Abstract
Eye movements are considered endophenotypes of schizophrenia. However, the genetic factors underlying eye movement are largely unknown. In this study, we explored the susceptibility loci for four eye movement scores: the scanpath length during the free viewing test (SPL), the horizontal position gain during the fast Lissajous paradigm of the smooth pursuit test (HPG), the duration of fixations during the far distractor paradigm of the fixation stability test (DF) and the integrated eye movement score of those three scores (EMS). We found 16 SNPs relevant to the HPG that were located in 3 genomic regions (1q21.3, 7p12.1 and 20q13.12) in the patient group; however, these SNPs were intronic or intergenic SNPs. To determine whether these SNPs occur in functional non-coding regions (i.e., enhancer or promoter regions), we examined the chromatin status on the basis of publicly available epigenomic data from 127 tissues or cell lines. This analysis suggested that the SNPs on 1q21.3 and 20q13.12 are in enhancer or promoter regions. Moreover, we performed an analysis of expression quantitative trait loci (eQTL) in human brain tissues using a public database. Finally, we identified significant eQTL effects for all of the SNPs at 1q21.3 and 20q13.12 in particular brain regions.
Similar content being viewed by others
Introduction
Schizophrenia is a psychiatric disease with a lifetime prevalence of 0.30–0.66%1. The diagnostic criteria for schizophrenia are based on subjective symptoms, including delusions, hallucinations and thought insertion, and objective symptoms, including disorganized speech and bewilderment. The age of onset is usually late adolescence. Nevertheless, it is difficult to diagnose schizophrenia in patients in late adolescence on the basis of clinical symptoms alone2,3. Although the development of a quantitative biomarker to aid in diagnosis is an attractive strategy, no such markers have been identified. Neuropsychological studies have shown that eye movement dysfunction is a clinical symptom of schizophrenia. Abnormalities in smooth pursuit eye movements (SPEMs)4,5,6, voluntary control of saccades7,8,9 and exploratory eye movement behaviors10,11,12,13 have been commonly observed. Studies comparing eye movement scores between non-schizophrenic individuals and patients with schizophrenia have distinguished the groups with accuracies of 80–90%14,15,16,17.
Several studies have reported that eye movement performance is limited not only in patients with schizophrenia but also in their siblings18,19, thus suggesting that eye movements may be a candidate endophenotype of schizophrenia. The heritability of smooth pursuit and anti-saccadic eye movement tasks was estimated as 0.4 to 0.6 in a family-based analysis of schizophrenia20,21. In particular, predictive pursuit gain in the smooth pursuit test has a very high heritability (heritability = 0.9)22, indicating that genetic factors underlie abnormalities in eye movement. However, it remains unknown whether patients with schizophrenia are predisposed to dysfunctional eye movements.
Genomic analyses have been performed to identify susceptibility loci for eye movement. The genetic loci related to SPEMs have been observed in the regions surrounding ERBB423, RANBP124, COMT25, ZDHHC826 and NRG127. In EEM, the eye fixations for the responsive search and cognitive search scores are associated with chromosomes 22q11.228 and 5q21.329, respectively. However, most previous studies focused on only specific gene polymorphisms. To extensively search for susceptibility loci in genomes, a genome-wide association study (GWAS) is more effective.
In this study, we performed a GWAS to identify SNPs associated with the following eye movement scores: the scanpath length during the free viewing test (SPL); the horizontal position gain during the fast Lissajous paradigm of the smooth pursuit test (HPG); and the duration of fixations during the far distractor paradigm of the fixation stability test (DF). Our previous study revealed that these eye movement scores distinguish patients with schizophrenia17. Our GWAS also included the integrated eye movement score (EMS), composed of these three measures, which showed discrimination ability with an accuracy of 82%. Our analyses revealed 17 susceptibility loci for HPG. Furthermore, chromatin state analysis showed that the SNPs at 1q21.3 and 20q13.12 are located in enhancer or promoter regions. The SNPs at 1q21.3 were located in intron or intergenic regions of the THEM4 or S100A10 genes, and the SNP at 20q13.12 was located upstream of the CDH22 gene. THEM4 is a negative regulator of the AKT1 gene, contributing to signal transduction in neurons30. S100A10 expression is lower in lymphoblastoid cell lines from patients with schizophrenia than in those from controls31. Cadherins such as CDH22 are involved in synaptic plasticity, which may contribute to learning and memory32. Recent studies have suggested that synaptic plasticity dysfunction is involved in schizophrenia33,34. To examine the functions of these SNPs in detail, our bioinformatics analyses showed that these SNPs were located in functional non-coding regions, such as enhancers or promoters, and affect the expression level of surrounding genes in specific brain regions.
Materials and Methods
Subjects
Eye movements were recorded in 60 patients with schizophrenia and 166 healthy control participants who were recruited at Osaka University. These subjects were included in previous studies35,36,37,38,39. All of the subjects were of Japanese descent and were biologically unrelated. They had no history of ophthalmologic disease or neurological/medical conditions that influence the central nervous system40,41. Specific exclusion criteria included atypical headaches, head trauma with loss of consciousness, chronic lung disease, kidney disease, chronic hepatic disease, thyroid disease, active cancer, cerebrovascular disease, epilepsy, seizures, substance-related disorders, and mental retardation. Healthy control participants were recruited through regional advertisements at Osaka University and were evaluated for psychiatric, medical, and neurological concerns by using the non-patient version of the Structured Clinical Interview for DSM-IV (SCID) to exclude individuals with current or past contact with psychiatric services and those who had received psychiatric medication.
Patients with schizophrenia were recruited from Osaka University Hospital and had been diagnosed by two or more trained psychiatrists according to the DSM-IV criteria on the basis of the SCID. The current symptoms of schizophrenia were evaluated. The total dose of their prescribed antipsychotics was calculated in chlorpromazine (CPZ) equivalents (mg/day)42. The current IQ was measured using the Japanese version of the Wechsler Adult Intelligence Scale-Third Edition (WAIS-III)43. Premorbid IQ was estimated using the Japanese version of the National Adult Reading Test44,45. The study was performed in accordance with the World Medical Association’s Declaration of Helsinki and was approved by the Research Ethics Committee of Osaka University. All of the participants provided written informed consent to participate in the study after a full explanation of the study procedures was provided. Anonymity was preserved for all participants.
Measurement of eye movement
The subjects faced a 19-in liquid crystal display monitor placed at a 70-cm distance from the observers’ eyes. Visual stimuli were presented using MATLAB (MathWorks, Natick, MA, USA) via the Psychophysics Toolbox extension46. The eye movements and pupil areas of the left eye were measured at 1 kHz using the EyeLink1000 (SR Research, Ontario, Canada) system. Measurements of the SPL, HPG and DF were collected as previously described16,17. The integrated EMS of the above three scores was calculated as in Morita et al.17.
Genotyping
Genotyping was performed using the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA) according to the manufacturer’s protocol. The genotypes were called from the CEL files by using Birdseed v2 for the 6.0 chip implemented in the Genotyping Console software (Affymetrix). We then applied the following QC criteria to exclude samples: (i) arrays with low QC (<0.4) according to Birdseed v2 (n = 0), (ii) samples for which <90% of the genotypes were called (n = 0) and (iii) samples in the same family according to pi-hat (>0.1, n = 0). Next, we excluded SNPs that (i) had low call rates (<0.97), (ii) were duplicated, (iii) were located on sex chromosomes, (iv) deviated from HWE in the controls (p < 1.0 × 10−5) or (v) had low MAF <0.01. After these QC exclusions, 554, 152 SNPs were retained for experimental analysis. Of the 554, 152 SNPs, we used SNPs for which the heterozygous genotype and homozygous minor allele genotype were present in more than 3 subjects for each measurement in each group. To test for the existence of genetic structure in the data, we performed a principal component analysis (PCA) using the EIGENSTRAT 3.0 software47. Twenty eigenvectors were calculated. Genotype information from the JPT (Japanese in Tokyo, Japan), CHB (Han Chinese in Beijing, China), CEU (Utah residents with ancestors from northern and western Europe) and YRI (Yoruba in Ibadan, Nigeria) in HapMap 3 was compared with our dataset to check for population stratification. Genotyping was performed as described in previous studies35,36,37,38,39.
Statistical analysis
A t-test was performed to measure statistical significance for all variables except sex by using R 3.3.0. Statistical significance for sex was calculated with the χ2 test. We performed linear regression analysis to identify SNP markers associated with the four eye movement scores using PLINK version 1.0748.
Analysis of functional non-coding regions
Functional non-coding regions, including enhancer and promoter regions, were searched by using the chromatin segmentation data from ChromImpute, which predicts chromatin states from a mathematical model based on the imputed data for 11 histone modifications and DNase from the Roadmap Epigenomics Project49,50. We used the chromatin segmentation data for 127 human cell lines and tissues (http://epigenomegateway.wustl.edu/browser/).
Cis-expression QTL (eQTL) analysis in brain tissues
We used the BRAINEAC database to perform cis-eQTL analysis in brain regions51. BRAINEAC is a database that includes SNPs associated with gene expression from 134 individuals in ten post-mortem brain regions: cerebellar cortex, frontal cortex, hippocampus, inferior olivary nucleus (sub-dissected from the medulla), occipital cortex, putamen (at the level of the anterior commissure), substantia nigra, temporal cortex, thalamus (at the level of the lateral geniculate nucleus) and intralobular white matter. The eQTL analysis used linear regression to delineate the effects of genotype against the expression level of the genes closest to the SNPs.
Results
Comparison of demographic data between healthy control participants and patients with schizophrenia
Our analyses included 226 subjects (60 patients with schizophrenia (SZ group) and 166 healthy control participants (HC group)). Participant demographics are provided in Table 1. The HC and SZ groups were not significantly different in sex (p = 0.280) but were significantly different in age (p = 2.46 × 10−4). We used the three eye movement scores reported in our previous study that best distinguished between healthy controls and schizophrenic individuals17. As expected, the three eye movement scores for the HC group were significantly higher than those for the SZ group (SPL: p = 2.78 × 10−13; HPG: p = 1.61 × 10−7; DF: p = 1.92 × 10−5). Similarly, the EMS of the HC group was also significantly higher than that of the SZ group (p = 2.20 × 10−16).
Whole-genome QTL analysis of eye movement scores
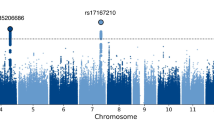
To search for SNP markers associated with the three eye movement scores and the integrated score, we performed a GWAS by using linear regression analysis in each group and in all of the samples (SZ, HC and ALL (the combined SZ and HC groups)). We found that several SNP markers yielded a genome-wide significant p-value (p = 5.0 × 10−8) for HPG in each group (1 SNP in the HC group, 16 SNPs in the SZ group and 1 SNP in the ALL group) (Fig. 1, Table 2 and Fig. S1), although there were no significant SNP markers for the other measurements (Figs S2–4). In the HC group, we found a SNP marker, rs17393065, that is located 15 kb 3′ of GALNT14. In the SZ group, we found 16 SNPs in 3 genomic regions (1q21.3, 7p12.1 and 20q13.12). Of the 14 SNPs on 1q21.3, 4 SNPs are in a THEM4 intron, and 10 SNPs are in an intergenic region between THEM4 and S100A10. The SNPs on 7p12.1 and 20q13.12 were 523 kb 3′ of POM121L12 and 2.6 kb 5′ of CDH22, respectively. In the ALL group, we found a SNP marker, rs1490191, located 34 kb 5′ of THEM4. Interestingly, this SNP marker was also significantly associated with HPG in the SZ group. The number of major alleles for all 18 identified SNPs was positively correlated with HPG, such that as the number of minor alleles increased, HPG decreased. To control for a significant difference between the HC and SZ groups in terms of age, we adjusted for the effect of age by linear regression analysis with age as an independent variable. Consequently, the 17 SNPs, except for one SNP at 7p12.1 in the SZ group, remained statistically significant.
Location of susceptibility loci for eye movement in functional non-coding regions
The SNPs found in this study were located in non-coding regions, such as introns and intergenic regions, which do not directly affect protein function. We examined the possibility of functional variants around identified SNPs. Most of the SNPs identified by GWAS do not affect the coding regions of genes. Functional variants may be in strong linkage disequilibrium (LD) with SNPs identified by GWAS. To identify variants in gene-coding regions genetically linked to the identified SNPs, we used the HaploReg database (Table S1). We extracted SNPs that were strongly linked to the identified SNPs based on the Asian population in the 1000 Genome Project (r2 > 0.8). Among them, we found 2 SNPs (rs3762427 and rs3748805) in the THEM4 coding region. rs3762427 and rs3748805 were synonymous and missense variants, respectively. To predict whether the missense variant rs3748805 is deleterious, we used three different programs (PolyPhen2, SIFT and PROVEAN). rs3748805 was “BENIGN” in the PolyPhen2 program, “Tolerated” in the SIFT program and “Neutral” in the PROVEAN program. These results showed that there were no functional variants in coding regions. Therefore, we next investigated the possibility that these SNPs may be located in functional non-coding regions (i.e., enhancers). To check them, we used the chromatin state from 127 human cell lines or tissues from the Roadmap Epigenomics Project. The chromatin states were predicted on the basis of 12 epigenomic markers (H3K4me1, H3K4me2, H3K4me3, H3K9ac, H3K27ac, H4K20me1, H3K79me2, H3K36me3, H3K9me3, H3K27me3, H2A.Z, and DNase) by using ChromHMM and ChromImpute49,50. The intergenic SNPs at 1q21.3 observed in the SZ group were near enhancer regions (yellow or orange segments in Fig. 2A). The SNP marker rs1490191, which was also identified in the ALL group, was in an active enhancer region (orange segments in Fig. 2A). These results suggested that the SNPs at 1q21.3 are located around enhancer regions and affect the expression of genes in the vicinity of the SNPs. We also found that the SNP rs6104543, upstream of CDH22 at 20q13.12, was located around bivalent promoters, which are regions that contain both a “repressive” and an “activating” chromatin modification52 (dark purple segments in Fig. 2B). We did not find functional non-coding regions around the other SNPs (7p12.1 in the SZ group; 2p23.1 in the HC group) (Fig. S5).
The state model of chromatin in 1q21.3 (A) and 20q13.12 (B). The chromatin was segmented into 25 states (i.e., promoter, enhancer) with the ChromHMM and ChromImpute algorithms45,46 by using data from the NIH Roadmap Epigenomics Consortium (http://www.roadmapepigenomics.org) and was visualized with the WashU Epigenome Browser (http://epigenomegateway.wustl.edu/browser/). Here, 25 states are summarized to 9 as shown in the color legend. The gray shaded stripe represents an active/weak enhancer (A) or bivalent promoter regions (B).
eQTL effect of susceptibility loci for eye movement
As shown above, the SNPs at 1q21.3 and 20q13.12 found in the SZ and ALL groups were around the THEM4, S100A10 and CHD22 genes. These genes are associated with neurotransmission and brain morphogenesis. THEM4 (thioesterase superfamily member 4) is also known as CTMP (carboxyl-terminal modulator protein) and is a negative regulator of the AKT1 gene, contributing to signal transduction in neurons30. S100A10 (S100 calcium binding protein A10) modulates the transport of neurotransmitters (e.g., calcium ion, serotonin)53. CDH22 (cadherin 22) is mainly expressed in the brain and is involved in brain morphogenesis as a cell-adhesion factor54,55. These genes may be involved because eye movements are closely linked to the activity of specific brain regions56,57,58.
To further examine whether the SNPs at 1q21.3 and 20q13.12 are associated with the expression levels of the surrounding genes in brain regions, we analyzed eQTL by using the BRAINEAC database, including 10 human brain regions (cerebellar cortex (CRBL), frontal cortex (FCTX), hippocampus (HIPP), medulla (specifically the inferior olivary nucleus, MEDU), occipital cortex (specifically the primary visual cortex, OCTX), putamen (PUTM), substantia nigra (SNIG), thalamus (THAL), temporal cortex (TCTX) and intralobular white matter (WHMT)). The eQTL analysis used linear regression to delineate the effects of genotype against the expression level of genes around the SNPs. Consequently, all of the SNPs at 1q21.3 were found to be significantly associated with THEM4 and/or S100A10 expression in at least one brain region. The significance threshold for the Bonferroni correction was set at 0.005 (=0.05/10 brain regions). As expected, 4 intronic SNPs at THEM4 were significantly associated with THEM4 but not S100A10 expression (Fig. 3). These SNPs satisfied a significance threshold in 4 brain regions (PUTM, MEDU, SNIG and THAL). In contrast, 7 intergenic SNPs showed a significant eQTL association with THEM4 in 2 brain regions (FCTX and PUTM). PUTM was common to all SNPs associated with THEM4. Additionally, 7 intergenic SNPs were significantly associated with S100A10 in the CRBL. The rs6104543 upstream of CDH22 was significant for OCTX. These results suggested that SNPs at 1q21.3 and 20q13.12 act as cis-eQTL and that these effects are different in each brain region.
Heatmap based on statistics from the eQTL analysis. Each color represents the significance (log10 (p-value)) of the association between the genotype of a SNP and the expression level of the gene that is shown for each line. P-values were provided by the BRAINEAC database (http://www.braineac.org/). An asterisk indicates that a SNP satisfies a significance threshold for the Bonferroni correction (0.05/10 brain regions = 0.005) in a brain region. This plot displays only significant eQTL associations in at least one brain region. CRBL, cerebellar cortex; FCTX, frontal cortex; PUTM, putamen; MEDU, medulla; SNIG, substantia nigra; THAL, thalamus; OCTX, occipital cortex; WHMT, intralobular white matter; TCTX, temporal cortex; HIPP, hippocampus.
Discussion
In this study, we explored susceptibility loci for three eye movement scores (SPL, HPG and DF) and the integrated eye movement score (EMS). We found 17 susceptibility loci for HPG in the SZ and ALL groups. Furthermore, our results showed that the SNPs at 1q21.3 and 20q13.12 are located in enhancer or promoter regions and that the genotypes of those SNPs are associated with the expression levels of the surrounding genes in specific brain regions.
To the best of our knowledge, our study is the first report using SNPs to explain the abnormal value of the horizontal position gain in the fast Lissajous paradigm of the smooth pursuit test in patients with schizophrenia. We found that the SNPs were only associated with HPG among four eye movement scores. Several studies have reported that SPEMs are inherited18,19,20,21. In particular, predictive pursuit gain, which is related to HPG, has very high heritability (heritability = 0.9)22. Therefore, we considered that the SNPs were associated with only HPG. In previous genomic studies of SPEMs, the natural logarithmic values of the signal/noise ratio (SNR)23,24,25,26 or velocity gain and saccadic frequency were used27 in the smooth pursuit test. In our study, we used the HPG as a measurement in the smooth pursuit test because our previous study showed that the HPG, rather than other measurements (i.e., SNR), has high discriminatory ability17. Our results may help elucidate the genetic association of eye movements from a different perspective in the smooth pursuit test.
We showed that the SNPs at 1q21.3 affect the expression of the THEM4 and S100A10 genes, which are near the SNP loci. THEM4 is a negative regulator of the AKT1 gene, which showed lower expression values in the frontal cortex of schizophrenic patients59. Our eQTL analyses showed that the SNPs on 1q21.3 were associated with the expression level of THEM4 in the frontal cortex, thus suggesting that THEM4 affects AKT1 expression in the frontal cortex. We have reported that the AKT1 genotype is related to gray matter volumes in schizophrenia patients in the frontostriatal region, which is part of the frontal cortex60. The frontal cortex contains the frontal eye field, an essential area for saccade and smooth pursuit eye movements61,62,63. The expression of THEM4 in the frontal cortex could be related to eye movements. We showed eQTL effects of S100A10 in the cerebellar cortex. S100A10 is up-regulated in Purkinje cells in a mouse model of inflammatory demyelinating diseases in the central nervous system64. The cerebellar cortex is involved in various motor activities and plays a role in smooth pursuit eye movement65. Moreover, we also identified CHD22 as the closest gene to a SNP on 20q13.12 in the SZ group. An intronic SNP on 20q13.12 was correlated with CDH22 expression in the occipital cortex. The occipital cortex is closely related to visual processing. V5, which is a region in the visual cortex, is responsible for the guidance of smooth pursuit, and lesions in V5 cause defects in smooth pursuit66.
We performed replication analysis for eye movement markers found in previous studies (Table S2). Consequently, 2 of 7 SNPs in the ERBB4, RANBP1, COMT and MAN2A1 genes showed significant associations with eye movement scores in our study (p < 0.05); however, these p-values were determined before correction for multiple tests. It is reasonable for some markers to be replicated in the Japanese population because these markers are also found in Korean and Chinese populations, which are genetically closely related to the Japanese population. Our results suggested that the effects of these markers on eye movement are constant, at least in East Asian populations.
Our study has several limitations. Our identified SNPs have small effect sizes. A power analysis based on α = 5 × 10−8 and 554, 152 SNPs estimated a power of approximately 0.2 for rs1490191 (MAF = 10% (dbSNP)) in the ALL group (n = 226). Although our study fortunately found some associated SNPs, our modest sample size likely resulted in our missing associations with other SNPs (type II error). To enhance the effect sizes and ensure the robustness of our findings, i.e., reduce type I error, we must collect more samples in an international multicenter study and must perform validation and replication. Another limitation is the significant difference between the HC and SZ groups in age (Table 1). Although we showed that the 17 SNPs, except for a SNP on 7p12.1 in the SZ group, remained statistically significant after linear regression analysis with age as an independent variable, analyses with age-matched controls are needed in future work. We suggested that three genes (THEM4, S100A10 and CDH22) are associated with eye movements. However, these associations are indirect. To support our results in future work, we need to identify whether the genes suggested in this study are associated with eye movements.
In conclusion, we have identified 17 potential susceptibility loci for HPG. Although these SNPs were found in non-coding regions, chromatin state analyses revealed that some of these SNPs were located in enhancer or promoter regions. Furthermore, eQTL analysis supported the finding that the SNPs in enhancer or promoter regions affected the expression levels of the surrounding genes. However, the sample size in our study is still too small to find established SNPs. In the future, an independent replication test should be performed to ensure the robustness of our findings.
Data availability
The datasets generated during and/or analyzed during the current study are not publicly available because they contain information that could compromise research participant privacy/consent but are available from the corresponding author on reasonable request.
References
van Os, J. & Kapur, S. Schizophrenia. Lancet 374, 635–45 (2009).
Menezes, N. M. & Milovan, E. First-episode psychosis: a comparative review of diagnostic evolution and predictive variables in adolescents versus adults. Can J Psychiatry 45, 710–716 (2000).
Werry, J. S., McClellan, J. M. & Chard, L. Childhood and Adolescent Schizophrenic, Bipolar, and Schizoaffective Disorders: A Clinical and Outcome Study. J. Am. Acad. Child Adolesc. Psychiatry 30, 457–465 (1991).
Clementz, B. A. & Sweeney, J. A. Is Eye Movement Dysfunction a Biological Marker for Schizophrenia? A Methodological Review. Psychol. Bull. 108, 77–92 (1990).
O’Driscoll, G. A. & Callahan, B. L. Smooth pursuit in schizophrenia: a meta-analytic review of research since 1993. Brain Cogn. 68, 359–70 (2008).
Levy, D. L., Sereno, A. B., Gooding, D. C. & O’Driscoll, G. A. Eye tracking dysfunction in schizophrenia: characterization and pathophysiology. Curr. Top. Behav. Neurosci. 4, 311–47 (2010).
Fukushima, J. et al. Disturbances of voluntary control of saccadic eye movements in schizophrenic patients. Biol. Psychiatry 23, 670–7 (1988).
Barton, J. J. S., Pandita, M., Thakkar, K., Goff, D. C. & Manoach, D. S. The relation between antisaccade errors, fixation stability and prosaccade errors in schizophrenia. Exp. brain Res. 186, 273–82 (2008).
Gooding, D. C. & Basso, M. A. The tell-tale tasks: a review of saccadic research in psychiatric patient populations. Brain Cogn. 68, 371–90 (2008).
Kojima, T. et al. Eye movements in acute, chronic, and remitted schizophrenics. Biol. Psychiatry 27, 975–89 (1990).
Kojima, T. et al. Stability of exploratory eye movements as a marker of schizophrenia–a WHO multi-center study. World Health Organization. Schizophr. Res. 52, 203–13 (2001).
Bestelmeyer, P. E. G. et al. Global visual scanning abnormalities in schizophrenia and bipolar disorder. Schizophr. Res. 87, 212–22 (2006).
Benson, P. J., Leonards, U., Lothian, R. M. St, Clair, D. M. & Merlo, M. C. G. Visual scan paths in first-episode schizophrenia and cannabis-induced psychosis. J. Psychiatry Neurosci. 32, 267–74 (2007).
Arolt, V., Teichert, H. M., Steege, D., Lencer, R. & Heide, W. Distinguishing schizophrenic patients from healthy controls by quantitative measurement of eye movement parameters. Biol Psychiatry. 44, 448–58 (1998).
Benson, P. J. et al. Simple viewing tests can detect eye movement abnormalities that distinguish schizophrenia cases from controls with exceptional accuracy. Biol Psychiatry. 72, 716–24 (2012).
Miura, K. et al. An integrated eye movement score as a neurophysiological marker of schizophrenia. Schizophr Res. 160, 228–9 (2014).
Morita, K. et al. Eye movement as a biomarker of schizophrenia: Using an integrated eye movement score. Psychiatry Clin Neurosci. 71, 104–14 (2017).
Takahashi, S. et al. Impairment of exploratory eye movement in schizophrenia patients and their siblings. Psychiatry Clin. Neurosci. 62, 487–493 (2008).
Ettinger, U. et al. Smooth pursuit and antisaccade eye movements in siblings discordant for schizophrenia. J. Psychiatr. Res. 38, 177–184 (2004).
Greenwood, T. A. et al. Initial heritability analyses of endophenotypic measures for schizophrenia: The Consortium on the Genetics of Schizophrenia. Arch. Gen. Psychiatry 64, 1242–50 (2007).
Katsanis, J., Taylor, J., Iacono, W. G. & Hammer, M. A. Heritability of different measures of smooth pursuit eye tracking dysfunction: a study of normal twins. Psychophysiology 37, 724–30 (2000).
Hong, L. E. et al. Familial aggregation of eye-tracking endophenotypes in families of schizophrenic patients. Arch. Gen. Psychiatry 63, 259–264 (2006).
Bae, J. S. et al. Genetic association analysis of ERBB4 polymorphisms with the risk of schizophrenia and SPEM abnormality in a Korean population. Brain Res. 1466, 146–151 (2012).
Cheong, H. S. et al. Association of RANBP1 haplotype with smooth pursuit eye movement abnormality. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 156, 67–71 (2011).
Park, B. L. et al. Association analysis of COMT polymorphisms with schizophrenia and smooth pursuit eye movement abnormality. J. Hum. Genet. 54, 709–712 (2009).
Shin, H. D. et al. Association of ZDHHC8 polymorphisms with smooth pursuit eye movement abnormality. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 153, 1167–1172 (2010).
Schmechtig, A. et al. Association of Neuregulin 1rs3924999 genotype with antisaccades and smooth pursuit eye movements. Genes, Brain Behav. 9, 621–627 (2010).
Takahashi, S. et al. Significant linkage to chromosome 22q for exploratory eye movement dysfunction in schizophrenia. Am. J. Med. Genet. 123B, 27–32 (2003).
Ma, Y. et al. Association of chromosome 5q21.3 polymorphisms with the exploratory eye movement dysfunction in schizophrenia. Sci. Rep. 5, 10299 (2015).
Miyawaki, T. et al. The endogenous inhibitor of Akt, CTMP, is critical to ischemia-induced neuronal death. Nat. Neurosci. 12, 618–626 (2009).
Sanders, A. R. et al. Transcriptome study of differential expression in schizophrenia. Hum. Mol. Genet. 22, 5001–5014 (2013).
Arikkath, J. & Reichardt, L. F. Cadherins and catenins at synapses: roles in synaptogenesis and synaptic plasticity. Trends in Neurosciences 31, 487–494 (2008).
Mei, L. & Xiong, W.-C. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat. Rev. Neurosci. 9, 437–452 (2008).
Stephan, K. E., Friston, K. J. & Frith, C. D. Dysconnection in Schizophrenia: From abnormal synaptic plasticity to failures of self-monitoring. Schizophrenia Bulletin 35, 509–527 (2009).
Hashimoto, R. et al. Genome-wide association study of cognitive decline in schizophrenia. Am J Psychiatry. 170, 683–4 (2013).
Hashimoto, R. et al. Common variants at 1p36 are associated with superior frontal gyrus volume. Transl. Psychiatry 4, e472 (2014).
Ohi, K. et al. Glutamate Networks Implicate Cognitive Impairments in Schizophrenia: Genome-Wide Association Studies of 52 Cognitive Phenotypes. Schizophr. Bull. 41, 909–918 (2015).
Ohi, K. et al. Genetic risk variants of schizophrenia associated with left superior temporal gyrus volume. Cortex. 58, 23–6 (2014).
Ohi, K. et al. Polygenetic Components for Schizophrenia, Bipolar Disorder and Rheumatoid Arthritis Predict Risk of Schizophrenia. Schizophr Res. 175, 226–9 (2016).
Fujino, H. et al. Performance on the Wechsler Adult Intelligence Scale-III in Japanese patients with schizophrenia. Psychiatry Clin Neurosci 68, 534–541 (2014).
Fukumoto, M. et al. Relation between remission status and attention in patients with schizophrenia. Psychiatry Clin. Neurosci. 68, 234–241 (2014).
Inada, T. & Inagaki, A. Psychotropic dose equivalence in Japan. Psychiatry Clin. Neurosci. 69, 440–447 (2015).
Japanese WAIS-III Publication Committee. Japanese Wechsler Adult Intelligence Scale, 3rd edn. Nihon Bunka Kagakusya, Tokyo (2006).
Matsuoka, K., Uno, M., Kasai, K., Koyama, K. & Kim, Y. Estimation of premorbid IQ in individuals with Alzheimer’s disease using Japanese ideographic script (Kanji) compound words: Japanese version of National Adult Reading Test. Psychiatry Clin. Neurosci. 60, 332–339 (2006).
Fujino, H. et al. Estimated Cognitive Decline in Patients With Schizophrenia: A Multicenter Study. Psychiatry Clin Neurosci. 60, 294–300 (2016).
Brainard, D. H. The psychophysics toolbox. Spat. Vis. 10, 433–6 (1997).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 38, 904–9 (2006).
Purcell, S. et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Ernst, J. & Kellis, M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–6 (2012).
Ernst, J. & Kellis, M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat. Biotechnol. 33, 364–376 (2015).
Ramasamy, A. et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 17, 1418–28 (2014).
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 125, 315–26 (2006).
Svenningsson, P. & Greengard, P. p11 (S100A10) - an inducible adaptor protein that modulates neuronal functions. Current Opinion in Pharmacology 7, 27–32 (2007).
Sugimoto, K. et al. Molecular cloning and characterization of a newly identified member of the cadherin family, PB-cadherin. J. Biol. Chem. 271, 11548–11556 (1996).
Kitajima, K., Koshimizu, U. & Nakamura, T. Expression of a novel type of classic cadherin, PB-cadherin in developing brain and limb buds. Dev. Dyn. 215, 206–214 (1999).
Pierrot-Deseilligny, C., Milea, D. & Müri, R. M. Eye movement control by the cerebral cortex. Curr Opin Neurol 17, 17–25 (2004).
Krauzlis, R. J. Recasting the smooth pursuit eye movement system. J. Neurophysiol. 91, 591–603 (2004).
Qiu, L. et al. Neuroanatomical circuitry associated with exploratory eye movement in schizophrenia: A voxel-based morphometric study. Plos One 6 (2011).
Emamian, E. S., Hall, D., Birnbaum, M. J., Karayiorgou, M. & Gogos, J. A. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet 36, 131–137 (2004).
Ohi, K. et al. The AKT1 gene is associated with attention and brain morphology in schizophrenia. World J Biol Psychiatry. 14, 100–13 (2013).
Petit, L. & Haxby, J. V. Functional anatomy of pursuit eye movements in humans as revealed by fMRI. J. Neurophysiol. 82, 463–71 (1999).
Rosano, C. et al. Pursuit and saccadic eye movement subregions in human frontal eye field: a high-resolution fMRI investigation. Cereb. Cortex 12, 107–15 (2002).
Tanaka, M. & Lisberger, S. G. Role of arcuate frontal cortex of monkeys in smooth pursuit eye movements. I. Basic response properties to retinal image motion and position. J. Neurophysiol. 87, 2684–99 (2002).
Craner, M. J. et al. Annexin II/PII is up-regulated in Purkinje cells in EAE and MS. Neuroreport 14, 555–558 (2003).
Robinson, F. R. & Fuchs, A. F. The Role of the Cerebellum in Voluntary Eye Movements. Annu. Rev. Neurosci. 24, 981–1004 (2001).
Dürsteler, M. R. & Wurtz, R. H. Pursuit and optokinetic deficits following chemical lesions of cortical areas MT and MST. J. Neurophysiol. 60, 940–65 (1988).
Acknowledgements
We appreciate the cooperation of all of the individuals who participated in this study. This work was supported by the Grants-in-Aid for Scientific Research (Grant Number 16K07222 (M.K. and A.N.), 17K15049 (M.K.), 25293250 (R.H.), 16H05375 (R.H.)) by MEXT. This work was supported by the Health and Labour Sciences Research Grants for Comprehensive Research on Persons with Disabilities and Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) from the Japan Agency for Medical Research and Development (AMED) (JP17dm0207006 and JP18dm0207006). The funders had no role in the study design, data collection and analyses, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
M.K. and R.H. contributed to study design. M.K., K. Miura and R.H. wrote the manuscript. M.K. analyzed genomic data. K. Miura analyzed eye movement tests. K. Miura and R.H. contributed to the interpretation of the data. M.K., K. Miura, K. Morita, H.Y., M.F., M.I., Y.Y., A.N. and R.H. contributed text to the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kikuchi, M., Miura, K., Morita, K. et al. Genome-wide Association Analysis of Eye Movement Dysfunction in Schizophrenia. Sci Rep 8, 12347 (2018). https://doi.org/10.1038/s41598-018-30646-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-30646-9
This article is cited by
-
On the Use of Eye Movements in Symptom Validity Assessment of Feigned Schizophrenia
Psychological Injury and Law (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.