Abstract
Calorie restriction (CR) extends lifespan and elicits numerous effects beneficial to health and metabolism in various model organisms, but the underlying mechanisms are not completely understood. Gut microbiota has been reported to be associated with the beneficial effects of CR; however, it is unknown whether these effects of CR are causally mediated by gut microbiota. In this study, we employed an antibiotic-induced microbiota-depleted mouse model to investigate the functional role of gut microbiota in CR. Depletion of gut microbiota rendered mice resistant to CR-induced loss of body weight, accompanied by the increase in fat mass, the reduction in lean mass and the decline in metabolic rate. Depletion of gut microbiota led to increases in fasting blood glucose and cholesterol levels independent of CR. A few metabolism-modulating hormones including leptin and insulin were altered by CR and/or gut microbiota depletion. In addition, CR altered the composition of gut microbiota with significant increases in major probiotic genera such as Lactobacillus and Bifidobacterium, together with the decrease of Helicobacter. In addition, we performed fecal microbiota transplantation in mice fed with high-fat diet. Mice with transferred microbiota from calorie-restricted mice resisted high fat diet-induced obesity and exhibited metabolic improvement such as alleviated hepatic lipid accumulation. Collectively, these data indicate that CR-induced metabolic improvement especially in body weight reduction is mediated by intestinal microbiota to a certain extent.
Similar content being viewed by others
Introduction
Calorie restriction (CR) is a dietary regimen that reduces calorie intake without incurring malnutrition. Countless researches since the 1930s have confirmed that CR is the only effective environmental intervention that is known to extend lifespan in many organisms including yeast, worms, flies, rodents and perhaps non-human primates1,2,3,4,5,6,7. Also, CR has been reported to prevent the occurrence of metabolic syndromes such as obesity and reduce the risk factors of age-associated diseases such as cancer, diabetes and atherosclerosis in many mammals, including humans8,9,10,11,12,13. However, the underlying mechanisms remain controversial.
It is widely accepted that metabolic syndromes and many age-associated disorders are intimately linked to diet. Furthermore, extensive research on gut microbiota represents clear evidence that diet modulates the composition and function of these microbes and the diet-microbiota interactions are pivotal moderators of metabolism14,15,16,17,18,19,20,21,22. Especially, emerging studies have demonstrated that gut microbiota can alter the absorption, metabolism and storage of calories23,24,25, although the actual mechanisms are difficult to elucidate. CR, as an important way of dietary intervention, could reshape the gut microbiota. Recent studies witnessed significant dynamic changes of mice and human gut microbiota as response to calorie-restricted diet26,27, although the overall bacterial phylogenetic alteration was not substantially affected by CR in humans28. However, it remains to be determined whether the phenomenon that CR alleviates metabolic syndromes is causally mediated by gut microbiota and if so, what are the underlying mechanisms. In this study, we used an antibiotic-induced microbiota-depleted (AIMD) mouse model combined with fecal microbiota transplantation (FMT) to investigate whether or not gut microbiota is causally involved in metabolic improvement associated with calorie restriction.
Results
Antibiotic treatment extensively depletes commensal gut microbiota
Following the schematic as described in Fig. 1A, we investigated four groups of mice in the experiment. In the control group (CTRL), the mice were continuously fed normal chow ad libitum. The calorie-restricted group (CR) was fed with a 70% of normal chow based on the food intake of CTRL group. Microbiota was depleted by antibiotic treatment in mice continuously fed ad libitum (AB) or fed with a 30% calorie-restriction diet (AB + CR). We started treating mice in AB and AB + CR groups with four nonabsorbable broad-spectrum antibiotics at the onset of calorie restriction. Fecal bacterial loads were examined by cultivation of anaerobic microbes using serial dilutions of resuspended fecal pellets on brain heart infusion (BHI) agar plates. Antibiotic treatment remarkably reduced culturable gut microbes by about one million-fold (Fig. 1B). Significant reduction of community diversity and richness of gut microbiota by the antibiotics were reflected by analyses of shannon index (Fig. 1C) and sobs index (Fig. 1D) via 16S rRNA gene sequencing, further indicating the effectiveness of antibiotic treatment.
Depletion of gut microbiota after antibiotic treatment. (A) Schematic design of the experiment. AL, ad libitum; Abx, antibiotic treatment. (B) Fecal bacterial loads in all groups of mice. Experiments were repeated 3 times. (C,D) Effects of antibiotic treatment on diversity and richness of fecal microbiota revealed respectively by shannon index (C) and sobs index (D). Data are expressed as means ± s.e.m, n = 9–10 per group.
Microbiota-depleted mice are resistant to CR-induced body weight loss
Upon antibiotic treatment and calorie restriction, the control mice and the microbiota-depleted mice responded differently to calorie restriction. As expected, CR was able to reduce body weight starting from the second week of application of CR diet (Fig. 2A). Since then, the mice without gut microbiota (AB + CR) experienced less body weight loss than the CR group (Fig. 2A), although the two groups of mice had equal levels of food intake (Fig. 2B). The significant discrepancy of body weight between the two groups stayed until the end of the experiment (Fig. 2A). In addition, at the end of the experiment, AB group gained more weight than the CTRL group (Fig. 2A), accompanied by an increase in food intake in the last few weeks (Fig. 2B). These results, therefore, indicated that gut microbiota plays an important role in CR-induced loss of body weight.
Body weight and metabolic alterations upon CR and gut microbiota depletion. (A) Body weight curves of the mice (n = 9–10 per group). **p < 0.01 between CR and AB + CR groups, #p < 0.05 and ###p < 0.001 between CTRL and AB groups. (B) Food intake (n = 9–10 per group). *p < 0.05 between AB and CTRL groups. (C,D) Quantification of body fat mass (C) and lean mass (D) by MRI scans (n = 9–10 per group). (E–H) Analyses with metabolic chamber to quantitate O2 consumption (E), CO2 production (F), respiratory exchange ratio (RER) (G) and energy expenditure (EE) (H) (n = 4 for each group). Data are expressed as means ± s.e.m.
Analyses of metabolic and blood parameters
We analyzed a few metabolic parameters of the mice. As compared to the mice without antibiotic treatment, microbiota-depleted mice were characterized by a significant increase in total body fat and a decrease in lean mass as determined by MRI (Fig. 2C,D). CR could slightly reduce the ratio of lean mass (Fig. 2D). However, the AB + CR group lost more lean mass than the CR group (Fig. 2D). In terms of metabolic rate or energy expenditure (EE) represented by O2 consumption, CO2 production and energy expenditure, the AB + CR group had the lowest metabolic rate among the four experimental groups, especially in the dark phase (Figs 2E–G and S1). In addition, CR was able to reduce metabolic rate during the dark period (Fig. 2E–G). Respiratory exchange ratio (RER) was reduced by either calorie restriction or antibiotic administration in light phase (Fig. 2H). In dark phase, calorie restriction was more effective to reduce RER in the mice treated with antibiotics (Fig. 2H). As decreased energy expenditure was commonly associated with weight gain and obesity29,30, our data suggested that the observed abrogation of CR-mediated body weight loss by microbiota deletion might be caused by a decrease in metabolic rate upon antibiotics administration. In other words, gut microbiota is likely important for the mice to maintain a relatively high level of metabolic rate so that depletion of the microbiota would result in a significant reduction of metabolic rate.
We also determined the fasting blood glucose and plasma levels of triglycerides (TG), total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C) and low-density lipoprotein cholesterol (LDL-C). Antibiotic treatment elevated fasting blood glucose level and plasma TC level in the mice (Fig. 3A,B), with mild alteration on plasma LDL-C level (Fig. 3C). These data indicated that gut microbiota depletion is able to increase the risk of metabolic dysregulation as the elevations of blood glucose and cholesterol levels are considered as the hallmarks of metabolic syndrome31,32. On the other hand, the four groups of mice had minimal change in plasma TG and HDL-C (Fig. 3D,E). In addition, the AB + CR mice had a significant elevation of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) as compared to other groups (Fig. 3F,G), indicating that liver damage might occur in response to calorie restriction in the absence of gut microbiota.
Impacts of CR and gut microbiota depletion on blood parameters. (A) Fasting blood glucose level in different groups of mice. (B–G) Blood levels of TC (B), LDL-C (C), TG (D), HDL-C (E), AST (F) and ALT (G) of the mice at the end of the experiment. Data are expressed as means ± s.e.m, n = 9–10 per group.
We also analyzed the histological and morphological changes of the mice. Antibiotic-treated mice were characterized by a significant increase in the length of the small intestine (Fig. S2A) and decreased liver/body weight ratio (Fig. S2B) but without apparent changes in the histology of the liver (figure not shown). Furthermore, under microscopical examination, hematoxylin-eosin staining of the jejunum sections in the antibiotic-treated mice revealed longer and thinner intestinal villi than the mice without antibiotic treatment (Fig. S2C).
Gut microbiota depletion and calorie restriction alters metabolism-modulating hormones
Interestingly, AB mice exhibited hyperphagia characterized by a significant increase in food intake (Fig. 2B). We thus assumed that the gut microbiota might be involved in the secretion of hormones that regulate body weight and appetite33,34. To verify our hypothesis, we determined the plasma levels of insulin, leptin, gastric inhibitory polypeptide (GIP) and peptide YY (PYY). The overall levels of these four hormones in all four groups were visualized by a heatmap shown in Fig. 4A. Leptin is a hormone that helps to regulate energy balance by suppressing hunger35 and leptin deficiency is commonly associated with obesity36. Calorie restriction could lower the plasma level of leptin, the effect of which was enhanced when the mice were exposed to antibiotic treatment (Fig. 4B). Insulin is known to regulate the metabolism of carbohydrates, lipids and proteins by aiding the body to store the glucose. Notably, similar to leptin, insulin is also an acute appetite suppressant37. Antibiotic treatment significantly decreased the plasma insulin level (Fig. 4C), consistent with the observed elevation of fasting blood glucose level upon antibiotics administration (Fig. 3A). GIP belongs to the family of incretins that are released by nutrients from the gastrointestinal tract to amply insulin secretion38. Besides its effects to induce insulin secretion upon glucose administration and to regulate fatty acid metabolism, GIP was recently found to be an obesity-promoting factor by acting on adipocytes39,40. Intriguingly, AB + CR mice had the highest level of GIP level among the four groups of mice (Fig. 4D). PYY is a hormone produced in the small intestine and helps to reduce appetite and limit food intake41. The plasma level of PYY in the AB + CR group was significantly higher than that of the CTRL group (Fig. 4E). Collectively, these data indicated that gut microbiota plays an important role in modulating hormones that regulate metabolism in the mice.
Calorie restriction alters the composition of gut microbiota
We then performed 16S rRNA gene sequencing with the mice feces collected before and after administration of calorie restriction and/or antibiotic treatment, aiming to find how gut microbiota responds to calorie restriction and mediates the changes of body weight and metabolism. By significance tests for differences in α diversity, the gut microbiota of CR mice featured a markedly increased shannon index (Fig. 5A) and sobs index (Fig. 5B). As shown by respective rarefaction curves (Fig. S3), these curves became much flatter to the right, indicating that a reasonable number of sequences were taken and the α diversity of the sampled community was sufficiently extrapolated. Collectively, these data indicate that calorie restriction could render the gut microbiota a more balanced and diversified ecosystem.
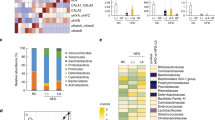
Structural rearrangement of gut microbiota in calorie-restricted mice. (A,B) Shannon index (A) and sobs index (B) of gut microbes as response to CR. (C) Composition of gut microbiota in CTRL and CR groups as shown in pie chart. (D) Partial least squares discriminant analysis (PLS-DA) of all the groups. (E) Generic differences between CTRL and CR groups with p-value < 0.1 in order of abundance. (F–H) Generic differences in Lactobacillus (F), Bifidobacterium (G) and Helicobacter (H) in the CTRL and CR groups at the 4th and 10th week of the experiment. Data are expressed as means ± s.e.m, n = 4–5 per group. *p < 0.05, **p < 0.01, ***p < 0.001.
The calorie restriction-induced difference in microbiota composition was illustrated in Fig. 5C. After calorie restriction, the bacterial phyla Bacteroidetes and Actinobacteria were slightly increased, while Firmicutes and Verrucomicrobia were slightly decreased. Also, a structural rearrangement of gut microbiota occurred after calorie restriction as illustrated by a supervised partial least squares discriminant analysis (PLS-DA) and hierarchical clustering (Figs 5D and S4). Notably, the antibiotic-treated mice exhibited no significant structural modulation under a calorie-restricted diet (Fig. 5D). Actually, based on the analysis of β diversity distance matrix, the bacterial structures in AB and AB + CR groups were more alike to each other after the experimental treatment (Fig. S5), as antibiotics extensively depleted resident gut microbiota.
We next performed linear discriminant analysis (LDA) effect size (LEfSe) analysis and identified a few bacterial genera that were significantly different between CTRL and CR groups (Figs 5E and S6). Lactobacillus and Bifidobacterium are widely approved probiotic genera with extensive health-promoting and immunomodulatory properties42,43,44. Compared to the CTRL group, the proportion of Lactobacillus and Bifidobacterium was increased in the CR group (p < 0.05 and p = 0.09 respectively, Fig. 5F,G). Helicobacter is a bacterial genus living mostly in the upper gastrointestinal tract which was often considered to be infectious and pathogenic45. We found that Helicobacter genus was significantly reduced by calorie restriction (Fig. 5H). In addition, a few other genera were altered by calorie restriction such as Lachnospiraceae NK4A136, Parasutterella, Clostridiales vadinBB60, Lachnoclostridium, Oscillibaster, Roseburia and Gordonibacter (Figs 5E and S6). Together, these data suggested that the composition and architecture of gut microbiota are altered by calorie-restricted diet, likely contributing to the metabolic benefits of CR.
Gut microbiota contribute to CR-mediated metabolic improvement
To further investigate whether gut microbiota alteration during CR is causally associated with metabolic improvement of the mice, we performed fecal microbiota transplantation (FMT) in diet-induced obesity (DIO) mice using microbiota collected respectively from mice fed normal chow ad libitum (AL) or 30% CR (Fig. 6A). Upon transplantation, the microbiota of the recipient mice resembled their corresponding donor groups to certain degrees as evaluated by OTU level based on 16s rRNA gene sequencing (Fig. S7). Notably, compared to the mice undergoing FMT from the control AL mice (HAL group), the mice that received FMT from the CR mice (HCR group) exhibited reduced body weight gain (Fig. 6B), accompanied by a decrease in body fat mass and an increase in lean mass (Fig. 6C,D). In addition, there was a slight improvement in glucose tolerance together with a significant reduction of fasting blood glucose level in the HCR group as compared to the HAL group (Figs 6E and S8A). In addition, HCR mice exhibited a significant decrease in blood leptin level (Fig. S8B), which was largely consistent with our observation with the CR mice (Fig. 4B). We also determined the level of plasma insulin level but no significant difference was found among the three HFD groups (Fig. S8C). This result was also concordant with our previous data that plasma insulin level had minimal change between CTRL and CR groups (Fig. 4C). Besides, some other metabolic and blood parameters such as plasma AST, ALT, TG, TC, HDL-C, LDL-C and hepatic TC, showed no significant changes among these groups of mice (Fig. S9). Furthermore, we examined histological changes of the liver by hematoxylin-eosin staining. As compared to the HBL group, mice in the HAL group exhibited similar morphology shown as a similar degree of hepatosteatosis induced by HFD (Fig. 6F). However, the HFD-induced hepatic steatosis appeared to be significantly alleviated in the HCR group (Fig. 6F). Consistently, the hepatic triglyceride level of the HCR group was significantly lower than those of HBL and HAL groups (Fig. 6G).
FMT from CR mice attenuated diet-induced obesity and partially ameliorated metabolic disturbances. (A) Schematic design of the FMT experiment. NC, normal chow; HFD, high fat diet. (B) Body weight curves of mice fed HFD (n = 6–8 per group). (C,D) Quantification of body fat mass (C) and lean mass (D) by MRI scans (n = 6–8 per group). (E) Oral glucose tolerance test of mice received FMT (n = 6–8 per group) with area under curve (AUC) shown in the inlet. (F) Representative H&E staining of the liver. Scale bar, 50 μm. (G) Hepatic triglyceride levels of the mice fed HFD (n = 6–8 per group). Data are expressed as means ± s.e.m, *p < 0.05, **p < 0.01.
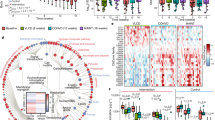
We also analyzed the microbial composition of the three groups of mice fed with HFD. Compared to the HBL group, FMT elevated the richness of microbiota reflected as sobs index (Fig. 7A). The diversity of microbial community shown by shannon index among the three groups had minimal changes (Fig. 7B). However, the HAL and HCR groups did have a structural discrepancy in the gut microbiota as illustrated by principal co-ordinates analysis (PCoA) and hierarchical clustering (Figs 7C and S10). We also applied LEfSe analysis and identified a few altered bacteria between the HAL and HCR groups (Fig. 7D,E). Compared to the HAL group, the microbial composition in the HCR group had increases in the abundances of Firmicutes (59.75% vs. 71.68%, p < 0.05) and Actinobacteria (1.25% vs. 4.13%, p = 0.06), together with a decrease in the abundance of Bacteroidetes (35.35% vs. 21.46%, p < 0.05). At genus level, the HCR group had a significant increase in genus Faecalibaculum, which significantly contributed to the overall differences (Fig. 7E). Besides, abundances of some other genera were different between the HAL and HCR groups such as Bacteroidales S24-7, Gordonibacter, Rikenella, Coriobacteriaceae UCG-002 and Coprococcus 1 (Figs. 7D,E).
The impacts of FMT from CR mice on the gut microbiota of DIO mice. (A,B) Shannon index (A) and sobs index (A) of fecal microbiota in DIO mice. (C) Principal co-ordinates analysis (PCoA) of the groups fed HFD. (D,E) Linear discriminant analysis (LDA) effect size (LEfSe) analysis (D) and corresponding LDA scores (E) in the HAL and HCR groups. p, phylum; c, class; o, order; f, family; g, genus. Data are expressed as means ± s.e.m, n = 6 for each group.
Discussion
In our study, we aimed to investigate the causal role of gut microbiota in the modulatory effects of calorie-restriction diet on metabolism. We hypothesized that reduced calorie intake could induce structural and functional changes in gut microbiota, which further impacts the physiological and metabolic profiles and determines the effect of calorie restriction. Our study reveals that antibiotic-induced gut microbiota depletion abrogates the body weight-lowering and metabolism-benefitting effects of calorie restriction. Furthermore, the FMT experiment clearly indicates that the gut microbiota from the CR mice is able to alleviate obesity in DIO mice. These data thus indicate for the first time that gut microbiota plays a causal role to mediate the beneficial effects of calorie restriction. Therefore, the gut microbiota plays an important role to ensure that calorie restriction can bring about various beneficial activities such as lowering body weight and maintaining a high level of basal metabolic rate, decreasing blood glucose level and reducing serum cholesterol level. Antibiotic treatment, therefore, largely abolishes the metabolism-regulatory functions of gut microbiota and abrogates the health-beneficial effect of gut microbiota.
Our study also suggests that gut microbiota is required to maintain a certain degree of basal metabolic rate (BMR). The gut microbiota could protect against metabolic disturbances especially by preventing a fast decline of BMR. Previous studies have demonstrated a phenomenon of “metabolic adaptation” or “adaptive thermogenesis” in which the obese people exhibit lower reduction in BMR than expectation after weight loss46,47,48. It was also believed that this phenomenon may predispose people to post-dieting weight regain49. Our study reveals that the microbiota-depleted and calorie-restricted mice had the lowest metabolic rate among all the experimental groups (Fig. 2). Thus, depletion of gut microbiota may lead to loss of the “metabolic buffering” function of the gut microbiota, leading to abrogation of CR-mediated body weight loss. In addition to lowering metabolic rate to compensate for calorie deficiency upon CR, adapting energy loss through fecal calories was also observed in mouse studies50. The alteration of the nutrient load induced rapid changes in the gut microbiota and correlated with stool energy loss likely through alteration of the proportion of Firmicutes and Bacteroidetes51. However, in our experiment, none of Firmicutes genus, Bacteroidetes genus, nor Firmicutes/Bacteroidetes ratio was significantly changed by CR (data not shown).
Our study also indicates that gut microbiota is required to maintain the morphology of the small intestine. Depletion of gut microbiota was able to increase the length of the small intestine together with elongated intestinal villi (Fig. S2). Interestingly, other studies have revealed that calorie restriction could promote the preservation and self-renewal of intestinal stem cells with coordination of Paneth cells through reducing mTORC1 signaling52. A longer small intestine length and extension of intestinal villi upon microbiota depletion would provide a greater absorptive area, leading to an increase of energy intake. Such antibiotic-mediated changes in the morphology of small intestine may partly explain our observation that CR-induced body weight loss is abrogated by depletion of gut microbiota.
We found that gut microbiota is also required for regulating the secretion of metabolism-modulating hormones. It is now widely accepted that gut microbiota serves as an endocrine organ53,54 and a number of hormones are secreted directly by intestinal microbes, such as serotonin and dopamine55,56. Also, it has been found that many hormones secreted by the gastrointestinal tract itself and brain are indirectly regulated by gut microbiota possibly mediated by short chain fatty acids (SCFAs)57. We found that the blood levels of leptin, insulin, GIP and PYY were all altered by depletion of gut microbiota (Fig. 4). Therefore, our data would favor a model in which the gut microbiota plays an active role in regulating metabolism-modulating hormones secreted by the gastrointestinal tract and other tissues, so that antibiotic treatment would perturb secretion of these hormones and result in detrimental effects to the metabolic health.
Additionally, calorie restriction can also act on gut microbiota itself through adjustments of the diversity and the proportion of beneficial and harmful bacteria. Previous research reported that life-long calorie-restricted mice represent a structural modulation of gut microbiota and exert health benefits probably via reduction of antigen load from the gut27. In our study, the 6-week calorie restriction is adequate to elicit a few changes in gut microbiota. In addition to an increase in diversity, CR is able to increase probiotic genera such as Lactobacillus and Bifidobacterium and meanwhile reduce harmful bacteria such as Helicobacter (Fig. 6). We therefore propose that calorie restriction may bring about the beneficial effects to the body by reshaping the structure of gut microbiota.
In the FMT experiments, we found that the CR-mediated metabolic improvements were, at least partially, contributed by gut microbiota. The previous study had demonstrated that 8-week FMT intervention with donor mice fed ad libitum could attenuate HFD-induced steatohepatitis in mice58. Here we found that CR-induced microbiota alteration was effective to alleviate certain HFD-mediated metabolic aberrations such as diet-induced obesity and hepatic lipid accumulation (Fig. 6). Interestingly, FMT to HFD-fed mice from calorie-restricted animals led to an increase in Faecalibaculum, which significantly contributed to the overall differences (Fig. 7E). Faecalibaculum, which is featured by F. rodentium, was recently found to produce lactic acid as a major metabolic end product59. As lactic acid-producing bacteria were believed to possess an anti-obesity effect60, the increase of Faecalibaculum might partially contribute to the loss of body weight upon FMT from the CR mice.
In summary, our study demonstrates the key role of gut microbiota in mediating the effects of calorie restriction. Our results also suggest that gut microbiota dysbiosis caused by unfavorable dietary habit or environmental stimuli such as antibiotics usage may impose a risk to the metabolism of the body. In addition, gut microbiota may serve as a target in the prevention and treatment of metabolic disorders.
Materials and Methods
Mice
All animals were maintained and used in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Institute for Nutritional Sciences, Chinese Academy of Sciences. All of the experimental procedures were carried out in accordance with the CAS ethics commission with an approval number 2010-AN-8. Male C57BL/6 mice at 4 wk of age, purchased from SLAC (Shanghai, China), were maintained in specific pathogen-free (SPF) conditions and kept on a 12 h light/dark cycle at the Institute for Nutritional Sciences. For our first animal experiment, all mice were weighed at the beginning and randomly allocated to one of the four groups so that the mean weights of each group were equal at the beginning of the experiment: (1) normal chow with free access to food and autoclaved water (CTRL, n = 10), (2) normal chow with 30% calorie restriction and autoclaved water (CR, n = 10), (3) normal chow with free access to food and antibiotic-treated water (AB, n = 11) and (4) normal chow with 30% calorie restriction and antibiotic-treated water (AB + CR, n = 10). For FMT experiment, all mice were weighed at the beginning and randomly allocated to one of the five groups: (1) normal chow ad libitum (AL, n = 5), (2) normal chow with 30% calorie restriction (CR, n = 5), (3) high-fat diet (Research Diet #D12492, with 60 kcal% fat) (HBL, n = 8), (4) high-fat diet undergoing FMT from AL group (HAL, n = 8) and (5) high-fat diet undergoing FMT from CR group (HCR, n = 7). All mice except for HFD mice were caged individually and maintained in sterile cages with autoclaved chip bedding. Mice fed HFD were caged 3–5 per cage.
Calorie restriction
Before the start of calorie restriction and antibiotic treatment, all mice were allowed ad libitum access to food sterilized by 60Co irradiation from SLAC (Shanghai, China) and autoclaved water for acclimation and monitoring of daily food intake in the first 2 weeks. Calorie restriction was initiated with a 15% food reduction (3.1 g/day) for 5 days. Mice in CR and AB + CR groups were administered 70% of the food intake of CTRL group from 10 wk of age onward (2.5 g/day at first). Food was given at 13:00~15:00 p.m. each day to avoid disturbance of the circadian clock. The daily consumption of food in CTRL and AB groups was recorded every day before food administration. The food intake of CTRL group was averaged every week to determine the amount of food for the following week for the CR and AB + CR groups.
Generation of antibiotic-induced microbiota-depleted (AIMD) mice
Antibiotic treatment was carried out together with calorie restriction. According to a previously published protocol61, 9-week old mice in AB and AB + CR groups received a combination of four nonabsorbable antibiotics: ampicillin, neomycin, metronidazole and vancomycin (Sangon Biotech, Shanghai, China) via oral gavage (0.2 mL) for 5 consecutive days (10 mg of each antibiotic per mouse per day) followed by administration in drinking water (ampicillin, neomycin and metronidazole: 1 g/L; vancomycin: 500 mg/L) which was renewed every week for the duration of the experiment. Mice in CTRL and CR groups received the same amount of sterile water by oral gavage in the first 5 days of treatment.
Body/tissue measurement
Body weight of all mice was recorded weekly and on the day of sacrifice. The weights of liver and lengths of the small intestine were measured after dissection and liver/body weight ratios were calculated for each mouse.
Mice fecal samples collection
Fresh fecal samples of all mice were collected at 15:00 ~ 17:00 p.m. to minimize possible circadian effects. Samples were collected into empty microtubes on ice and stored at -80 °C within 1 h for future use.
Cultivation of anaerobic fecal microbiota
Freshly collected fecal pellets were weighed and suspended in 0.9% NaCl (10 mg/mL). After homogenization with a tissue grinder, the suspensions were performed a 10-fold serial dilution and plated on brain heart infusion (BHI) agar (Hopebio, China). All plates were incubated at 37 °C for 48 ~ 72 hours in anaerobic culture bags (Hopebio, China) with oxygen-reducing, carbon dioxide-generating sachets (Oxoid, Thermo Fisher Scientific, UK) and anaerobic indicators (Mitsubishi Gas Chemical, Japan). The number of CFU was counted and only anaerobic culturable fecal microbes could be quantified by this method.
Body composition analysis
Mice body composition was assessed at 12 wk of age by echoMRI (Houston, USA) and the data of total body fat mass, lean mass, per cent body fat and per cent body lean were collected for each mouse, according to manufacturer’s directions.
Measurement of metabolic rate and physical activity
Mice at 12 wk of age were randomly chosen (n = 4 for each group) for determination of metabolic rate using the comprehensive laboratory animal monitoring system (CLAMS-16, Columbus Instruments, USA) according to the manufacturer’s instructions. Mice were allowed to acclimate to the system for the first 24 h. Oxygen uptake (VO2), carbon dioxide production (VCO2) and respiratory exchange ratio (RER) were recorded in the following 24 h. All parameters above were collected with every period of 16 min.
Measurement of plasma and liver parameters
Mice were euthanized and blood was immediately collected from the heart into EDTA-K2-treated microtubes (Kangjian Medical, China). Then the microtubes were centrifuged at 3,000 rpm for 15 min and the supernatant plasma was divided into 3 portions for different uses. All plasma samples excluding those for immediate uses were stored at −80 °C and multiple (>2) freeze/thaw cycles were avoided. Hepatic lipids were extracted with a previously reported method62. Plasma levels of aspartate transaminase (AST) and alanine transaminase (ALT) were determined by ALT/AST Determination Kit (ShenSuo UNF, China). Plasma and hepatic levels of triglycerides (TG), total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C) and low-density lipoprotein cholesterol (LDL-C) were determined by the colorimetric method with corresponding kits (ShenSuo UNF, China) according to manufacturer’s instructions.
Measurement of plasma hormones
Mice metabolic hormones were determined using either specialized ELISA kit or Milliplex MAP Magnetic Bead Mouse Metabolic Hormone Panel (Millipore MMHMAG-44K, Merck, Germany). Plasma leptin and insulin were determined respectively by mouse leptin ELISA Kit (MultiSciences, China) and mouse insulin ELISA Kit (Beyotime, China) according to manufacturer’s instructions. The Milliplex panel was custom-designed for the simultaneous quantification of GIP (total), insulin, leptin and PYY (total). Plasma samples (10 μL) were assayed using a Luminex MAGPIX analyzer (Merck Millipore, Germany) and the data were processed by Milliplex Analyst 5.1 software and R v3.3.2 (R Core Team, 2016).
Hematoxylin-eosin staining of liver and small intestine samples
Mice livers and small intestines were dissected and washed in PBS. Liver and small intestine samples were fixed in 4% polyformaldehyde for 48 h and embedded in paraffin. Paraffin-embedded sections (4 μm) were subjected to standard hematoxylin-eosin staining.
Fecal microbiota transplantation
Before FMT, mice were given a combination of ampicillin, neomycin, metronidazole and vancomycin (Sangon Biotech, China) in drinking water (ampicillin, neomycin and metronidazole: 1 g/L; vancomycin: 500 mg/L) for 10 consecutive days to remove indigenous gut microorganisms. After a 3-day recovery, FMT was operated twice a week. In brief, 200–300 mg of fresh stool was collected respectively from AL and CR group and was homogenated in 5 ml of PBS, settled by gravity for 2 min and the supernatant was gavaged 200 μl to each recipient mouse.
Oral glucose tolerance test (OGTT)
Mice were fasted overnight and subsequently given 2 g/kg of glucose solution (Sigma-Aldrich, USA) by oral gavage. Blood glucose was determined at 0, 15, 30, 60, 90 and 120 min after glucose administration (Abbott, USA).
Fecal DNA extraction, PCR amplification and Illumina MiSeq sequencing
Fecal DNA was extracted using the E.Z.N.A. stool DNA Kit (Omega Bio-tek, USA) according to the manufacturer’s instructions. DNA concentration was determined by a NanoDrop 2000 (Thermo Scientific, USA) and the integrity was examined by 1% agarose gel electrophoresis. The V3-V4 region of the bacterial 16S ribosomal RNA gene was amplified using primers 338 F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806 R (5′-GGACTACHVGGGTWTCTAAT-3′). The primers were incorporated with an 8-mer barcode sequence at the 5′ end which was unique to each sample. For PCR assays, 4 μL of 5 × Fast Pfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μΜ), 0.4 μL of FastPfu Polymerase (TransGen, China), 0.2 μL of BSA, 10 ng of template DNA and ddH2O were mixed to 20 μL in total. All PCR amplifications were performed in a thermocycler PCR system (ABI GeneAmp 9700, USA) in triplicate and following these steps: 95 °C for 3 min, followed by 30 cycles each consisting of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 45 s, followed by a final extension of 72 °C for 10 min. Amplicons were examined and extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, USA) according to the manufacturer’s instructions and quantified using QuantiFluor-ST (Promega, USA). The purified amplicons from different samples were mixed at equal molar ratios and ligated with 300-bp paired-end adapters by TruSeq DNA Sample Prep Kit (Illumina, USA) then sequenced on an Illumina MiSeq platform (Illumina, USA) according to standard protocols.
Bioinformatics and statistical analysis of sequencing data
The raw sequencing data were analyzed on the free online platform of Majorbio I-Sanger Cloud Platform (www.i-sanger.com, Majorbio, China). The raw paired-end reads were quality trimmed and filtered using Trimmomatic v0.32 (http://www.usadellab.org/cms/?page=trimmomatic)63. Then the optimized reads were merged with FLASH v1.2.11 (http://ccb.jhu.edu/software/FLASH/)64 according to their overlap sequences. The merged reads were divided by samples based on the barcode sequences. The average number of reads per sample was over 37,000 and the average length per read was 442 bp in our first animal model. In the FMT experiments, the average number of reads per sample was over 57,000 and the average length per read was 437 bp. After dereplication (http://drive5.com/usearch/manual/dereplication.html) and discarding the singletons (http://drive5.com/usearch/manual/singletons.html), the sequences were clustered into operational taxonomic units (OTUs) at 97% similarity using UPARSE v7.1 (http://drive5.com/uparse/) in QIIME (Quantitative Insights Into Microbial Ecology, http://www.qiime.org)65 while the chimeric sequences were identified and removed using UCHIME. The taxonomical classification was performed using RDP-classifier v2.2 (http://sourceforge.net/projects/rdp-classifier/) based on the SILVA 16S rRNA database (Release 128, http://www.arb-silva.de, Max Planck Institute, Germany) with a confidence threshold of 70%. Rarefaction curves and alpha diversity (Shannon) were determined using mothur v1.30.1 (http://www.mothur.org/wiki/Schloss_SOP#Alpha_diversity) and beta diversity was determined using QIIME. Unweighted UniFrac distance matrix analysis was performed in FastUniFrac (http://UniFrac.colorado.edu/) using OTUs for each sample. Partial least squares discriminant analysis (PLS-DA) is a supervised pattern recognition method and was achieved in R tools using package mixOmics. Significant changes in the abundance of OTUs or other levels between two groups were assessed using Student’s t test with bonferroni correction for multiple tests (α = 0.05). Linear discriminant analysis (LDA) coupled with effect size (LEfSe) was performed using LEfSe program (http://huttenhower.sph.harvard.edu/galaxy/root?tool_id = lefse_upload). A phylogenetic tree was built using FastTree v2.1.3 (http://www.microbesonline.org/fasttree/). All the graphs of bioinformatics unless otherwise stated, were generated by R v3.3.2 (R Core Team, 2016).
Statistical analysis
Data are expressed as mean ± s.e.m. The unpaired Student’s t test with two tails was used to determine the significance of the differences between two groups. For data that showed a normal distribution and homogeneity of variance, a one-way ANOVA was performed for comparisons among more than two groups using an FDR post hoc analysis. To determine if two different factors have an effect on a measured variable, a two-way ANOVA was used, followed by FDR post hoc test. Comparisons of medians between non-normally distributed groups were performed using the Kruskal-Wallis H test for simultaneous comparisons of more than two groups. Statistical tests were performed using Microsoft Excel (Microsoft, USA), R v3.3.2 (R Core Team, 2016), or Prism 7 (GraphPad Software).
References
McCay, C. M., Crowell, M. F. & Maynard, L. A. The effect of retarded growth upon the length of life span and upon the ultimate body size. J Nutr 10, 63–79 (1935).
Lane, M. A., Ingram, D. K. & Roth, G. S. Beyond the rodent model: Calorie restriction in rhesus monkeys. Age (Omaha) 20, 45–56, https://doi.org/10.1007/s11357-997-0004-2 (1997).
Lakowski, B. & Hekimi, S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci USA 95, 13091–13096 (1998).
Lin, S. J. et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418, 344–348, https://doi.org/10.1038/nature00829 (2002).
Mair, W. & Dillin, A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem 77, 727–754, https://doi.org/10.1146/annurev.biochem.77.061206.171059 (2008).
Colman, R. J. et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 325, 201–204, https://doi.org/10.1126/science.1173635 (2009).
Fontana, L., Partridge, L. & Longo, V. D. Extending healthy life span–from yeast to humans. Science 328, 321–326, https://doi.org/10.1126/science.1172539 (2010).
Lane, M. A. et al. Diet restriction in rhesus monkeys lowers fasting and glucose-stimulated glucoregulatory end points. Am J Physiol 268, E941–948 (1995).
Hursting, S. D., Lavigne, J. A., Berrigan, D., Perkins, S. N. & Barrett, J. C. Calorie restriction, aging and cancer prevention: mechanisms of action and applicability to humans. Annu Rev Med 54, 131–152, https://doi.org/10.1146/annurev.med.54.101601.152156 (2003).
Fontana, L., Meyer, T. E., Klein, S. & Holloszy, J. O. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. P Natl Acad Sci USA 101, 6659–6663, https://doi.org/10.1073/pnas.0308291101 (2004).
Heilbronn, L. K. et al. Effect of 6-month calorie restriction on biomarkers of longevity, metabolic adaptation and oxidative stress in overweight individuals: a randomized controlled trial. JAMA 295, 1539–1548, https://doi.org/10.1001/jama.295.13.1539 (2006).
Fontana, L. & Klein, S. Aging, adiposity and calorie restriction. JAMA 297, 986–994, https://doi.org/10.1001/jama.297.9.986 (2007).
Longo, V. D. & Fontana, L. Calorie restriction and cancer prevention: metabolic and molecular mechanisms. Trends Pharmacol Sci 31, 89–98, https://doi.org/10.1016/j.tips.2009.11.004 (2010).
Ley, R. E. et al. Evolution of mammals and their gut microbes. Science 320, 1647–1651, https://doi.org/10.1126/science.1155725 (2008).
De Filippo, C. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 107, 14691–14696, https://doi.org/10.1073/pnas.1005963107 (2010).
Muegge, B. D. et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974, https://doi.org/10.1126/science.1198719 (2011).
Wu, G. D. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108, https://doi.org/10.1126/science.1208344 (2011).
Walker, A. W. et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5, 220–230, https://doi.org/10.1038/ismej.2010.118 (2011).
Cotillard, A. et al. Dietary intervention impact on gut microbial gene richness. Nature 500, 585–588, https://doi.org/10.1038/nature12480 (2013).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563, https://doi.org/10.1038/nature12820 (2014).
Kovatcheva-Datchary, P. et al. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab 22, 971–982, https://doi.org/10.1016/j.cmet.2015.10.001 (2015).
Sonnenburg, E. D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215, https://doi.org/10.1038/nature16504 (2016).
Backhed, F. et al. The gut microbiota as an environmental factor that regulates fat storage. P Natl Acad Sci USA 101, 15718–15723, https://doi.org/10.1073/pnas.0407076101 (2004).
Koren, O. et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150, 470–480, https://doi.org/10.1016/j.cell.2012.07.008 (2012).
Sommer, F. et al. The Gut Microbiota Modulates Energy Metabolism in the Hibernating Brown Bear Ursus arctos. Cell Rep 14, 1655–1661, https://doi.org/10.1016/j.celrep.2016.01.026 (2016).
Griffin, N. W. et al. Prior Dietary Practices and Connections to a Human Gut Microbial Metacommunity Alter Responses to Diet Interventions. Cell Host Microbe 21, 84–96, https://doi.org/10.1016/j.chom.2016.12.006 (2017).
Zhang, C. et al. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat Commun 4, 2163, https://doi.org/10.1038/ncomms3163 (2013).
Ott, B. et al. Effect of caloric restriction on gut permeability, inflammation markers and fecal microbiota in obese women. Sci Rep 7, 11955, https://doi.org/10.1038/s41598-017-12109-9 (2017).
Ravussin, E. et al. Reduced rate of energy expenditure as a risk factor for body-weight gain. N Engl J Med 318, 467–472, https://doi.org/10.1056/NEJM198802253180802 (1988).
Rogers, N. H., Perfield, J. W. II., Strissel, K. J., Obin, M. S. & Greenberg, A. S. Reduced energy expenditure and increased inflammation are early events in the development of ovariectomy-induced obesity. Endocrinology 150, 2161–2168, https://doi.org/10.1210/en.2008-1405 (2009).
Barish, G. D., Narkar, V. A. & Evans, R. M. PPAR delta: a dagger in the heart of the metabolic syndrome. J Clin Invest 116, 590–597, https://doi.org/10.1172/JCI27955 (2006).
Kahn, R. et al. The metabolic syndrome: time for a critical appraisal: joint statement from the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care 28, 2289–2304 (2005).
Bauer, P. V., Hamr, S. C. & Duca, F. A. Regulation of energy balance by a gut-brain axis and involvement of the gut microbiota. Cell Mol Life Sci 73, 737–755, https://doi.org/10.1007/s00018-015-2083-z (2016).
Woods, S. C. & D’Alessio, D. A. Central control of body weight and appetite. J Clin Endocrinol Metab 93, S37–50, https://doi.org/10.1210/jc.2008-1630 (2008).
Friedman, J. M. & Halaas, J. L. Leptin and the regulation of body weight in mammals. Nature 395, 763–770, https://doi.org/10.1038/27376 (1998).
Montague, C. T. et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature 387, 903–908, https://doi.org/10.1038/43185 (1997).
Woods, S. C., Stein, L. J., McKay, L. D. & Porte, D. Jr. Suppression of food intake by intravenous nutrients and insulin in the baboon. Am J Physiol 247, R393–401 (1984).
Efendic, S. & Portwood, N. Overview of incretin hormones. Horm Metab Res 36, 742–746, https://doi.org/10.1055/s-2004-826157 (2004).
Miyawaki, K. et al. Inhibition of gastric inhibitory polypeptide signaling prevents obesity. Nat Med 8, 738–742, https://doi.org/10.1038/nm727 (2002).
Yamada, Y. & Seino, Y. Physiology of GIP–a lesson from GIP receptor knockout mice. Horm Metab Res 36, 771–774, https://doi.org/10.1055/s-2004-826162 (2004).
le Roux, C. W. & Bloom, S. R. Peptide YY, appetite and food intake. Proc Nutr Soc 64, 213–216 (2005).
Delzenne, N. M., Neyrinck, A. M., Backhed, F. & Cani, P. D. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat Rev Endocrinol 7, 639–646, https://doi.org/10.1038/nrendo.2011.126 (2011).
Fukuda, S. et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547, https://doi.org/10.1038/nature09646 (2011).
Fukuda, S., Toh, H., Taylor, T. D., Ohno, H. & Hattori, M. Acetate-producing bifidobacteria protect the host from enteropathogenic infection via carbohydrate transporters. Gut Microbes 3, 449–454, https://doi.org/10.4161/gmic.21214 (2012).
Hua, J. S., Zheng, P. Y. & Bow, H. Species differentiation and identification in the genus of Helicobacter. World J Gastroenterol 5, 7–9 (1999).
Lowell, B. B. & Spiegelman, B. M. Towards a molecular understanding of adaptive thermogenesis. Nature 404, 652–660, https://doi.org/10.1038/35007527 (2000).
Rosenbaum, M., Hirsch, J., Gallagher, D. A. & Leibel, R. L. Long-term persistence of adaptive thermogenesis in subjects who have maintained a reduced body weight. Am J Clin Nutr 88, 906–912 (2008).
Rosenbaum, M. & Leibel, R. L. Adaptive thermogenesis in humans. Int J Obes (Lond) 34(1), S47–55, https://doi.org/10.1038/ijo.2010.184 (2010).
Camps, S. G., Verhoef, S. P. & Westerterp, K. R. Weight loss, weight maintenance and adaptive thermogenesis. Am J Clin Nutr 97, 990–994, https://doi.org/10.3945/ajcn.112.050310 (2013).
Hambly, C. & Speakman, J. R. Contribution of different mechanisms to compensation for energy restriction in the mouse. Obesity research 13, 1548–1557, https://doi.org/10.1038/oby.2005.190 (2005).
Jumpertz, R. et al. Energy-balance studies reveal associations between gut microbes, caloric load and nutrient absorption in humans. Am J Clin Nutr 94, 58–65, https://doi.org/10.3945/ajcn.110.010132 (2011).
Yilmaz, O. H. et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495, https://doi.org/10.1038/nature11163 (2012).
Clarke, G. et al. Minireview: Gut microbiota: the neglected endocrine organ. Mol Endocrinol 28, 1221–1238, https://doi.org/10.1210/me.2014-1108 (2014).
Evans, J. M., Morris, L. S. & Marchesi, J. R. The gut microbiome: the role of a virtual organ in the endocrinology of the host. J Endocrinol 218, R37–47, https://doi.org/10.1530/JOE-13-0131 (2013).
Cryan, J. F. & Dinan, T. G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 13, 701–712, https://doi.org/10.1038/nrn3346 (2012).
Tsavkelova, E. A., Botvinko, I. V., Kudrin, V. S. & Oleskin, A. V. Detection of neurotransmitter amines in microorganisms with the use of high-performance liquid chromatography. Dokl Biochem 372, 115–117 (2000).
Dockray, G. J. Gastrointestinal hormones and the dialogue between gut and brain. J Physiol 592, 2927–2941, https://doi.org/10.1113/jphysiol.2014.270850 (2014).
Zhou, D. et al. Total fecal microbiota transplantation alleviates high-fat diet-induced steatohepatitis in mice via beneficial regulation of gut microbiota. Sci Rep 7, 1529, https://doi.org/10.1038/s41598-017-01751-y (2017).
Chang, D. H. et al. Faecalibaculum rodentium gen. nov., sp. nov., isolated from the faeces of a laboratory mouse. Antonie Van Leeuwenhoek 108, 1309–1318, https://doi.org/10.1007/s10482-015-0583-3 (2015).
Tsai, Y. T., Cheng, P. C. & Pan, T. M. Anti-obesity effects of gut microbiota are associated with lactic acid bacteria. Appl Microbiol Biotechnol 98, 1–10, https://doi.org/10.1007/s00253-013-5346-3 (2014).
Kuss, S. K. et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334, 249–252, https://doi.org/10.1126/science.1211057 (2011).
Xu, D. et al. PAQR3 modulates cholesterol homeostasis by anchoring Scap/SREBP complex to the Golgi apparatus. Nat Commun 6, 8100, https://doi.org/10.1038/ncomms9100 (2015).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120, https://doi.org/10.1093/bioinformatics/btu170 (2014).
Magoc, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963, https://doi.org/10.1093/bioinformatics/btr507 (2011).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336, https://doi.org/10.1038/nmeth.f.303 (2010).
Acknowledgements
This work was supported by research grants from National Natural Science Foundation of China (31630036 and 81390350 to Y.C.), Ministry of Science and Technology of China (National Key R&D Program of China 2016YFA0500103 to Y.C.), Chinese Academy of Sciences (XDA12010102 and QYZDJ-SSW-SMC008 and ZDRW-ZS-2016-8 to Y.C.).
Author information
Authors and Affiliations
Contributions
S.W. and Y.C. designed the experiment, wrote the paper and prepared the figures. S.W., M.H., X.Y., J.Z. and L.C. performed the experiment. L.W. and Y.L. provided technical assistance.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, S., Huang, M., You, X. et al. Gut microbiota mediates the anti-obesity effect of calorie restriction in mice. Sci Rep 8, 13037 (2018). https://doi.org/10.1038/s41598-018-31353-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-31353-1
This article is cited by
-
Gut microbiota influence frailty syndrome in older adults: mechanisms and therapeutic strategies
Biogerontology (2024)
-
Differential effects of plant-based flours on metabolic homeostasis and the gut microbiota in high-fat fed rats
Nutrition & Metabolism (2023)
-
The antitumour effects of caloric restriction are mediated by the gut microbiome
Nature Metabolism (2023)
-
Grape seed and skin extract, a potential prebiotic with anti-obesity effect through gut microbiota modulation
Gut Pathogens (2022)
-
Gut microbiota predicts body fat change following a low-energy diet: a PREVIEW intervention study
Genome Medicine (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.