characteristics of the participants for the association study
The participants of the study were recruited from the epidemiologic. The Table 1 showed the general demographic characteristic of the study participants. There was no statistically significant difference about age and gender between ARCs and controls (P > 0.05).
Bioinformatics selection of candidate SNPs
Ten SNPs in 3′-UTR region of seven genes were selected for genotyping. Their basic information and predicted miRNAs were listed in Table 2.
Association between SNPs and risk of ARC
Among the ten SNPs, the allele frequency of XPC-2229090 of ARC cases was significantly different from those of controls before and after multiple comparison correction (Bonferroni correction) (P < 0.0001, Pa < 0.001) (Table 3). We then further performed stratification analysis to explore the SNP involvement in subtypes of ARC. The results showed that frequency of the minor alleles of XPC-2229090 were significantly lower in the C, N and M type of ARCs than in the controls (P = 0.0372; P = 0.0001; P = 0.008) (Table 4). However, the significances of the SNPs was only present between ARNC and controls after Bonferroni correction.
The genetic model analysis found that rs2229090 were associated with the relevant types of ARC in the dominant model and the recessive model. The associations still exist after Bonferroni correction (P < 0.05) (Table 5).
The effects of rs2229090 on the mRNA levels of XPC in biopsy samples
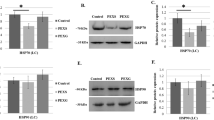
As shown in Fig. 1A, the mRNA expression of XPC was lower in LECs of ARNC group than that of the controls. Moreover, individuals carrying the minor G allele in all subjects compared with the CC genotype (CG versus CC, P < 0.05; GG versus CC, P < 0.01) (Fig. 1B).
The correlation of rs2229090 with DNA breaks and ARC risk evaluated by Comet assay
There were prominent comets indicating lots of DNA breaks in the LECs and peripheral lymphocytes of ARNCs and few in LECs and peripheral lymphocytes of the controls (Fig. 2A,B). The %Tail DNA and OTM by Comet assay in lymphocytes and LECs of ARNCs and controls are shown in Table 6. The assay showed there were much more DNA damage in ARNCs than controls (P < 0.001) (Fig. 2C). The %Tail DNA and OTM in LECs was positive correlation with those in lymphocytes (Fig. 2D). However, there no correlation of DNA breaks of peripheral lymphocytes and LECs with different genotypes of rs2229090 was found (Fig. 2E).
Representative images of comet assay: (A) controls and (B) ARNCs. (C) DNA breaks measured by Comet assay in LECs and peripheral lymphocytes, ARNCs had more DNA breaks than the controls. (D) The correlation between lymphocytes and LECs in %Tail DNA and OTM. (E) the DNA damage between different genotypes showed no difference. *P < 0. 001. Scale bars: 50 μm.
Functional analysis of the rs2229090
The SNP rs2229090 is located in a putative 3′-UTR binding site of hsa-miR-589-5p, and The C allele of rs2229090 was predicted to bind more efficiently than the G allele to the miRNA (Fig. 3A).
The effect of hsa-miR-589-5p on miRSNP rs2229090. (A) Constructs of different alleles of miRNA-binding sites. (B) MiR-589-5p mimics or inhibiters were co-transfected with the reporter constructs containing C allele or G allele into SRA01/04 cells. *P < 0.05, compared with G allele group. (C) Levels of XPC mRNA expression of the SRA01/04 cells transfected with hsa-miR-589-5p mimics or inhibitors. *P < 0.05, compared with control group.
To test whether there is an allele-specific effect of rs2229090 on XPC expression using a surrogate report gene in the presence of hsa-miR-589-5p, we transfected the miRNA mimics or inhibiters and constructed reporter plasmids (rs2229090-C and rs2229090-G) to SRA01/04 cell lines. Co-transfection with hsa-miR-589-5p mimics, the relative luciferase activity was lower in the reporter constructs containing rs2229090 C allelic than of the G allele in the cell lines. Meanwhile, co-transfection with hsa-miR-589-5p inhibitors, the relative luciferase activity was higher than the controls without hsa-miR-589-5p in the experiment using rs2229090 C allelic reporter constructs but not the G allelic reporter constructor (P > 0.05). (Fig. 3B), indicating that the interaction between hsa-miR-589-5p and the C allele of mRNA is more robust.
We further examined whether hsa-miR-589-5p could inhibit XPC expression in the cell line. We measured XPC mRNA after transfecting SRA01/04 cells (CC genotype) with has-miR-589-5p mimics and inhibitors., The XPC mRNA decreased when mimics were added as shown in Fig. 3C (P < 0.05).
Discussion
DNA oxidative damage may lead to ARC, and its timely repair can maintain the healthy status in LECs11. DNA oxidative damage caused by various factors can be repaired through the DNA damage repair enzymes. Once the function of these repair genes is in malfunction, it would be a serious problem for cells and organisms10.
Many reasons can lead to the inefficiency of DNA repair, one of the reasons is the variation of DNA repair genes34. SNP are the most abundant form of DNA variation13,14. We have proved the roles of some vital genes of DSBR and NER in ARC4,26. In this study, we selected other genes of NER and DSBR pathways10,19 to explore whether they are also related to ARC formation. In the current research, we only found XPC -rs2229090 was associated with risk of ARNC with the C allele as a risk and G allele as a protection. This SNP is located in 3′-UTR region of the gene and in the binding sequence of hsa-miR-589-5p by in silico prediction. Hsa-miR-589-5p can reduce luciferase activity in an allele-specific manner (C allele) in vitro was found by luciferase reporter assays. We also found that the expression of XPC was lower in samples carry with the C allele in ARC and control groups. The mechanism of this genetic component to ARC pathogenesis could be described as: assuming that individuals have similar level of hsa-miR-589-5p in lens tissue; those with the C allele would have stronger interaction with hsa-miR-589-5p, resulting in lower XPC expression and DNA repair capability than the individuals carrying G allele.
NER is one of the most and well-established DNA repair mechanisms in maintaining genomic stability and integrity35. XPC is a vital part of the NER pathway and plays a vital role in the early steps of global genome NER36,37, which plays vital role for removal of oxidative damage35 and regulation of the cell cycle for DNA damage response38. The SNPs in the coding region of XPC have been associated with many diseases in some studies39,40.
MiRNAs participate in regulation of genes expression through the binding the 3′-UTR of target mRNA leading to mRNA degradation or translational repression41. Our data added new evidence on the biology of SNP and miRNA interaction in the context of gene expression Indeed, GG or CG genotype of rs2229090 was associated with significantly increased mRNA levels of XPC compared with CC genotype; the rs2229090 SNP has functional consequences on miRNAs targeting. We proved that the G allele of rs2229090 altered the expression of mRNA of XPC, which were highly possibly due to changes in binding free energy with hsa-miR-589-5p. In our previous study, compared with controls, ARC patients have more DNA damage in peripheral lymphocytes and in LECs. These damage of two locales were positively correlated5. In this study, the degree of DNA damage in peripheral lymphocytes and LECs assessed by Comet assay was significantly higher in ARNCs regardless of the genotypes. But the DNA damage between different genotypes showed no difference. Therefore, we believe that DNA damage in peripheral lymphocytes and LECs is common phenomenon and not associated with different allele in ARC group.
Currently, we could not explain why rs2229090 is exclusively associated with N types of ARC. Previous reports XPC binds to a wide variety of damage such as UV-induced photoproducts42. Our previous study showed that UV-induced DNA damage lead to the formation of N types of ARC4. In current study, we found the expression of XPC with C allele is lower in N types of ARCs than that of controls. We speculate that the lower expression of XPC leads to deficiency in repairing UV-induced damage, thus increasing the risk of ARNC.
In conclusion, our study focused on the importance of SNPs located in 3′-UTR of DNA repair genes to increase understanding of ARC pathogenesis. The results suggested that miRSNP rs2229090 of XPC may influence an individual’s susceptibility to ARNC in Han Chinese population. The rs2229090 C allele of XPC gene may be possive correlation with the risk of ARNC, and the mechanism may be that the free energy of hsa-miR-589-5p binging C allele is excessive than G allele of rs222900 of XPC, further destroy the post-transcription of XPC, thus leading to ARNC. This finding provides a novel approach and potential therapeutic target for ARNC management.
Data Availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Khairallah, M. et al. Number of People Blind or Visually Impaired by Cataract Worldwide and in World Regions, 1990 to 2010. Invest Ophthalmol Vis Sci. 56, 6762–6769 (2015).
Zhou, J. et al. Thioredoxin Binding Protein-2 Regulates Autophagy of Human Lens Epithelial Cells under Oxidative Stress via Inhibition of Akt Phosphorylation. Oxid Med Cell Longev. 2016, 4856431 (2016).
Asbell, P. A. et al. Age-related cataract. Lancet. 365, 599–609 (2005).
Wang, Y., Li, F., Zhang, G. W. & Guan, H. J. Ultraviolet-B induces ERCC6 repression in lens epithelium cells of age-related nuclear cataract through coordinated DNA hypermethylation and histone deacetylation. Clin Epigenetics. 8, 62 (2016).
Liu, X. C., Liu, X. F., Hu, Z. D. & Li, Z. H. Polymorphisms of DNA repair genes XPD (Lys751Gln) and XRCC1 (Arg399Gln), and the risk of age-related cataract: a meta-analysis. Curr Eye Res. 40, 676–682 (2015).
Wang, Y., Li, F., Zhang, G. W. & Guan, H. J. Altered DNA Methylation and Expression Profiles of 8-Oxoguanine DNA Glycosylase 1 in Lens Tissue from Age-related Cataract Patients. Curr Eye Res. 40, 815–821 (2015).
Li, B., Zhou, J., Zhang, G. W. & Guan, H. J. Relationship Between the Altered Expression and Epigenetics of GSTM3 and Age-Related Cataract. Invest Ophthalmol Vis Sci. 57, 4721–4732 (2016).
Aggarwal, N. et al. The Association of Low-Penetrance Variants in DNA Repair Genes with Colorectal Cancer: A Systematic Review and Meta-Analysis. Clin Transl Gastroenterol. 8, e109 (2017).
Wilson, D. M., Sofinowski, T. M. & McNeill, D. R. Repair mechanisms for oxidative DNA damage. Front Biosci. 8, d963–981 (2003).
Wood, R. D., Mitchell, M. & Lindahl, T. Human DNA repair genes. Mutat Res. 577, 275–283 (2005).
Su, S., Yao, Y., Zhu, R. & Guan, H. J. The associations between single nucleotide polymorphisms of DNA repair genes, DNA damage, and age-related cataract: Jiangsu Eye Study. Invest Ophthalmol Vis Sci. 54, 1201–1207 (2013).
Zheng, L. R. et al. Association between DNA repair genes (XPD and XRCC1) polymorphisms and susceptibility to age-related cataract (ARC): a meta-analysis. Graefes Arch Clin Exp Ophthalmol. 252, 1259–1266 (2014).
Momtaz, R., Ghanem, N. M., El-Makky, N. M. & Ismail, M. A. Integrated Analysis of SNP, CNV and Gene Expression Data in Genetic Association Studies. Clin Genet. 93, 557–566 (2018).
Liu, M. E. et al. A functional polymorphism of PON1 interferes with microRNA binding to increase the risk of ischemic stroke and carotid atherosclerosis. Atherosclerosis. 228, 161–167 (2013).
Liang, L. et al. MicroRNA-223 enhances radiation sensitivity of U87MG cells in vitro and in vivo by targeting ataxia telangiectasia mutated. Int J Radiat Oncol Biol Phys. 88, 955–960 (2014).
Sambandan, S. et al. Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science. 355, 634–637 (2017).
Golden, R. J. et al. An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature. 542, 197–202 (2017).
Rong, H., Gu, S. S., Zhang, G. & Guan, H. J. MiR-2964a-5p binding site SNP regulates ATM expression contributing to age-related cataract risk. Oncotarget. 49, 84945–84957 (2017).
Vymetalkova, V. et al. Polymorphisms in microRNA binding sites of mucin genes as predictors of clinical outcome in colorectal cancer patients. Carcinogenesis. 38, 28–39 (2017).
Wilkins, O. M. et al. Genome-scale identification of microRNA-related SNPs associated with risk of head and neck squamous cell carcinoma. Carcinogenesis. 38, 986–993 (2017).
Assie, G. et al. Integrated genomic characterization of adrenocortical carcinoma. Nat Genet. 46, 607–612 (2014).
Huang, Y. et al. Circulating miRNA29 family expression levels in patients with essential hypertension as potential markers for left ventricular hypertrophy. Clin Exp Hypertens. 39, 119–125 (2017).
Zeng, J. Y. et al. Molecular mechanisms in microRNA-mediated TRB3 gene and hypertension left ventricular hypertrophy. Exp Ther Med. 13, 1907–1911 (2017).
Kabaria, S. et al. Inhibition of miR-34b and miR-34c enhances alpha-synuclein expression in Parkinson’s disease. FEBS Lett. 589, 319–325 (2015).
Gorucu Yilmaz, S., Erdal, M. E., Avci Ozge, A. & Sungur, M. A. SNP Variation in MicroRNA Biogenesis Pathway Genes as a New Innovation Strategy for Alzheimer Disease Diagnostics: A Study of 10 Candidate Genes in an Understudied Population From the Eastern Mediterranean. Alzheimer Dis Assoc Disord. 30, 203–209 (2016).
Gu, S. S., Rong, H., Zhang, G. W. & Guan, H. J. Functional SNP in 3′-UTR MicroRNA-Binding Site of ZNF350 Confers Risk for Age-Related Cataract. Hum Mutat. 37, 1223–1230 (2016).
Klein, B. E., Klein, R. & Linton, K. L. Prevalence of age-related lens opacities in a population. The Beaver Dam Eye Study. Ophthalmology. 99, 546–552 (1992).
Chylack, L. T. et al. The Lens Opacities Classification System III. The Longitudinal Study of Cataract Study Group. Arch Ophthalmol. 111, 831–836 (1993).
Hu, J. et al. Age-related macular degeneration-susceptibility single nucleotide polymorphisms in a han chinese control population. Ophthalmic Epidemiol. 18, 137–142 (2011).
Yu, D. et al. MicroRNA hsa-miR-29a-3p modulates CYP2C19 in human liver cells. Biochem Pharmacol. 98, 215–223 (2015).
Knox, B., Wang, Y., Rogers, L. J. & Ning, B. T. A functional SNP in the 3′-UTR of TAP2 gene interacts with microRNA hsa-miR-1270 to suppress the gene expression. Environ Mol Mutagen. 59, 134–143 (2018).
Zhang, J., Wu, J., Yang, L. & Guan, H. J. DNA damage in lens epithelial cells and peripheral lymphocytes from age-related cataract patients. Ophthalmic Res. 51, 124–128 (2014).
Wang, Y., Zhang, J. F., Wu, J. & Guan, H. Expression of DNA repair genes in lens cortex of age-related cortical cataract. Exp Mol Pathol. 102, 219–223 (2017).
Vural, P. et al. Genetic polymorphisms in DNA repair gene APE1, XRCC1 and XPD and the risk of pre-eclampsia. Eur J Obstet Gynecol Reprod Biol. 146, 160–164 (2009).
Leibeling, D., Laspe, P. & Emmert, S. Nucleotide excision repair and cancer. J Mol Histol. 37, 225–238 (2006).
Min, J. H. & Pavletich, N. P. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 449, 570–575 (2007).
Brown, K. L. et al. Binding of the human nucleotide excision repair proteins XPA and XPC/HR23B to the 5R-thymine glycol lesion and structure of the cis-(5R, 6S) thymine glycol epimer in the 5′-GTgG-3′ sequence: destabilization of two base pairs at the lesion site. Nucleic Acids Res. 38, 428–440 (2010).
Chen, Z. et al. Attenuated expression of xeroderma pigmentosum group C is associated with critical events in human bladder cancer carcinogenesis and progression. Cancer Res. 67, 4578–4785 (2007).
Qiao, B. et al. In vitro functional effects of XPC gene rare variants from bladder cancer patients. Carcinogenesis. 32, 516–521 (2011).
Campayo, M. et al. Single nucleotide polymorphisms in tobacco metabolism and DNA repair genes and prognosis in resected non-small-cell lung cancer. J Surg Res. 167, e5–12 (2011).
Kotoula, V. et al. Expression of DNA repair and replication genes in non-small cell lung cancer (NSCLC): a role for thymidylate synthetase (TYMS). BMC Cancer. 12, 342 (2012).
Trego, K. S. & Turchi, J. J. Pre-steady-state binding of damaged DNA by XPC-hHR23B reveals a kinetic mechanism for damage discrimination. Biochemistry. 45, 1961–1969 (2006).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81470616 and 81500706).
Author information
Authors and Affiliations
Contributions
X.Z. and L.H.K. performed the experiments. X.Z. and M.Y. analyzed the data. X.Z. and J.W. wrote the manuscript. H.J.G. reviewed the manuscript and interpreted the data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zou, X., Kang, L., Yang, M. et al. MicroRNA binding mediated Functional sequence variant in 3′-UTR of DNA repair Gene XPC in Age-related Cataract. Sci Rep 8, 15198 (2018). https://doi.org/10.1038/s41598-018-33071-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-33071-0
Keywords
This article is cited by
-
Circ_HIPK3 Inhibits H2O2-Induced Lens Epithelial Cell Injury in Age-Related Cataract Depending on the Regulation of miR-495-3p/HDAC4 Pathway
Biochemical Genetics (2023)
-
CELF1 Selectively Regulates Alternative Splicing of DNA Repair Genes Associated With Cataract in Human Lens Cell Line
Biochemical Genetics (2023)
-
Exosomal microRNA-222-3p increases UVB sensitivity of lens epithelium cells by suppressing MGMT
International Ophthalmology (2022)
-
microRNA-199a-5p regulates epithelial-to-mesenchymal transition in diabetic cataract by targeting SP1 gene
Molecular Medicine (2020)
-
MicroRNAs, DNA damage response and ageing
Biogerontology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.