Abstract
Several studies have been undertaken with the goal of understanding how bacterial transcriptomes respond to the human spaceflight environment. However, these experiments have been conducted using a variety of organisms, media, culture conditions, and spaceflight hardware, and to date no cross-experiment analyses have been performed to uncover possible commonalities in their responses. In this study, eight bacterial transcriptome datasets deposited in NASA’s GeneLab Data System were standardized through a common bioinformatics pipeline then subjected to meta-analysis to identify among the datasets (i) individual genes which might be significantly differentially expressed, or (ii) gene sets which might be significantly enriched. Neither analysis resulted in identification of responses shared among all datasets. Principal Component Analysis of the data revealed that most of the variation in the datasets derived from differences in the experiments themselves.
Similar content being viewed by others
Introduction
Space travel inside human-rated spacecraft exposes living organisms to a number of stressors including microgravity, ionizing radiation, vibration, and altered atmospheric composition1. During the history of human spaceflight, many studies have been performed in low-Earth orbit within the protection of Earth’s magnetic field, to understand how spaceflight factors, particularly microgravity, affect macroscopic organisms. As a result we have a fairly detailed mechanistic understanding of how humans2, animals3, and plants4 respond when exposed to microgravity. In contrast, we have a relatively poor understanding of how single-celled microorganisms respond to the microgravity environment.
In order to understand how microbes sense and respond to stresses encountered during human spaceflight, a number of experiments have been conducted inside various human space habitats (e.g. Shuttle, Mir, ISS) using model microbial systems. In early studies, various phenotypic outputs from microbes grown in space were measured, such as: growth rate and yield; virulence; biofilm formation and architecture; and resistance to antibiotics or abiotic stresses, to name but a few [reviewed in5,6,7]. In more recent years, with the advent of the genomics and post-genomics revolutions in biology, there have been efforts to understand more fundamental molecular aspects of microbial spaceflight responses, by performing global-scale “-omics” analyses of the transcriptome, proteome, or metabolome of microbes cultivated in spaceflight. Such studies have yielded insights into the molecular responses of certain individual microbes to the spaceflight environment8,9,10,11, but a common “spaceflight response” has not emerged. Collectively the data rather seem to indicate that each individual organism mounts its own unique response to the microgravity environment; to date, however, this notion has not been subjected to rigorous testing.
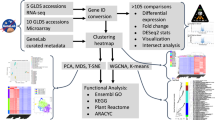
Datasets from spaceflight biology experiments have been deposited into a common repository, NASA’s GeneLab Data System (GLDS) (genelab.nasa.gov). The GLDS contains a collection of -omics data obtained from NASA-funded spaceflight and ground-based spaceflight simulation experiments. The GLDS was designed to consolidate and archive such datasets in a publicly accessible manner to facilitate the testing of novel research questions and new hypotheses, beyond those for which the spaceflight experiments were originally conducted. Often, exposure of a microorganism to a particular environmental stress (heat, cold, high salt, etc.) will provoke a stereotypic and reproducible response to that particular exposure (i.e., heat-shock, cold-shock, osmotic-shock, etc. response) which can be visualized by alterations in the organism’s global pattern of transcription, i.e., its transcriptome. We therefore sought to investigate spaceflight-induced changes in bacterial transcriptomes with an eye toward identifying commonalities in their response to spaceflight exposure. Transcriptome datasets were collected from the GLDS and analyzed using a standardized bioinformatics pipeline to detect and quantify differential gene expression. The results from each dataset were then compared to identify possible effects from spaceflight as well as spaceflight-independent effects such as those arising from different hardware configurations or growth conditions. We report here that our meta-analysis of the current transcriptome datasets deposited in the GLDS failed to find any significant commonalities in the response of diverse bacteria to the spaceflight environment.
Results
Examination of the data summarized in Table 1 revealed that the transcriptome datasets deposited in the GLDS were derived from spaceflight experiments utilizing different hardware, media, incubation times, and transcript measurement technologies. In all cases, flight (FL) cultures were compared with a set of ground control (GC) cultures incubated under matched conditions of hardware, growth medium, and temperature. Five different bacterial species were examined, 3 Gram-negative species (P. aeruginosa, S. enterica, and R. rubrum) and 2 Gram-positive species (B. subtilis and S. aureus). Two species (P. aeruginosa and S. aureus) have been flown only once, whereas 3 species (B. subtilis, R. rubrum, and S. enterica) have each been flown in space on two separate occasions (Table 1). The raw data from each experiment were extracted from the GLDS, converted into a common format, and analyzed using the bioinformatics pipeline as described in Methods.
Principal Component Analysis
Datasets for the 3 organisms flown on 2 separate occasions presented an opportunity to better understand the possible sources of variation among the datasets. We therefore performed Principal Component Analysis (PCA) and plotted the results (Fig. 1). Examination of the plots revealed some interesting features. First, the tightness of clustering of replicates within each experiment is an indication of reproducibility among the replicates. For example, in the B. subtilis data, BRIC-21 FL and GC samples clustered relatively tightly, whereas BRIC-23 FL and GC samples showed greater dispersion (Fig. 1A). The R. rubrum datasets showed reasonably tight clustering of replicates (Fig. 1B), as did the S. enterica datasets, with the exception of an outlier in the STS-123 GC replicates (Fig. 1C). Second, examination of Principal Component 1 (PC1) revealed that for all three organisms the greatest source of variation in the datasets was derived from differences in the two experimental trials (Fig. 1). Third, examination of Principal Component 2 (PC2) revealed variation between FL and GC samples within the same experiment. Examination of PC2 showed that in the BRIC-21, BRIC-23 (B. subtilis), and MESSAGE 2 (R. rubrum) datasets the FL and GC samples formed well-separated groups, indicating differences in gene expression patterns (Fig. 1A and B). In contrast, FL vs. GC samples in the BASE A (R. rubrum), STS-115 and STS-123 (S. enterica) experiments clustered closely together, indicating very little difference in gene expression patterns between FL and GC samples from these missions (Fig. 1B and C).
Differential Expression Analysis: Gram-negative species
The Gram-negative species examined included the α-proteobacterium R. rubrum, and two γ-proteobacteria, P. aeruginosa and S. enterica; the raw data for these organisms were originally derived from fluorescence microarray technology using three different platforms (GenePix, Affymetrix, and QuantArray) (Table 1). In order to make meaningful comparisons among the datasets, they were imported into a common bioinformatics pipeline as described in Methods and analyzed to discover significantly differentially expressed transcripts.
P. aeruginosa. Crabbé et al.8 cultivated P. aeruginosa strain PAO1 in Lennox broth in Fluid Processing Apparatus (FPA) hardware on the STS-115 mission. They reported the differential expression of 167 genes between FL and GC samples (52 up-regulated and 115 down-regulated in FL samples)8. After the raw datasets from that experiment were run through our bioinformatics pipeline and the resulting P values adjusted for multiple testing bias12, we found no significantly differentially expressed transcripts between FL vs. GC samples from that experiment (Table 2).
S. enterica. Wilson et al.9,10 reported on the transcriptomic response to spaceflight of S. enterica serovar Typhimurium strain χ3339 on two separate missions, STS-1159 and STS-12310. In the STS-115 experiment, cells were cultivated in liquid Lennox broth in FPA hardware, and 167 transcripts were reported to be significantly differentially expressed (69 up-regulated and 98 down-regulated in FL samples)9. Processing of the raw data from this experiment through our bioinformatics pipeline resulted in identification of fewer (104 total) differentially expressed transcripts, and most of the discrepancy resided in genes up-regulated in FL samples (6 up- and 98-down regulated in FL samples) (Table 2). In the STS-123 experiment10, cells were cultivated in liquid M9 medium in FPA hardware, and 38 total transcripts were reported to be significantly differentially expressed (14 up-regulated and 24 down-regulated in FL samples)10. However, our processing and analysis of the raw data from this experiment resulted in identification only 1 significantly up-regulated transcript in FL samples and 0 significantly down-regulated transcripts (Table 2).
R. rubrum. Mastroleo et al.11 reported on the transcriptomic response to spaceflight of R. rubrum strain S1H on two separate experiments called MESSAGE 2 and BASE A. In the MESSAGE 2 experiment, R. rubrum was cultivated on Sistrom-peptone-yeast agar in Petri plates for 8 days, and 218 total transcripts were reported to be significantly differentially expressed (191 up-regulated and 27 down-regulated in FL samples)11. Our own processing of the raw data from the MESSAGE 2 experiment resulted in close agreement; we found 220 total transcripts differentially expressed (191 up- and 29 down-regulated in FL samples) (Table 2). In the BASE A experiment, R. rubrum was cultivated on Sistrom-succinate agar in Petri plates for 12 days, and 64 total transcripts were reported to be significantly differentially expressed (53 up-regulated and 11 down-regulated in FL samples)11. Our own processing of the raw data from the BASE A experiment resulted in reasonable agreement; we identified 50 differentially expressed genes (41 up-regulated and 9 down regulated in FL samples) (Table 2).
We next compared the S. enterica STS-115 and the R. rubrum MESSAGE 2 and BASE A datasets using KEGG Orthology (KO) terms to identify differentially expressed genes in common. As depicted in the resulting Venn diagrams (Fig. 2), we failed to find any genes in common among the up-regulated (Fig. 2A) or down-regulated (Fig. 2B) transcripts from all three experiments, and in pairwise comparisons only 2 genes were found to be up-regulated in common, both from the R. rubrum MESSAGE 2 and BASE A experiments (Fig. 2A). The first of these genes belonged to KEGG Orthology K00341 and was annotated as nuoL, encoding NADH-quinone oxidoreductase subunit L, which shuttles electrons from NADH to quinones in the respiratory electron transport chain13. The second gene belonged to KEGG Orthology K02897 and was annotated as rplY encoding large subunit ribosomal protein L2514 (Table 3). In all other cases, genes were either significantly expressed but in the opposite direction or were not significantly differentially expressed (Table 3). Functional enrichment analysis of the differentially expressed genes found statistically significant enrichment of 4 up-regulated pathways from only one dataset (R. rubrum MESSAGE 2)—including butanoate metabolism, TCA cycle, oxidative phosphorylation, and ribosomes (denoted by asterisks in Fig. 3A). Among down-regulated pathways, no significantly enriched pathways were found (Fig. 3B).
Functional characterization of the differentially expressed genes identified in the datasets for Gram-negative organisms. Depicted are numbers of genes belonging to the indicated biological pathways (KEGG Orthologies) found to up-regulated (A) or down-regulated (B) in FL samples from the R. rubrum BASE A and MESSAGE 2 experiments (gray and purple bars, respectively), and the S. enterica STS-115 experiment (yellow bars) experiment. Asterisks denote pathways deemed to be significantly enriched (adjusted P < 0.05, Fisher’s exact test).
Differential Expression Analysis: Gram-positive species
Datasets for Gram-positive species included the Firmicutes B. subtilis (BRIC-21 and BRIC-23 missions) and S. aureus (BRIC-23 mission) (Table 1). It should be noted here that all three missions were performed using the same hardware (BRIC-PDFU), medium (liquid TSYG), and transcriptome mapping method (RNA-seq), thus minimizing these potential confounding factors15. The three raw datasets were run through our bioinformatics pipeline and compared to identify differentially expressed genes in common (Fig. 4). B. subtilis samples had the highest number of shared differentially expressed genes between two datasets. We identified 55 up-regulated and 36 down-regulated genes in common, resulting in a 33% concordance between the B. subtilis BRIC-21 and BRIC-23 datasets. Detailed analysis of the differentially expressed B. subtilis genes in the BRIC-21 and BRIC-23 missions will be described in detail in a separate publication (Morrison et al., submitted).
KEGG Orthology comparisons identified only one gene, KEGG Orthology K00318 annotated as proline dehydrogenase encoded by the putB gene16, as being significantly up-regulated during spaceflight in all three experiments. In pairwise comparisons, 25 S. aureus genes were found to be differentially expressed in at least one of the B. subtilis spaceflight experiments (Table 4). Eight genes were significantly down-regulated in both B. subtilis datasets, but significantly up-regulated in the S. aureus datasets, including nitrate reductase (narG, narH, narJ), nitrite reductase (nasD, nasE), arginosuccinate synthase (argG), lactate dehydrogenase (ldh) and lactate permease (lctP) (Table 4). Functional enrichment analysis of the Gram-positive datasets found significant enrichment of 3 up-regulated KEGG pathways only in B. subtilis BRIC-21 FL samples (the pathways for alanine, aspartate, and glutamate metabolism; arginine and proline metabolism; and siderophore biosynthesis) (Fig. 5A). B. subtilis BRIC-23 FL samples were significantly enriched in up-regulated chemotaxis and two-component systems, and the up-regulated KEGG pathways for biotin metabolism and non-ribosomal peptide antibiotic biosynthesis were significantly enriched in both B. subtilis BRIC-21 and BRIC-23 experiments (Fig. 5A). No up-regulated pathways were significantly enriched in the S. aureus BRIC-23 experiment (Fig. 5A). Regarding down-regulated pathways, functional enrichment analysis identified significant enrichment of ABC transporters in the B. subtilis BRIC-21 dataset, and the pathways of nitrogen metabolism and two-component systems in both the B. subtilis BRIC-21 and BRIC-23 datasets (Fig. 5B). Again, no down-regulated pathways were significantly enriched in the S. aureus BRIC-23 experiment (Fig. 5B).
Functional characterization of the differentially expressed genes identified in the datasets for Gram-positive organisms. Depicted are numbers of genes belonging to the indicated biological pathways (KEGG Orthologies) found to up-regulated (A) or down-regulated (B) in FL samples from the B. subtilis BRIC-21 (red bars) and BRIC-23 (blue bars) experiments, and the S. aureus BRIC-23 (green bars) experiment. Asterisks denote pathways deemed to be significantly enriched within the up- or down-regulated genes (adjusted P < 0.05, Fisher’s exact test).
Gene Set Enrichment Analysis
The analytical method used above (i.e., differential expression analysis of single genes) failed to uncover any commonalities in the response of all bacteria tested to the spaceflight environment. However, because these organisms belong to various taxa which have evolved divergent mechanisms for dealing with stress, it is possible that commonalities in their response to spaceflight might not be revealed by single-gene analyses. Therefore, we turned to Gene Set Enrichment Analysis (GSEA), an analytical method that derives its power by focusing on groups of genes that share common biological functions, chromosomal locations, or regulation17.
GSEA of Gram-negative datasets
GSEA results of the Gram-negative datasets are presented in Table 5 and Fig. 6. Raw microarray fluorescence images were processed and normalized using the same pipeline used for differential expression analysis as described in Methods. Normalized expression value files were converted into the required format for GenePattern and GSEA was performed using the default settings.
P. aeruginosa. Roy et al.18 previously conducted a GSEA on the STS-115 P. aeruginosa dataset. Their P. aeruginosa GSEA identified 36 enriched KEGG gene sets (8 enriched in FL and 28 enriched in GC), whereas our GSEA identified 31 enriched gene sets (6 enriched in FL and 25 enriched in GC), for a concordance rate of 60% between our results and those of Roy et al.18.
S. enterica. Roy et al.18 also performed a GSEA on the STS-115 and STS-123 S. enterica datasets. For the STS-115 experiment Roy et al. reported 19 enriched KEGG gene sets (1 enriched in FL and 18 enriched in GC samples). Our bioinformatic normalization and GSEA identified 21 enriched KEGG gene sets (7 in FL and 14 in GC), and a pair-wise comparison of the results resulted in a 38% concordance between the two GSEA results. GSEA of the STS-123 experiment by Roy et al. reported a total of 13 enriched KEGG gene sets (3 enriched in FL and 10 enriched in GC)18. However, when the STS-123 data was processed using our pipeline, GSEA found no enriched KEGG gene sets in FL or GC samples (Table 5).
R. rubrum. GSEA of the MESSAGE 2 experiment identified 13 enriched KEGG gene sets (12 enriched in FL and 1 enriched in GC) (Table 5 and Fig. 6). GSEA of the BASE A experiments identified only 1 enriched KEGG gene set in the FL samples and no enriched gene sets in the GC samples (Table 5). Pairwise comparisons of the two R. rubrum experiments found that the KEGG gene set “protein export” was enriched in both MESSAGE 2 and BASE A experiments (Table 5), suggesting a possible organism-specific response.
Cross-comparison of Gram-negative datasets revealed no gene sets that were enriched in all datasets (Fig. 6). Pairwise comparisons of the datasets found 9 KEGG enriched gene sets (1 in FL and 8 in GC) (Table 5) in the STS-115 P. aeruginosa and S. enterica experiments (Fig. 6). Pairwise comparisons of the two STS-115 experiments and the two R. rubrum experiments did not uncover any common enriched gene sets in FL (Fig. 6A) or GC (Fig. 6B) samples.
GSEA of Gram-positive datasets
GSEA results of the Gram-positive datasets are presented in Table 5 and Fig. 7. As noted with the Gram-negative datasets, there were no KEGG gene sets that were enriched in all three Gram-positive datasets (Fig. 7). Pair-wise comparisons between the datasets identified a total of 11 shared gene sets (8 enriched in FL and 3 enriched in GC). Seven of these gene sets (5 in FL and 2 in GC) were enriched in both B. subtilis datasets resulting in a 27% concordance rate between the two B. subtilis experiments (Fig. 7). Two gene sets, “arginine biosynthesis” and “quorum sensing”, were enriched in BRIC-21 B. subtilis and BRIC-23 S. aureus FL samples (Table 5), and two gene sets, “TCA cycle” in FL and “pyrimidine metabolism” in GC, were enriched in the BRIC-23 B. subtilis and S. aureus datasets (Table 5).
Discussion
In order to determine the effect that cultivation in the human spaceflight environment has on global gene expression in bacteria, several transcriptome studies have been performed, and have reported that spaceflight alters the transcript levels of genes involved in primary and secondary metabolism, ribosomal proteins, and virulence factors8,9,10,11,15. For the most part, these studies were performed independently with little or no direct comparison across different experiments. In this communication we describe the first meta-analysis comparing all publicly available transcriptome profiles from bacteria exposed to the human spaceflight environment. Using a standardized bioinformatics pipeline, the datasets were normalized and analyzed both for differentially expressed single genes and enriched gene sets. Neither single-gene analysis nor GSEA uncovered any genes or gene sets that were significantly differentially expressed across all datasets examined, a finding which does not support the notion of a shared bacterial “spaceflight response” at the level of the transcriptome.
There are several potential reasons why the meta-analysis described here did not uncover a common spaceflight response. First may be due to the disparate collection of bacteria tested, consisting of both Gram-negative and Gram-positive species. It has been well-documented that microbes belonging to widely dispersed taxonomic groups can respond to the same environmental stress using partially overlapping and partially distinct mechanisms19,20,21. A second reason may derive from the diverse experimental setups used; the eight datasets were derived from experiments using 5 different types of growth media (2 semisolid and 3 liquid) and 4 different types of spaceflight hardware (Table 1). Such variation could be visualized by PCA of different datasets utilizing the same organisms (Fig. 1). The highest percentage of explained variance between datasets (PC1) indicated that differences in cultivation conditions (e.g., media, hardware, incubation time) exerted a greater effect on global gene expression than differences in FL vs. GC samples (PC2) (Fig. 1). In most bacterial studies conducted in space, experimenters chose growth conditions and medium according to the historical precedent of the particular bacterium being used. While this is a perfectly valid justification when designing a single experiment, such confounding factors make it difficult to separate spaceflight effects from procedural, hardware, or media-specific effects. Third, the Gram-negative datasets were obtained several years ago using traditional fluorescence microarrays, necessitating conversion of the data into a format compatible with modern RNA-seq technology for statistical comparison. When statistical correction for multiple testing bias was applied, 2 of the 5 Gram-negative datasets (STS-123 S. enterica and STS-115 P. aeruginosa) demonstrated essentially no statistically significant difference in their transcriptome patterns in FL vs. GC samples. Fourth, the final phenotype that a bacterium would exhibit is the product of its physiological response to the spaceflight environment. Measuring the transcriptome captures only one aspect of physiology; it does not take into account the numerous post-transcriptional processes (translation, protein processing and modification, metabolic regulation of enzyme activity, assembly of supramacromolecular structures, etc.) which must take place in order for a microbe to manifest its final phenotype.
A slightly more consistent situation was encountered in the Gram-positive datasets used in this study, as these 3 experiments were designed and executed using the same liquid medium (TSYG) and hardware (BRIC-PDFU) (Table 1). However, a notable difference in the 3 experiments were the times of incubation, hence growth phase at which cells were frozen for subsequent RNA extraction. According to pre-flight ground validation experiments15, BRIC-21 B. subtilis samples were grown to late exponential phase, whereas BRIC-23 B. subtilis and S. aureus samples were more likely experiencing the transition from exponential to stationary phase15. By PCA, this difference was revealed in PC1 between the BRIC-21 and BRIC-23 B. subtilis samples (Fig. 1A). Cross-comparison of the B. subtilis and S. aureus datasets found only one gene, proline dehydrogenase, up-regulated in all three experiments, and there were no significantly enriched gene sets common among all 3 datasets (Table 5, Fig. 7). In fact, we noticed that several genes associated with growth under oxygen limitation (encoding nitrate/nitrite reductases and lactate permease) were down-regulated in B. subtilis FL samples but up-regulated in S. aureus FL samples (Table 4). Oxygen availability should have been consistent across all samples, due to their being cultivated in the same medium (TSYG) and hardware (BRIC-PDFUs); thus it is possible that these differences may be pointing towards organism-specific responses to the spaceflight environment.
Interestingly, we noted that the B. subtilis BRIC-21 and BRIC-23 datasets exhibited the highest concordance (~33%) in shared differentially expressed transcripts of any of the datasets examined (Fig. 4). Of all the datasets examined, these two were generated under the closest approximation of identical conditions. This observation reinforces the notion that a necessary first step towards distinguishing true spaceflight effects from experimental noise is the strict control and replication of experimental conditions. In order to develop an accurate understanding of how bacteria respond and adapt to the human spaceflight environment, we suggest that future spaceflight experiments should attempt to utilize standardized experimental conditions, and to perform flight experiments on at least two independent missions15. This would greatly reduce experimental noise and, in the long run, more rigorously address the question of whether microorganisms mount common and consistent responses to spaceflight.
Methods
Dataset selection
For this study, we investigated transcriptomes from bacterial cultures grown in the human spaceflight environment and their corresponding ground control cultures. Spaceflight experiments and organisms included: MESSAGE 2 and BASE A (Rhodospirillum rubrum strain S1H)11; STS-115 (Salmonella enterica serovar Typhimurium strain χ3339 and Pseudomonas aeruginosa strain PAO1)8,9; STS-123 (S. enterica serovar Typhimurium strain χ3339)10; BRIC-21 (Bacillus subtilis strain 168)22,23; and BRIC-23 (B. subtilis strain 168 and Staphylococcus aureus strain UAMS-1)15. All of the datasets used are openly available through NASA’s GeneLab Data System (GLDS) repository (genelab.nasa.gov) and are listed in Table 1. It should be noted that some of the GLDS datasets also contained transcriptome profiles obtained from simulated microgravity (clinostat) experiments, but only samples exposed to actual spaceflight and their corresponding ground control samples were analyzed in this study. One dataset, GLDS-9524 (Escherichia coli strain ATCC 4157), was excluded from this study because transcriptome effects caused by antibiotic stress could not be distinguished from possible effects of spaceflight.
Microarray processing and normalization
All datasets were preprocessed and normalized individually. Non-Affymetrix microarray files were imported and normalized using the R package limma25. The normexp method26 with an offset of 50 was used for background correction, and expression values were normalized within and between arrays using the loess and quantile27 methods, respectively. Normalization of Affymetrix files was done using a second R package affy28. Background correction of Affymetrix files was accomplished using the Robust Multichip Average (RMA) algorithm29 and normalization of Affymetrix arrays was accomplished using the same quantile method used in limma25.
RNA-seq read alignment, quantification, and normalization
Raw Illumina RNA-seq FASTQ files were transferred directly without preprocessing from the GLDS into the University of Florida’s High-Performance Research Computing system HiPerGator v. 2.0 (https://www.rc.ufl.edu/services/hipergator/). Samples were trimmed using FASTQ Trimmer30 and the read quality for each file was determined by FASTQC31. Reads were aligned to their appropriate genomes using Bowtie232 and read alignment quality was checked using the program SAMstat33. Finally, gene count quantification was performed using HTSeq34. The trimmed mean of M-values (TMM) normalization of gene counts was performed in limma25,35, and the normalized counts were transformed using the built-in ‘voom’36 conversion. Reference genomes and annotation files used throughout the RNA-seq analysis were acquired from the National Center for Biotechnology Information Genome page (https://www.ncbi.nlm.nih.gov/genome/).
Principal Component Analysis
Normalized microarray expression values and RNA-seq gene counts derived from the same organism were combined for principal component analysis (PCA). PCA was performed on each organism individually using the built-in stat package in R37. The calculated loading scores and explained variance for the first two principal components were plotted in R to visualize sample clustering.
Differential Expression Analysis
Normalized microarray expression values and RNA-seq gene counts were run through limma to identify differentially expressed genes. For a gene to be considered differentially expressed, it had to exhibit at least a 2-fold change with a P value < 0.05 between flight (FL) and ground (GC) samples. To reduce false positive results, P values were adjusted in limma using the Benjamini-Hochberg method12. Differentially expressed genes were annotated using the NCBI database. Functional enrichment analysis of differentially expressed genes was carried out using the STRING database38.
Comparison of Differentially Expressed Genes
To compare the differentially expressed genes found in each dataset we used a method based on functional similarity. Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO) identifiers are genome annotations within the KEGG database39,40 which are assigned according to the molecular function of a gene. KO information for all differentially expressed genes was acquired from the Universal Protein Resource (UniProt)41. KO numbers shared between multiple datasets were collected for manual inspection. Common KO numbers with opposing magnitude directions (i.e. up in FL for dataset A and down in FL for dataset B) were not considered similar responses to spaceflight and were left out when discussing common responses between differing datasets.
Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA) was conducted using the multitool platform GenePattern42 following a previously published protocol comparing datasets GLDS-11 and GLDS-1518. The gene sets for all organisms used in this study were obtained from the KEGG database. Normalized microarray expression values and RNA-seq gene counts were converted into the format recommended by GenePattern, and GSEA was performed using the default settings. Gene sets with a P value < 0.05 and q-value < 0.25 were considered to be enriched. Enriched gene sets among FL and GC samples were compared across datasets to identify any similarities.
References
Thirsk, R., Kuipers, A., Mukai, C. & Williams, D. The space-flight environment: the International Space Station and beyond. CMAJ 180, 1216–1220, https://doi.org/10.1503/cmaj.081125 (2009).
Pietsch, J. et al. The effects of weightlessness on the human organism and mammalian cells. Curr. Mol. Med. 11, 350–364, https://doi.org/10.2174/156652411795976600 (2011).
Lychakov, D. V. Behavioural and functional vestibular disorders after space flight: 2. Fish, amphibians and birds. J. Evol. Biochem. Physiol. 52, 1–16, https://doi.org/10.1134/s0022093016010014 (2016).
Vandenbrink, J. P. & Kiss, J. Z. Space, the final frontier: A critical review of recent experiments performed in microgravity. Plant Science 243, 115–119, https://doi.org/10.1016/j.plantsci.2015.11.004 (2016).
Rosenzweig, J. A. et al. Spaceflight and modeled microgravity effects on microbial growth and virulence. Appl Microbiol Biotechnol 85, 885–891, https://doi.org/10.1007/s00253-009-2237-8 (2010).
Rosenzweig, J. A., Ahmed, S., Eunson, J. & Chopra, A. K. Low-shear force associated with modeled microgravity and spaceflight does not similarly impact the virulence of notable bacterial pathogens. Appl Microbiol Biotechnol https://doi.org/10.1007/s00253-014-6025-8 (2014).
Horneck, G., Klaus, D. M. & Mancinelli, R. L. Space microbiology. Microbiol Mol Biol Rev 74, 121–156, https://doi.org/10.1128/MMBR.00016-09 (2010).
Crabbé, A. et al. Transcriptional and proteomic responses of Pseudomonas aeruginosa PAO1 to spaceflight conditions involve Hfq regulation and reveal a role for oxygen. Appl Environ Microbiol 77, 1221–1230, https://doi.org/10.1128/AEM.01582-10 (2011).
Wilson, J. W. et al. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc. Natl. Acad. Sci. USA 104, 16299–16304, https://doi.org/10.1073/pnas.0707155104 (2007).
Wilson, J. W. et al. Media ion composition controls regulatory and virulence response of S almonella in spaceflight. Plos One 3, 10, https://doi.org/10.1371/journal.pone.0003923 (2008).
Mastroleo, F. et al. Experimental design and environmental parameters affect Rhodospirillum rubrum S1H response to space flight. ISME Journal 3, 1402–1419, https://doi.org/10.1038/ismej.2009.74 (2009).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Statist. Soc. B 57, 289–300 (1995).
Ito, M., Morino, M. & Krulwich, T. A. Mrp antiporters have important roles in diverse bacteria and archaea. Frontiers Microbiol. 8, 12, https://doi.org/10.3389/fmicb.2017.02325 (2017).
Uemura, Y., Isono, S. & Isono, K. Cloning, characterization, and physical location of the rplY gene which encodes ribosomal protein L25 in Escherichia coli K12. Mol. Gen. Genet. 226, 341–344, https://doi.org/10.1007/bf00273625 (1991).
Fajardo-Cavazos, P. & Nicholson, W. L. Establishing standard protocols for bacterial culture in Biological Research in Canister (BRIC) hardware. Gravitational and Space Research 4, 58–69 (2016).
Bender, H. U. et al. Functional consequences of PRODH missense mutations. Am J Hum Genet 76, 409–420, https://doi.org/10.1086/428142 (2005).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550, https://doi.org/10.1073/pnas.0506580102 (2005).
Roy, R., Shilpa, P. P. & Bagh, S. A systems biology analysis unfolds the molecular pathways and networks of two Proteobacteria in spaceflight and simulated microgravity conditions. Astrobiology 16, 677–689, https://doi.org/10.1089/ast.2015.1420 (2016).
Schumann, W. Regulation of bacterial heat shock stimulons. Cell Stress & Chaperones 21, 959–968, https://doi.org/10.1007/s12192-016-0727-z (2016).
Barria, C., Malecki, M. & Arraiano, C. M. Bacterial adaptation to cold. Microbiology-SGM 159, 2437–2443, https://doi.org/10.1099/mic.0.052209-0 (2013).
Michiels, C., Bartlett, D. H., Aertsen, A. & (eds). High-Pressure Microbiology. (ASM Press, 2008).
Morrison, M. D., Fajardo-Cavazos, P. & Nicholson, W. L. Cultivation in space flight produces minimal alterations in the susceptibility of Bacillus subtilis cells to 72 different antibiotics and growth-inhibiting compounds. Appl. Environ. Microbiol. 83, e01584–01517, https://doi.org/10.1128/AEM.01584-17 (2017).
Fajardo-Cavazos, P., Leehan, J. D. & Nicholson, W. L. Alterations in the spectrum of spontaneous rifampicin-resistance mutations in the Bacillus subtilis rpoB gene after cultivation in the human spaceflight environment. Frontiers Microbiol. 9, 192, https://doi.org/10.3389/fmicb.2018.00192 (2018).
Zea, L. et al. A molecular genetic basis explaining altered bacterial behavior in space. PLoS One 11, e0164359, https://doi.org/10.1371/journal.pone.0164359 (2016).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47, https://doi.org/10.1093/nar/gkv007 (2015).
Silver, J. D., Ritchie, M. E. & Smyth, G. K. Microarray background correction: maximum likelihood estimation for the normal-exponential convolution. Biostatistics 10, 352–363, https://doi.org/10.1093/biostatistics/kxn042 (2009).
Smyth, G. K. & Speed, T. Normalization of cDNA microarray data. Methods 31, 265–273 (2003).
Gautier, L., Cope, L., Bolstad, B. M. & Irizarry, R. A. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315, https://doi.org/10.1093/bioinformatics/btg405 (2004).
McGee, M. & Chen, Z. X. Parameter estimation for the exponential-normal convolution model for background correction of affymetrix GeneChip data. Stat. Appl. Genet. Mol. Biol. 5, 26, https://doi.org/10.2202/1544-6115.1237 (2006).
Blankenberg, D. et al. Manipulation of FASTQ data with Galaxy. Bioinformatics 26, 1783–1785, https://doi.org/10.1093/bioinformatics/btq281 (2010).
FastQC: a quality control tool for high throughput sequence data. v. 0.11.5 (Babraham Bioinformatics (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), 2010).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25, https://doi.org/10.1186/gb-2009-10-3-r25 (2009).
Lassmann, T., Hayashizaki, Y. & Daub, C. O. SAMStat: monitoring biases in next generation sequencing data. Bioinformatics 27, 130–131, https://doi.org/10.1093/bioinformatics/btq614 (2011).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, https://doi.org/10.1093/bioinformatics/btu638 (2015).
Liu, R. et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res 43, e97, https://doi.org/10.1093/nar/gkv412 (2015).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29, https://doi.org/10.1186/gb-2014-15-2-r29 (2014).
R: A language and environment for statistical computing v. 3.3.0 (R Foundation for Statistical Computing, Vienna, Austria, 2016).
Szklarczyk, D. et al. STRINGv10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452, https://doi.org/10.1093/nar/gku1003 (2015).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 (2000).
Kotera, M., Hirakawa, M., Tokimatsu, T., Goto, S. & Kanehisa, M. The KEGG databases and tools facilitating omics analysis: latest developments involving human diseases and pharmaceuticals. Methods Mol Biol 802, 19–39, https://doi.org/10.1007/978-1-61779-400-1_2 (2012).
UniProt Consortium, T. UniProt: the universal protein knowledgebase. Nucleic Acids Res 46, 2699, https://doi.org/10.1093/nar/gky092 (2018).
Reich, M. et al. GenePattern 2.0. Nat Genet 38, 500–501, https://doi.org/10.1038/ng0506-500 (2006).
Acknowledgements
We thank Patricia Fajardo-Cavazos for critical reading of the manuscript. This work was supported by grant NNX17AD51G from NASA Research Opportunities in Space Biology.
Author information
Authors and Affiliations
Contributions
M.D.M. and W.L.N. conceived the study. M.D.M. performed the bioinformatics data processing and W.L.N. performed the biological analysis. Both authors wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Morrison, M.D., Nicholson, W.L. Meta-analysis of data from spaceflight transcriptome experiments does not support the idea of a common bacterial “spaceflight response”. Sci Rep 8, 14403 (2018). https://doi.org/10.1038/s41598-018-32818-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-32818-z
Keywords
This article is cited by
-
Whole genome-scale assessment of gene fitness of Novosphingobium aromaticavorans during spaceflight
BMC Genomics (2023)
-
Microgravity and evasion of plant innate immunity by human bacterial pathogens
npj Microgravity (2023)
-
The influence of spaceflight and simulated microgravity on bacterial motility and chemotaxis
npj Microgravity (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.