Abstract
We present the first empirical treatment of the northernmost breeding dragonfly, Somatochlora sahlbergi. We sequenced populations from United States, Canada, Finland, Sweden and Norway for cytochrome oxidase I (COI) and D2 region of 28s. We found that, despite geographic barriers across its vast arctic range, S. sahlbergi is a single species. Not only does it appear to interbreed across its entire range, there also seems to be almost no variation among European and North American populations in their COI gene fragment (the barcode gene), which is usually extremely variable. We further found that characters thought to be diagnostic for the larvae of S. sahlbergi were absent in our European samples. We review and re-describe the habitat of this species based on new findings from recent field observations. Finally, we report for the first time the likely presence of this species in Japan. We hope our findings will encourage further study of this species and other under-studied insect taxa that inhabit the remote Arctic.
Similar content being viewed by others
Introduction
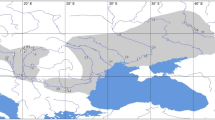
Somatochlora sahlbergi Trybom is an enigmatic dragonfly that is distributed at high latitudes across the Holarctic (see Fig. 1). First described by F. Trybom in 1889, due to its extreme habitat this species remains understudied. Adults of this species are mid-sized, dark metallic green dragonflies, much like other members of the genus Somatochlora. They have hyaline wings and a dense coat of setae on their body1,2. The shape of the cerci in the male is distinctive from other members of this genus2. The larva is dark reddish brown or orange-brown with a relatively pale and yellowish venter3. Like the adult, the larva also has a dense coat of setae. Larvae are suggested to have long lateral spines on abdominal segments 8 and 94; however, this character has not been well assessed across the geographic range of the taxon.
Somatochlora sahlbergi has the northernmost breeding range of any of the world’s approximately 6000 odonate species5,6,7. The southern limit of most of its populations in Europe and North America is at higher latitudes than the northern limits of most Holarctic dragonfly species. The species has been reported from about 71 locations across Russia, Norway, Finland, Sweden, Canada and the USA (Fig. 1, Supp. Table S1, also see8). The southernmost record in North America is at 63°N in western Alaska; the range continues east into the Yukon and Northwest Territories of Canada but as yet no population has been recorded east of the Mackenzie River3. Donnelly9 reports a single exception from 68.75°N 134.25°W (OdonataCentral.org record no. 247913), which has not been confirmed as a S. sahlbergi locale by the present authors (this location is probably a regional centroid rather than an exact locality). In Europe, the southernmost limit is 68°N, across Finland, Norway and Sweden. In Asia, this species occurs farther south than in North America and Europe, extending to 51.65°N along the Mongolian border and in the Kamchatka Peninsula of Russia. Although S. sahlbergi has relatively restricted distribution in North America and Europe, in Russia according to Belyshev10, it ranges throughout the Siberia. He recorded populations along the Yana, Indigirka, and Yenisey Rivers and farther south along the Amur and Shilka Rivers, and at Lake Baikal close to the Mongolian border. Additionally, there are populations along the rivers Ob and Lena that transect Russia from north to south.
Holarctic distribution of S. sahlbergi — locations with exact coordinates are shown in blue, while locations estimated from literature are shown in orange. The approximate location of a novel potential population of S. sahlbergi in Japan (informed by NCBI data), is shown in red. See Supp. Table S1 for record details.
S. sahlbergi is usually associated with peatlands in permafrost (specifically in Canada, Finland, Sweden and Norway), which have low mean annual temperatures and precipitation3,7,8,11. Adults fly around these peatlands where their larvae live in cold, deep3 and flowing water10 (pers. obs. MKK and WRK). In most of its North American and European range, S. sahlbergi occurs at the latitudinal treeline3, north of which trees are unable to grow due to low moisture and extreme temperature. Schröter8 suggested that S. sahlbergi is closely associated with palsa mires in the region of Fennoscandia (i.e., the Scandinavian Peninsula, Finland, Karelia, and the Kola Peninsula). In Canada and Alaska, larvae are found in deep, cold ponds surrounded by sedges and aquatic mosses and underlain with permafrost soil7. These locations are also characterized by annual mean temperatures of less than 0 °C and mean annual rainfall <450 mm7.
Although it was described 120 years ago, S. sahlbergi is still relatively rare in collections3,12,13,14,15,16 and information about its evolutionary history and ecology is poor8. It is a fascinating species with a broad and perplexing distribution in some of the planet’s most extreme habitats. In this study, we explore the population structure of this species across its range. We believe that because of its geography S. sahlbergi might be isolated into Old World and New World clades without gene flow instead of a single panmictic population. We test the hypothesis that there is distinct genetic structure among populations in North America and Eurasia. In addition, we discuss the utility of lateral spines for taxonomic identification of northern congeneric Somatochlora and speculate how the natural history of S. sahlbergi may have been influenced by Quaternary glaciation, as is the case with many other northern species.
Methods
Taxon Sampling
Specimens were collected in Scandinavia and Finland in September 2014 (Sweden, Norway, Finland, by JLW and GS) and July 2015 (Finland, by Magnus Billqvist). Samples from the Yukon Territory in Canada were collected in August 2015 (by MKK and WRK). In Scandinavia and Finland, we visited known S. sahlbergi sites, and also explored potential habitat (see Fig. 1). During the Yukon Territory sampling, we visited all the previously known localities7 (see Fig. 1). At each location, we sampled both larvae and adults and collected 28 samples in total that we identified as S. sahlbergi from Europe and Canada. Four samples were adults from the Yukon and the rest were larvae from Europe. To supplement our North American sampling, we borrowed 6 North American specimens from the Beaty Biodiversity Museum, University of British Columbia, Vancouver, bringing our specimen count to 34. Besides S. sahlbergi, we also collected Somatochlora albicincta (Burmeister), which were included in our analysis. See Supp. Table S2 for further information on the samples.
DNA isolation, gene selection and PCR amplifications
DNA was extracted from legs of the 34 S. sahlbergi samples using a Qiagen Blood and Tissue Extraction Kit (Qiagen Sciences, Germantown, MD). We followed the protocol specified by the manufacturer but extended the incubation time to 24 h. Each sample was amplified for the mitochondrial gene cytochrome oxidase I (COI) and a fragment of the nuclear ribosomal 28 S subunit, D2. We used a COI primer pair (Supp. Table S3) which amplifies approximately 516 bp fragment found 214 bp downstream of 5′ region of COI gene. COI, universally regarded as the barcode gene, is a fast-evolving gene that has been repeatedly demonstrated to vary among individuals in a population (see17,18,19,20). Therefore, this gene fragment should provide enough resolution to discern the genetic structure among the populations. The nuclear gene D2 was chosen to provide an independent dataset to compare to the mitochondrial COI. It contains five hypervariable stem and loop regions, which have been shown to be useful for distinguishing inter- and intrageneric species relationships21. The amplified PCR product was purified and sequenced by Macrogen (Macrogen USA, Rockville, MD). We successfully amplified 28 sequences for COI and 25 for D2. All sequences were deposited in the NCBI GenBank database (see Supp. Table S2 for accession numbers).
Sequence alignment
Initial alignments were performed in ClustalX22 and then manually checked for incongruencies in Mesquite23. The D2 region was largely invariant among the species in our dataset, hence secondary structural alignment was not necessary. We created two data matrices: one for each of the genes. The final COI matrix consisted of 28 samples sequenced de novo and an additional 20 samples amplified at the Environmental Protection Agency (EPA) in Cincinnati, OH (E. Pilgrim, unpublished) bringing our S. sahlbergi total to 48 (Supp. Table S2). To determine whether S. sahlbergi is a monophyletic species, we included sequences from other Somatochlora species, S. alpestris (Selys), S. arctica (Zetterstedt), S. clavata Oguma, S. dido Needham, S. elongata (Scudder), S. exuberata Bartenev, S. franklini (Selys), S. graeseri Selys, S. hudsonica (Hagen), S. metallica (Vander Linden), S. minor Calvert, S. semicircularis (Selys), S. septentrionalis (Hagen), S. uchidai Förster, S. viridiaenea (Uhler), S. whitehousei Walker, S. williamsoni Walker, and several unknown Somatochlora specimens (Supp. Table S2). These sequences were downloaded from GenBank. Helocordulia uhleri (Selys) and Cordulia amurensis Selys were used as more distant outgroups.
Our final D2 matrix included a total of 42 individuals, of which 25 were S. sahlbergi and the rest were S. albicincta (Supp. Table S2). This table includes only those specimens that were successfully sequenced (at the Ware Lab) for both genes.
To compare S. sahlbergi to other northern odonate species, we downloaded nucleotide sequences from the Barcode of Life Data System24 for dragonflies with similar ranges to S. sahlbergi in North America and Europe. We included 10 species in addition to S. albicincta and S. sahlbergi (Supp. Table S4): Aeshna canadensis Walker, A. eremita Scudder, A. interrupta Walker, A. juncea (Linnaeus), A. septentrionalis Burmeister, A. subarctica Walker, A. umbrosa Walker, Leucorrhinia glacialis Hagen, Libellula quadrimaculata Linnaeus, and Sympetrum danae (Sulzer). For each of these species, alignments were created in ClustalX and then manually investigated for ambiguities in Mesquite. All alignment files made in this study are available from the corresponding author upon request.
Phylogenetic and population analysis
We conducted a phylogenetic analysis to check the validity of S. sahlbergi as a monophyletic species within the genus Somatochlora. We based our phylogenetic reconstruction of the genus Somatochlora on COI for which we had a more exhaustive sequence sampling compared to D2. We also reconstructed the relationships among populations of S. sahlbergi using a concatenated matrix of COI and D2 data. Phylogenetic analyses for the datasets (COI and COI + D2) were done using maximum likelihood inference via iq-tree25. For the COI + D2 dataset, we partitioned the dataset by gene fragment. Substitution models for both the data sets were determined in iq-tree prior to tree reconstruction26. Bootstrap values for reconstructed trees from the two datasets were obtained using the ultrafast bootstrap approach in iq-tree, with 1000 replications27. The iq-tree analysis was run on the iq-tree web server28.
Haplotype analysis of the COI data for all species were performed in popart29 using the minimum-spanning network algorithm30. Because our preliminary tree reconstruction suggested that 3 Somatochlora alpestris sequences taken from GenBank were S. sahlbergi, we included these specimens also in our haplotype reconstruction of the latter species, giving a total of 51 samples.
We used mega (v7.0.14)31 for a comparative analysis of intraspecific genetic diversity among the 12 northern odonate species, including S. sahlbergi. We estimated the percentage of intraspecific diversity (referred to as PID here after) for each of the species using both Jukes-Cantor (JC) and Kimura-2-parameter (K2P) nucleotide evolution models.
Larval morphology
Larvae were inspected and identified under a Nikon SMZ-745T zoom stereo dissecting microscope. We counted the number of S. sahlbergi specimens with and without lateral spines on abdominal segments 8 and 9.
Results
Phylogenetic reconstruction
To test the monophyly of S. sahlbergi we reconstructed a phylogenetic tree from COI dataset, comprising a total of 19 Somatochlora species, shown in Fig. 2. Helocordulia uhleri and Cordulia amurensis were used as outgroups. Somatochlora graeseri + S. uchidai form a clade (99% bootstrap support) that is sister to the remaining Somatochlora. At the next most apical split, S. sahlbergi is supported as a valid species with a bootstrap value of 87%. It is recovered as sister to the remaining species in Somatochlora, which form several multi-species clades.
COI ML tree for genus Somatochlora — S. sahlbergi is recovered as a monophyletic group within Somatochlora. Numbers at the nodes represent the bootstrap values recovered using the ultrafast bootstrap method in IQTREE. For details on geographic sampling within S. sahlbergi see Fig. 3. Asterisks (“*”) on branches indicates bootstrap support lower than 50%.
Somatochlora albicincta, S. whitehousei, S. septrentionalis, and S. hudsonica are recovered in one clade with a bootstrap support of 99%. Somatochlora minor, S. elongata and S. williamsoni are recovered as a monophyletic clade with strong support (88%), as are S. metallica and S. exuberata (82% bootstrap support). S. franklini is also recovered as a monophyletic group with 99% support.
Interestingly, three NCBI samples, identified by accession numbers AB708910.1, AB708909.1 and AB708908.1, are labelled as “S. alpestris” on GenBank, yet were consistently recovered within the monophyletic S. sahlbergi clade (Figs 2 and 3) in our analysis. These sequences originated from Karube et al.32, who published a total of five sequences from Hokkaido, Japan, each identified as S. alpestris. We included all 5 sequences32 in our tree reconstruction: three were recovered within the S. sahlbergi clade while the other two were recovered as a separate clade with 100% bootstrap support (Fig. 2).
COI haplotype map of S. sahlbergi — different S. sahlbergi populations are colored according to their geographic location of origin. The haplotype map represents the relationship of all the populations of S. sahlbergi, distributed in four different haplotypes; H0, H1, H2 and H3 and S. albicincta. Within S. sahlbergi, 45 out of 51 samples share haplotype H0. The remaining 6 samples fall in 3 different haplotypes. The phylogenetic position of all the S. sahlbergi samples are color coded according to their geographic affinities.
For the CO1 + D2 dataset, we included S. sahlbergi and S. albicincta sequenced de novo from the Yukon and Europe. Helocordulia uhleri was used as the outgroup in this analysis. Somatochlora albicincta was recovered as a valid species with 86% bootstrap support, while S. sahlbergi was recovered as a valid species with 74% bootstrap support (Supp. Fig. S1). Within S. sahlbergi, samples were recovered in a polytomy with short branch lengths, suggesting that these sequences have only few nucleotide differences between them.
We conclude from our phylogenetic analysis that S. sahlbergi, sampled across its Holarctic range, comprises a single species from its well-supported monophyly in the tree; however, we hesitate to draw further conclusions regarding the phylogenetic relationships among other members of the genus Somatochlora. We recommend additional gene and taxon sampling among Somatochlora before drawing such conclusions.
Haplotype analysis
Results from our haplotype analysis of S. sahlbergi using the gene COI are shown in Fig. 3. These results are congruent with the phylogenetic analysis, showing very little variation among populations of S. sahlbergi. The analysis included 51 samples collected from Sweden (3), Finland (5), Norway (14), the Yukon (23), Alaska (3) and Japan (3: GenBank samples that had been recovered as S. sahlbergi in our phylogenetic reconstruction). We recovered a total of four different haplotypes, labeled H0, H1, H2, and H3. Haplotype H0 is shared by 45 out of 51 samples that were used in the analysis. These 45 samples were collected across the three continents, which leads us to conclude that S. sahlbergi do not show any signature of genetic differentiation by distance. H1 and H2 each comprise a single sample collected in the Yukon Territory. H3 contains 4 samples collected in Yukon territory. H1, H2, and H3 each differ from H0 by a single nucleotide difference.
Intraspecific diversity
Levels of intraspecific diversity were lowest in S. sahlbergi and S. albicincta (PID = 0.1%) among the 12 northern species (Fig. 4, Supp. Table S5). Aeshna juncea (PID = 1.9%), A. subarctica (PID = 2.2%), L. quadrimaculata (PID = 1.0%) and S. danae (PID = 3.6%) each had almost tenfold higher intraspecific diversity levels than S. sahlbergi. Aeshna umbrosa showed 0.7% intraspecific divergence, A. canadensis and A. interrupta showed 0.5% divergence while Leucorrhinia glacialis and A. eremita showed 0.4% and 0.3% divergence respectively. A. septentrionalis showed the second lowest sequence divergence at 0.2%. We got the same values of intraspecific diversity using KP2 and JC nucleotide evolution models.
Comparative haplotype networks of northern odonates — seven northern species compared to S. sahlbergi, which shows less variation in its COI gene than do the other species. Circles represent haplotypes and slashes over a line connecting two haplotypes represent the number of nucleotide differences between them. See Supp. Tables S4 and S5 for more details.
Larval morphology
All but three S. sahlbergi larvae lacked lateral spines on segments 8 and 9 (with: N = 3, without: N = 15). This lack of spines did not appear to be associated with larval stadium.
Discussion
Here, we present the first comprehensive phylogenetic reconstruction of Somatochlora sahlbergi and double its known localities from approximately 718 to around 140 (Supp. Table S1). Our results support three main conclusions regarding S. sahlbergi: (1) it is a single species across its entire geographic range, with low levels of intraspecific genetic diversity; (2) the presence/absence of lateral spines is not a reliable character for distinguishing its larvae from those of other Somatochlora species; and (3) its range seems to include Japan.
Somatochlora sahlbergi is a single species across its geographic range
Despite populations in North America and Europe being divided by oceans, our results suggest that S. sahlbergi populations may be panmictic across their Holarctic range. We recovered four different haplotypes among all the populations of S. sahlbergi, but most of the individuals, sampled from different continents, shared one single haplotype (Fig. 3). The additional haplotypes (H1, H2 and H3) differ from the main one (H0) by a single nucleotide difference each, but this does not indicate any significant difference within the populations because a one-nucleotide difference can be captured in a sequence just by chance (e.g., due to a sequencing error). Additionally, in comparison to other northern odonates, S. sahlbergi shows the lowest amount of intraspecific divergence (PID = 0.1%) in the COI. The only other species to show intraspecific sequence divergence of 0.1% is S. albicincta; however, this value can be expected in S. albicincta because all the samples used in the analysis were collected from a small geographic location. In fact, most of the northern species, despite having smaller geographic ranges, show more intraspecific variation across their populations in comparison to S. sahlbergi (Supp. Table S5).
Along with low intraspecific diversity, S. sahlbergi also shows low haplotypic diversity. Figure 4 shows haplotype networks for some northern species in comparison to S. sahlbergi. Aeshna juncea, for example, a circumboreal dragonfly that overlaps with S. sahlbergi in its northern range and exhibits a large haplotypic diversity, indicating much higher genetic variation than that seen in S. sahlbergi (sample size of A. juncea: 57; sample size of S. sahlbergi: 51). Similarly, Libellula quadrimaculata, which is found in both Europe and North America, also shows high haplotype diversity with a complex network connecting these haplotypes. We found that the D2 region of ribosomal 28S, known for being highly variable due to the presence of loops in its secondary structure, was extremely invariable in S. sahlbergi. Below we discuss four hypotheses that either alone or in a combination may explain the low genetic variation seen in the COI and D2 genes in S. sahlbergi.
First, gene flow might be prevalent among the North American and European populations. It would follow that individuals of S. sahlbergi are moving (likely through flight) between North America and Eurasia and mating. There are two possible directions of this movement: individuals may travel from western North America, through eastern North America (where they do not appear to live), and into Europe and Russia over the Atlantic Ocean; or they may fly the shorter distance over the Bering Sea (See Fig. 1). In either case, these insects would be dispersing for thousands of kilometers. Anax junius Drury33 and A. ephippiger (Burmeister)34 have been recorded travelling large distances and crossing vast water-bodies; however, the longest transoceanic migration ever recorded for a dragonfly (and any insect for that matter) is accomplished by Pantala flavescens (Fabricius). This dragonfly is capable of migrating a remarkable distance of 3,500 km across Indian Ocean35,36. Troast et al.17 concluded that P. flavescens is a panmictic species, with gene flow among its various populations across the globe. S. sahlbergi shows a similar trend, with little genetic variation across its global populations. However, there is a difference in the possible mode of dispersion for these dragonflies: P. flavescens is a passive flier, dependent on wind systems like intercontinental trade winds to carry it across oceans17, whereas S. sahlbergi, with its comparatively narrow wings would have to rely more on active flight, engaging in more flapping flight than gliding to stay airborne. There are examples of active flying, migratory dragonfly species (e.g., A. junius37); however, none of them are known to fly as far as P. flavescens, or over the distances that S. sahlbergi individuals would need to fly to maintain gene flow between populations. Based on a few observations of patrolling behaviour exhibited by males, S. sahlbergi are considered to be “flyers”, which spend the majority of their active time flying rather than perching3,14,38. Yet, due to the elusive nature of this species, there are no empirical records of the distance they are capable of travelling. Radio-transmitters have been successfully used to track dispersal distance and direction of A. junius39, but the smaller S. sahlbergi may not be able to support the weight of these devices. Alternatively, an isotope analysis40 of wings may help determine whether individuals are migrating across the Northern Atlantic Ocean or Bering Sea. To test this hypothesis, we are now extending our sampling to the Russian population.
Second, the time since divergence between the populations may be relatively short, indicating that populations S. sahlbergi were connected until relatively recently. Assuming this species had the same geographic range in its past as it does now, then during the last glaciation these populations would have been connected to each other through Beringia. The Beringian land bridge, which spanned the modern-day Bering Strait, disappeared relatively recently when the sea level rose around 11,500 cal BP41. Perhaps members of S. sahlbergi were able to maintain migration along that route for a while after the bridge disappeared. This hypothesis can be tested by estimating the divergence time between populations using coalescence models42,43. However, results from coalescence analysis using our current data set will not be reliable since the lack of genetic variation in our DNA sequences will lead to recovering artificially low substitution rates.
Third, long generation times in S. sahlbergi may explain their lack of genetic diversity. Molecular evolution rates in species are thought to be correlated with generation time: the smaller the generation time, the faster the rate of molecular evolution. This correlation has been found in several groups of organisms, including invertebrates44. In temperate regions, odonate life cycles become increasingly long with increasing latitude as larval growth can be arrested by a reduction in photoperiod and temperature6. Individuals of S. sahlbergi survive in places where the water is frozen for at least eight months out of the year (observations by GS). Therefore, perhaps this species takes several years to complete its life cycle. Ulf Norling studied life cycle length in two northern odonate species, Leucorrhinia dubia (Vander Linden) and Coenagrion hastulatum Charpentier, at 67°N. He found that L. dubia had an approximately 4-year development period45 while C. hastulatum took 3–4 years to develop46. Based on these observations, it is reasonable to assume that the very short summers in the north add at least one extra year to the life cycle for S. sahlbergi, suggesting 4–5 years of total larval development time, which is supported by analyses of various larval instars by Cannings and Cannings3. Thus, a relatively long generation time, combined with a relatively short time since divergence (as discussed above) might have led to very low levels of genetic variation in S. sahlbergi.
Fourth and finally, S. sahlbergi may have suffered a genetic bottleneck at some point in its evolutionary history. Genetic bottlenecks, resulting in a dramatic decrease in genetic diversity, can be caused by events such as the introduction of new pathogens or predators, or habitat loss (perhaps caused by a change in climate). Due to their proximity to glacial ice sheets, populations of S. sahlbergi have likely experienced many climatic fluctuations in their evolutionary past. During the last glacial termination (17.7–11.5 kBP), the Northern hemisphere saw periods of cold/warm oscillations as the glaciers were receding47. This was marked by an intense period (Younger Dryas: 12.9-11.7 kBP) marked by a cold and dry climate48. The relatively stable Holocene climate has also seen several intervals of rapid climate change (e.g., the Little Ice Age and Medieval Warm Period) that affected the Northern Hemisphere49. These climatic fluctuations might have affected some populations of S. sahlbergi more severely than other, perhaps more southern populations, especially in North America and Europe, leading to their local extinction. Additionally, the Holocene fluctuations after the last glaciation may have made recolonization difficult for the few remaining individuals.
Larval morphology
Lateral spines
Larvae of S. sahlbergi are described as having dense and fairly-long setae covering their abdomens, with prominent lateral spines on segments 8 and 92; however, we found that these characteristics were not always present, especially in the case of lateral spines. Out of the total 24 larvae from northern Europe that genetically group as S. sahlbergi, only 3 had lateral spines. This is in contrast to samples from Norway examined previously by Norling and Sahlén4, as well as the description of the larva from Canada3 where all larvae possessed lateral spines. Absence of these lateral spines in larvae makes S. sahlbergi difficult to distinguish from other sympatric Somatochlora species (S. albicincta in the Yukon and S. alpestris in Europe, which sometimes occupy similar habitats).
Color
All the 24 larval specimen that were collected by GS and JLW in Europe were dark brown in color (Fig. 5), unlike the orange-brown suggested by Cannings and Cannings3. The underside of these larvae were greyish/green-brown and we did not see any specimen with a yellow underside. We suspect that the reddish/orange color observed by both Cannings and Cannings3 and Valle50 might be an artefact of the preservation method rather than the original color of a freshly caught larvae. Mud or mineral deposits (for example iron) can cause larvae to appear a different color than the original.
Japan: A new location for Somatochlora sahlbergi
S. sahlbergi was described from a sample collected in Siberia51. Since then, it has been recorded around the Arctic and subarctic regions (Fig. 1). Based on our results, we propose that this species may also be present in Japan. S. sahlbergi has not been recorded in Japan before, but has been found in the Kamchatka peninsula in Russia, which is ~2,700 km from Japan. Three samples that have been identified as S. alpestris on GenBank can either be a case of misidentification or may demonstrate hybridization between S. sahlbergi and S. alpestris. S. sahlbergi has been known to hybridize with other congeneric Somatochlora species like S. hudsonica and S. albicincta3,7. Therefore, it is important to further study the Somatochlora that are found in northern Japan to gain further insight into the history of this group and of S. sahlbergi.
Habitat notes
Based on previous literature, S. sahlbergi has been considered a habitat specialist5,8, often recorded in or near a rather specialized Arctic habitat called a “palsa mire”; however, our recent observations in northwestern Canada, Sweden, Norway and Finland and records from Kamchatka indicate that S. sahlbergi may not be restricted to palsa mires7,52. We suggest instead that this species inhabits a variety of habitat types.
In Sweden, Norway and Finland, larvae were taken from small to large bodies of slowly-seeping water, which were covered in mosses and directly connected to deep, cold springs (pers. obs. by GW and JLW). In northwestern Canada, S. sahlbergi has been associated with pools and small lakes that have “aquatic moss as the dominant vegetation and deep, cold water”3.
In August 2015, MKK and WRK surveyed 70 distinct sites in the Yukon and Northwest Territories along the Dempster Highway between the Ogilvie Mountains to the south and the Mackenzie River to the northeast; these included eight localities in the Yukon Territory where S. sahlbergi had previously been spotted or collected between 1980 and 2010 (SG Cannings, pers. comm.). S. sahlbergi was found in only two of these localities, and only one those two was previously recorded by Cannings. The first S. sahlbergi site (Supp. Table S1, site code CA150812-03) was a pond, surrounded by tall trees, shrubs and sedges, with moss on its bottom. The water was 39–54 cm deep and approximately 14 °C. A single S. sahlbergi larva was collected along with several aeshnid larvae. The second site (site code CA150815-05) was a pair of connected ponds, with a shallow region of sedges between them, surrounded by trees. Three male and one female S. sahlbergi adults were collected, primarily flying over this sedge area, as well as one S. albicincta larva. It is interesting, and perhaps alarming, to note that S. sahlbergi had apparently disappeared (or at least were undetected) from seven of Cannings’ previous sites.
Given this evidence, we conclude that S. sahlbergi is perhaps not as much of a specialist as previously suggested5,8 in the literature. It is important to come to a consensus on the habitat of this species because we may not have the opportunity to study this system for long, as S. sahlbergi inhabits the edge of a climatic zone that is at immediate risk from climate change. Observations from a recent expedition to the Yukon (by MKK and WRK) and recent studies in the Scandinavian region5 suggest that S. sahlbergi might already be facing habitat loss in these locations because of temperature increase caused by global climate change53,54. Additionally, there is evidence that S. sahlbergi is facing competition from S. metallica in Eurasia, which is moving north due to global warming5. This study is a first step toward exploring the evolutionary and biogeographical history of S. sahlbergi, a single species that shows surprisingly low genetic diversity in its global distribution and is likely under stress from warming and competition.
Conclusion
S. sahlbergi is a single, panmictic species across the North American and European section of its range. Populations of S. sahlbergi show very little genetic variation in comparison to other dragonfly species from northern latitudes. It is presently unclear whether S. sahlbergi individuals are dispersing long distances and if so, whether they cross the ocean. We also conclude that lateral spines are not an identification character for S. sahlbergi compared to other congenerics. Future work should examine material from the Asian part of the geographic range of this remarkable Arctic dragonfly.
Data Availability Statement
Sequences used in the study are available on GenBank (see Supp. Table S2 for accession numbers). Alignments created during the analysis are available on request from the corresponding author.
References
Walker, E. The North American Dragonflies of the Genus Somatochlora (University of Toronto, 1925).
Needham, J. G., Westfall, M. J. & May, M. L. Dragonflies of North America (Scientific Publishers, 2000).
Cannings, S. & Cannings, R. The larva of Somatochlora sahlbergi Trybom, with notes on the species in the Yukon Territory, Canada (Anisoptera: Corduliidae). Odonatologica 14, 319–330 (1985).
Norling, U. & Sahlén, G. Odonata, dragonflies and damselflies in The aquatic insects of North Europe Vol. 2 (ed. Nilsson, A.) 13–65 (Apollo Books, 1997).
Schröter, A., Schneider, T., Schneider, E., Karjalainen, S. & Hämäläinen, M. Observations on adult Somatochlora sahlbergi- a species at risk due to regional climate change?(Odonata: Corduliidae). Libellula 31, 41–60 (2012).
Corbet, P. S. Dragonflies: behavior and ecology of Odonata. (Comstock Publishing Associates, 1999).
Cannings, S. G. & Cannings, R. A. Dragonflies (Odonata) of the Yukon in Insects of the Yukon (eds Danks, H. V. & Downes, J. A.) 169–200 (Biological Survey of Canada (Terrestrial Arthropods), 1997).
Schröter, A. Review of the distribution of Somatochlora sahlbergi (Odonata: Corduliidae). International Dragonfly Fund Report 41, 1–27 (2011).
Donnelly, T. Distribution of North American Odonata. Part II: Macromiidae. Cordullidae, and Libellulidae. Bull. Am. Odonatol. 8, 1–32 (2004).
Belyshev, B. The dragonflies of Siberia (Odonata). Vol. 1, Part 2 (Nauka, 1973).
Hilton, D. F. Odonata of peatlands and marshes in Canada. Mem. Entomol. Soc. Can. 119, 57–63 (1987).
Sahlén, G. A new site for Somatochlora sahlbergi Trybom in Inari Lapland (Odonata, Corduliidae). Notulae. Entomol. 67, 3–4 (1987).
Kosterin, O. New distribution records of Somatochlora sahlbergi Trybom (Odonata, Corduliidae). Acta Hydroentomol. Lativica 2, 22–26 (1992).
Sahlén, G. Tundratrollsländan Somatochlora sahlbergi funnen i nordligaste Sverige. Entomol. Tidskrift 115, 137–142 (1994).
Pedersen, H. Somatochlora sahlbergi Trybom. 1899 (Odonata: Corduliidae)- a new species to Norway. Fauna Norvegica Ser. B 39, 22 (1992).
Olsvik, H. & Dolmen, D. Distribution, habitat, and conservation status of threatened Odonata in Norway. Fauna Norvegica, Ser. B 39, 1–21 (1992).
Troast, D., Suhling, F., Jinguji, H., Sahlén, G. & Ware, J. A global population genetic study of Pantala flavescens. PLOS One 11, e0148949 (2016).
Lim, P.-E., Tan, J., Suana, I. W., Eamsobhana, P. & Yong, H. S. Distinct genetic lineages of Bactrocera caudata (Insecta: Tephritidae) revealed by COI and 16S DNA sequences. PLOS One 7, e37276 (2012).
Theissinger, K. et al. Glacial survival and post‐glacial recolonization of an arctic–alpine freshwater insect (Arcynopteryx dichroa, Plecoptera, Perlodidae) in Europe. J. Biogeography 40, 236–248 (2013).
Dinsdale, A., Cook, L., Riginos, C., Buckley, Y. & Barro, P. D. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann. Entomol. Soc. Amer. 103, 196–208 (2010).
Ware, J., May, M. & Kjer, K. Phylogeny of the higher Libelluloidea (Anisoptera: Odonata): an exploration of the most speciose superfamily of dragonflies. Mol. Phylogenet. Evol. 45, 289–310 (2007).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Maddison, W. P. & Maddison, D. R. Mesquite: a modular system for evolutionary analysis. Version 3.2 http://mesquiteproject.org (2017).
Ratnasingham, S. & Hebert, P. D. A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLOS One 8, e66213 (2013).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. Iq-tree: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14, 587–589 (2017).
Minh, B. Q., Nguyen, M. A. T. & von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195 (2013).
Trifinopoulos, J., Nguyen, L.-T., von Haeseler, A. & Minh, B. Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235 (2016).
Leigh, J. W. & Bryant, D. popart: full‐feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116 (2015).
Bandelt, H.-J., Forster, P. & Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Karube, H., Futahashi, R., Sasamoto, A. & Kawashima, I. Taxonomic revision of Japanese odonate species, based on nuclear and mitochondrial gene genealogies and morphological comparison with allied species. Part I. Tombo, Fukui 54, 75–106 (2012).
Corbet, P. S. The first recorded arrival of Anax junius, Drury (Anisoptera: Aeshnidae) In Europe: A Scientist’s Perspective. Int. J. Odonatol. 3, 153–162 (2000).
Ólafsson, E. Drekaflugan Hemianax ephippiger (Burm.)(Odonta), óvæntur gestur á Íslandi. Náttúrufræðingurinn 45, 209–212 (1975).
Samways, M. & Osborn, R. Divergence in a transoceanic circumtropical dragonfly on a remote island. J. Biogeography 25, 935–946 (1998).
Anderson, R. C. Do dragonflies migrate across the western Indian Ocean? J. Trop. Ecol. 25, 347–358 (2009).
May, M. L. & Matthews, J. H. Migration in Odonata: a case study of Anax junius in Dragonflies and Damselflies: Model Organisms for Ecological and Evolutionary Research (ed. Córdoba-Aguilar, A.) 63–77 (Oxford University Press, 2008).
Wildermuth, H. Die Falkenlibellen Europas. Vol. 653 (2008).
Wikelski, M. et al. Simple rules guide dragonfly migration. Biol. Letters 2, 325–329 (2006).
Hobson, K. A., Anderson, R. C., Soto, D. X. & Wassenaar, L. I. Isotopic evidence that dragonflies (Pantala flavescens) migrating through the Maldives come from the northern Indian subcontinent. PLOS One 7, e52594 (2012).
Jakobsson, M. et al. Post-glacial flooding of the Bering Land Bridge dated to 11 cal ka BP based on new geophysical and sediment records. Climate of the Past 13, 991 (2017).
Skoglund, P., Götherström, A. & Jakobsson, M. Estimation of population divergence times from non-overlapping genomic sequences: examples from dogs and wolves. Mol. Biol. Evol. 28, 1505–1517 (2010).
Ho, S. Y. & Duchêne, S. Molecular‐clock methods for estimating evolutionary rates and timescales. Mol. Ecol. 23, 5947–5965 (2014).
Thomas, J. A., Welch, J. J., Lanfear, R. & Bromham, L. A generation time effect on the rate of molecular evolution in invertebrates. Mol. Biol. Evol. 27, 1173–1180 (2010).
Norling, U. Photoperiodic control of larval development in Leucorrhinia dubia (Vander Linden): a comparison between populations from northern and southern Sweden (Anisoptera: Libellulidae). Odonatologica 13, 529–550 (1984).
Norling, U. The life cycle and larval photoperiodic responses of Coenagrion hastulatum (Charpentier) in two climatically different areas (Zygoptera: Coenagrionidae). Odonatologica 13, 429–449 (1984).
Dansgaard, W. et al. Evidence for general instability of past climate from a 250-kyr ice-core record. Nature 364, 218–220 (1993).
Benson, L., Burdett, J., Lund, S., Kashgarian, M. & Mensing, S. Nearly synchronous climate change in the Northern Hemisphere during the last glacial termination. Nature 388, 263–265 (1997).
Mayewski, P. A. et al. Holocene climate variability. Quaternary Res. 62, 243–255 (2004).
Valle, K. Materialien zur Odonatenfauna Finnlands. II. Somatochlora sahlbergi Trybom. Notulae Entomol. 9, 41–51 (1931).
Trybom, F. Trollsländor (Odonata), insamlade unter Svenska expeditionen till Jenisei 1876. Bihang till Kungliga Svenska vetenskapsakademiens handlingar 154, 1–21 (1889).
Mäkinen, J. T. (Somatochlora sahlbergi) kilpisjärvellä. Suomen sudenkorentoseuran julkaisuja – CRENATA 8, 14–17 (2015).
Fronzek, S., Luoto, M. & Carter, T. R. Potential effect of climate change on the distribution of palsa mires in subarctic Fennoscandia. Climate Res. 32, 1–12 (2006).
Luoto, M., Heikkinen, R. K. & Carter, T. R. Loss of palsa mires in Europe and biological consequences. Environ. Conserv. 31, 30–37 (2004).
Acknowledgements
We thank Robert and Sydney Cannings for all their input since the conception of this project. We would especially like to thank Sydney Cannings for his invaluable help and guidance in the organisation of the field work in northwestern Canada, extremely helpful discussions on the ecology of S. sahlbergi, and for providing many of the localities presented in Supp. Table S1. We would like to thank Erik Pilgrim for his valuable contribution of Yukon S. sahlbergi sequences and helpful input in manuscript writing. We thank Magnus Billqvist for the Finnish 2015 sample. We thank Karen Needham from the Beaty Museum at the University of British Columbia for arranging the loan of Alaska material. We thank Hans Olsvik for providing locality data for Norway. This work was supported by the Rutgers University (JLW funding), the Carl Tryggers Foundation (GS funding), and by a Postdoctoral Research Fellowship in Biology from the National Science Foundation (grant no. 1611642, WRK funding).
Author information
Authors and Affiliations
Contributions
M.K.K.: Project conception, sample collection in Yukon, data generation and analysis and wrote and revised the manuscript. G.S.: Project conception, sample collection in Finland, Sweden and Norway, revised the manuscript. W.R.K.: Sample collection in Yukon, generated distribution map, wrote and revised the manuscript. J.L.W.: Project conception, sample collection in Finland, Sweden and Norway, revised the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kohli, M.K., Sahlén, G., Kuhn, W.R. et al. Extremely low genetic diversity in a circumpolar dragonfly species, Somatochlora sahlbergi (Insecta: Odonata: Anisoptera). Sci Rep 8, 15114 (2018). https://doi.org/10.1038/s41598-018-32365-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-32365-7
Keywords
This article is cited by
-
Are subcortical rove beetles truly Holarctic? An integrative taxonomic revision of north temperate Quedionuchus (Coleoptera: Staphylinidae: Staphylininae)
Organisms Diversity & Evolution (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.