Abstract
Human echovirus 12 (E-12) belongs to the enterovirus B species. To date, only one full-length genome sequence of E-12 (prototype strain Travis) is available in the GenBank database. This study determined the complete sequence of three E-12 strains, which were isolated from the stools of three healthy children in Yunnan, China, in 2013. We revealed that the three Yunnan E-12 strains had only 80.8–80.9% nucleotide identity and 96.4–96.8% amino acid identity with the Travis strain based on pairwise comparisons of the complete genome nucleotide and amino acid sequences. The three Yunnan strains shared 99.7% nucleotide identity and 99.1–99.5% amino acid similarity. Phylogenetic and similarity plot analyses showed that intertypic recombination occurred in the non-structural regions of the three Yunnan E-12 strains. This is the first report of the complete genome sequence of E-12 in China and it enriches the complete genome sequences of E-12 in the GenBank database.
Similar content being viewed by others
Introduction
Enteroviruses (EVs) are members of the genus Enterovirus, family Picornaviridae, and are further divided into 12 species: enterovirus A–H, J and rhinovirus A–C1, comprising more than 100 serotypes. EVs are single-stranded, positive, non-enveloped small RNA viruses. The genome of EV contains approximately 7,400 nucleotides including a long open reading frame (ORF). It encodes a polyprotein that is cleaved into 4 structural proteins (VP4, VP2, VP3, and VP1) and 7 non-structural proteins (2A, 2B, 2C, 3A, 3B, 3C, and 3D) flanked by a 5′-untranslated region (UTR) and a 3′-UTR1,2. EVs can cause a wide range of diseases, such as fever, hand-foot-and-mouth disease (HFMD), aseptic meningitis, acute flaccid paralysis (AFP), encephalitis, and myocarditis. However, most EV infections are asymptomatic or result in mild symptoms. Enterovirus B (EV-B) species include 6 coxsackievirus B (CV-B) serotypes, coxsackievirus A9 (CV-A9), 28 echovirus (E) serotypes, enterovirus B69 and 25 new types3.
Echoviruses are the major cause of aseptic meningitis4,5,6. They share a similar genome structure to other EVs. They are commonly associated with asymptomatic or mild infections but can also cause serious diseases such as AFP7. However, E-12 is not a prevalent EV serotype and was previously isolated through AFP surveillance8. There is currently only one genome sequence of E-12 in the GenBank database, which was isolated from the intestine of a patient with diarrhoea and aseptic meningitis9. In the present study, we report the complete genome sequences of three additional E-12 isolates that were recovered from three healthy children aged 3–6 years in the Yunnan Province of China in 2013. The determination and analysis of the complete genome sequence of E-12 will help to elucidate the origin and evolutionary characteristics of E-12 in China and provide beneficial information for related disease surveillance.
Results
Isolation and typing
The eight isolates (K603/YN/CHN/2013, K605/YN/CHN/2013, K606/YN/CHN/2013, K624/YN/CHN/2013, K629/YN/CHN/2013, K630/YN/CHN/2013, K646/YN/CHN/2013 and K1529/YN/CHN/2013) were isolated from KMB17 (human embryonic lung diploid fibroblast) cells. The eight strains were confirmed to be E-12 by the amplification of the partial VP1 sequences. The sequences were analysed using an online enterovirus genotyping tool. They were isolated from six males and two females with ages ranging from 3 to 6 years.
VP1 sequence analysis
The complete VP1 sequences of the eight Yunnan strains had the highest similarity with other E-12 strains in the GenBank database and had a percentage reaching 98%. The eight strains were identified as E-12 using an EV molecular typing criteria.
The eight isolates had 79.7–80.6% nucleotide and 93.8–96.3% amino acid similarity with the complete VP1 coding region of the E-12 prototype strain Travis. They shared 60.7–79.1% nucleotide and 60.5–85.2% amino acid similarity with the prototype of other EVs. The complete VP1 sequence and amino acid similarities among the eight Yunnan strains were 98.4–100% and 96.6–100%, respectively.
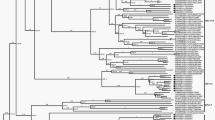
Because of the lack of a complete VP1 sequence in GenBank, 53 E-12 partial VP1 sequences (including the eight Yunnan isolates from the present study) were used for sequence analysis. Sequence analysis was based on the 657-nucleotide (positions 2617–3273) partial VP1 sequences of E-12 (Fig. 1). In alignment with all 53 E-12 VP1 sequences available in GenBank, the eight isolates had 81.4–95.0% nucleotide similarity with those of the E-12 strains, and all E-12 strains were divided into seven genotypes (A-G) according to the generally accepted criterion for enterovirus genotype demarcation (the nucleotide sequence variation in the VP1 region within an enterovirus genotype should be less than 11% and the nucleotide sequence variation in the same region between enterovirus genotypes should be more than 11%)10. Genotype A consisted of the eight isolates from the present study, the Yunnan isolated strains, alongside three strains from the Shandong Province of China and a Mongolian strain. The other four Yunnan strains isolated between 2008 and 2009 were clustered into genotype E. Three Indian E-12 strains belonged to genotype B, and genotype D was represented by the other four Indian strains. All of the strains from genotype C were from Madagascar. The French strain and prototype strain belonged to genotypes F and G, respectively. Estimates of the average evolutionary divergence of the sequence pairs within and between genotypes are shown in Table 1. The mean nucleotide (amino acid) sequence divergence within genotype A was 9.1% (1.0%), which was below the overall mean 17.5% (1.7%) cut-off divergence value and indicated the homology between the strains isolated in the study and other Chinese E-12 strains.
The phylogenetic tree based on partial VP1 sequences of global E-12 isolates. The triangle indicates E-12 isolates used in the present study and squares indicate other Chinese isolates. The circle indicates the prototype strain. The tree is constructed from the 657-nucleotide (positions 2617–3273) partial VP1 sequence.
Complete genome analysis
The complete genome sequences of three strains named K624/YN/CHN/2013, K605/YN/CHN/2013, and K1529/YN/CHN/2013 (abbreviated as K624, K605, and K1529, respectively) isolated from Yunnan Province in 2013 were determined. The three isolates were chosen from the east, north and south of Kunming, respectively. The complete genome nucleotide sequence and amino acid similarities among the three isolates were 99.7% and 99.1–99.5%, respectively. In addition, the complete VP1 nucleotide sequence and amino acid similarities among the eight Yunnan strains were 98.4–100% and 96.6–100%, respectively. Thus, we did not determine the sequence of the 5 remaining isolates in the other genomic regions and considered the five remaining isolates to be very similar to the three fully sequenced isolates.
The genome sequences among these three strains were found to be 7,417–7,424 nucleotides in length and included a single ORF of 6,579 nucleotides encoding a single polypeptide of 2,193 amino acids. The sequences were flanked by a 5′-UTR of 743 nucleotides and a 3′-UTR of 92–99 nucleotides. The complete genome nucleotide sequence and amino acid similarities among the three Yunnan isolates were 99.7% and 99.1–99.5%, respectively. Since the complete genome nucleotide homology of these three strains was as high as 99.7%, one of them (K624) was selected for detailed analysis.
The full-length genome of the K624 strain contains 7,424 nucleotides including a 5′-UTR of 743 nucleotides followed by an ORF that encoded a polyprotein of 2,193 amino acids and a 3′-UTR of 99 nucleotides. Nucleotide and deduced amino acid sequence identities between strain K624 and echovirus prototype strains are shown in Table 2. K624 had 80.9% similarity with the complete genome of the E-12 prototype strain and the deduced amino acid sequence had 96.8% similarity.
The K624 strain shares 80.8%, 82.3%, and 79.1% nucleotide identity with the P1, P2, and P3 coding regions of the E-12 prototype strain, respectively, and 67.7–72.7%, 79.3–82.4%, and 77.6–80.2% nucleotide identity with the P1, P2, and P3 regions of the other E prototype strains, respectively.
Phylogenetic analysis of P1, P2, and P3 coding regions
Phylogenetic analysis was based on the P1, P2, and P3 region of the three Yunnan isolates and all of the available EV-B strains (Fig. 2). In the P1 capsid coding region, three Yunnan isolates were clustered together with the E-12 prototype strain Travis (X77708), which is in consonance with the molecular typing results. In the P2 region, the Yunnan isolates did not form a lineage with the E-12 prototype strain and grouped together with other EV-B serotypes such as E-7 strain DH22G/JS/2012 and two CVB4 strains (CVB4/GX/10 and CVB4/BM24G/NM/CHN/20100). Among these, the nucleotide sequence of the P2 genomic region was the most homologous (85.5%) to the corresponding sequence of the E-7 strain DH22G/JS/2012. However, in the P3 region, the Yunnan isolates did not form a lineage with the E-12 prototype strain and grouped together with other EV-B serotypes, especially the E-6 strain E6SD11CHN, which was the most homologous (94.0%). From the above results, this suggests that one or more putative recombination events had occurred between the E-12 Yunnan isolates and other EV-B types.
Phylogenetic relationships based on the P1, P2, and P3 coding sequences of 381 EV-B strains. Three E-12 strains and 378 EV-B strains were analysed by nucleotide sequence alignment using the neighbour-joining algorithms implemented in the MEGA 6.06 program. The numbers at the nodes indicate bootstrap support for that node (percentage of 1,000 bootstrap replicates). The scale bars represent the genetic distance. Only high bootstrap values (>75%) are shown. ▲ indicates the strain isolated in this investigation, and ● indicates the prototype strain. The red nodes denote the other E-12 strains. The complete trees are available in Supplementary Fig. S1.
Recombination analysis
BLAST was used to select the available sequences with high homology to K624 (Table 3). The P2 and P3 regions of strain K624 had the highest identity with E6SD11CHN, whereas the Yunnan strain had the highest identities (94%-100%) with the CV-B4 strain CVB4/GX/10, CV-B5 strain CVB5/PMKA1315/THA/2011, and E-6 strain E6SD11CHN in the 3A, 3B, 3C and 3D coding regions of P3, respectively. Among these, the nucleotide sequence of the 3B genomic region was consistent with the corresponding sequence of CVB5/PMKA1315/THA/2011 (100%). In addition, the Yunnan strain had the highest identity (97% and 93%) with CV-B5 strain 19CSF and EV-79 strain NH95-0601 in the 3′-UTR and 5′-UTR regions, respectively.
The sequences with the highest homology were used to conduct the recombination analysis. The similarity plot analysis and bootscanning analysis both suggested multiple recombination events in the genomic sequence of strain K624/YN/CHN/2013 (Fig. 3). In the P1 coding region, strain K624/YN/CHN/2013 had the highest identity with the E-12 prototype strain. Whereas in the 2A to 2C coding regions, it was most closely related to the CV-B4 strain Cph1, E-30 strain KM/A363/09 and E-6 strain E6SD11CHN. In the 3A to 3D coding regions, it was most homologous with E-6 strain SD11/CHN and CV-B4 strain GX/10, respectively.
Similarity plots and bootscanning analysis of the three Chinese E-12 strains with closely related strains. The analyses were conducted via Simplot 3.5.1 using a sliding window of 200 nt moving in 20-nt steps. The genome of strain K624/YN/CHN/2013 (a), K605/YN/CHN/2013 (b), and K1529/YN/CHN/2013 (c) serves as a query sequence.
Discussion
Following the eradication of polio, diseases caused by non-polio enteroviruses (NPEVs) have been recognized as a major public health concern in Asian countries. Among EVs, polioviruses (PVs) are the main cause of AFP. Although PV has been eradicated in China due to an intensive oral polio vaccination (OPV) program, non-polio AFP cases have been reported. NPEV genotypes associated with AFP had been reported as E-11, E-12, EV-76, E-20, E-7, E-3, E-9, CV-B1 and CV-B311,12. However, most of them were collected from stool samples of AFP patients (EV-A, B, C and even D). This does not mean that these NPEVs were the causative agents of AFP. Other samples from the CNS, serum and other sites of lesions of the patients are required to elucidate the relationship.
Yunnan is not only a frontier province but also a tourist province. The population of Yunnan frequently exchanges with neighbouring Southeast or South Asian countries where AFP cases are often reported. Thus, to date, all Yunnan E-12 strains in the GenBank database accounted for the majority (41.7%) of the total isolates. In the study, all of the Yunnan strains were isolated from the stool samples of healthy populations. However, E-12 could also be isolated from the stools of patients with AFP, HFMD and diarrhoea13,14,15. Thus, the pathogenic potential of the Yunnan E-12 strains needs additional study. Furthermore, an epidemiological investigation of the association between E-12 and its related diseases needs to be conducted.
Except for four strains, the Yunnan E-12 isolates from the present study clustered with the other E-12 strains isolated from the Yunnan Province of China and these strains had the highest identity with each other, which indicated that the Yunnan strains had existed for a long time.
Recombination in enteroviruses has been reported as a major mechanism participating in the evolution of these viruses, especially EV-B16,17. We performed a preliminary screen with BLAST for strains that possessed a high similarity with the E-12 strains in different coding regions. Then, we proposed recombination events in different coding regions that were observed between the Yunnan isolates and the E-6 strain E6SD11CHN, CV-B4 strains (CVB4/GX/10 and CVB4/CB4 Cph1), EV-79 strain NH95-0601, CV-B5 strains (19CSF, CVB5/China/GD237/2009, and CVB5/PMKA1315/THA/2011), CV-A9 strain EBMSV0003/THA/2010, E-30 strain KM/A363/09, and E-12 strain 281-YN-CHN-2016HC. The results indicate that all of these viruses probably shared common ancestors that provided some non-structural genomic regions through recombination. Among them, the CV-A9 strain EBMSV0003/THA/2010 and the CV-B5 strain CVB5/PMKA1315/THA/2011 were isolated from patients with respiratory infections, respectively18; E6SD11CHN, CVB4/GX/10, and CVB5/China/GD237/2009 were isolated from patients with HFMD19,20,21; the CV-B5 strain 19CSF was isolated from patients with aseptic meningitis22. HFMD, which was first reported in New Zealand in 1957, is a common infectious disease mainly occurring in children under 5 years of age. Since 2008, HFMD has become a pressing issue for public health in China. It was incorporated into the C-class infectious diseases and has been ranked as one of the infectious diseases with the highest incidence in China for nearly 10 years. EVs are the causative agent of HFMD. CV-A16, EV-A71, and CV-A6 have been identified as the most frequent pathogens of HFMD, and other EVs, such as CV-B1, CV-B2, CV-B3, CV-B4 and CV-B5, can also cause HFMD23,24,25. In addition, although the surveillance system of aseptic meningitis is very limited, many studies have shown that EVs were the predominant pathogen of aseptic meningitis, and EVs associated with aseptic meningitis outbreaks often focus on EV-B serotypes such as E-30, E-6, CV-A9, CV-B3, and CV-B526,27,28,29. The causative agents for HFMD and aseptic meningitis often overlap. In addition, multi-serotypes were not only co-circulated and co-infected during HFMD outbreaks and sporadic cases or asymptomatic infections but also during cases of aseptic meningitis, especially EV-B strains30, and the epidemiologic feature may provide ample opportunity for the virus to mingle and recombine. Thus, we suggest that Yunnan strains co-circulated with other EV-B strains and underwent extensive genetic exchanges with a few of the EV-B strains.
In conclusion, this is the first report on the entire genome of E-12 in China. Sequence analysis indicates that the Yunnan E-12 strains have a high genetic diversity compared with the other E-12 strains and intertypic recombination has occurred in the non-structural regions. E-12 strains were reported in many countries, such as the Central African Republic, Cameroon, India, Nigeria, Romania, Pakistan, and the USA. Therefore, E-12 is probably widespread (worldwide), and multiple recombination events may drive the evolution of E-12. Although these Yunnan E-12 strains were isolated from healthy children, its pathogenic potential cannot be excluded and its pathogenicity needs further research and study. Epidemiological surveillance should continue to evaluate the association between E-12 and its related diseases.
Materials and Methods
Ethics statement
Parents and/or legal guardians of the participates in this study gave written informed consent. The protocol was in accordance with the Helsinki Declaration and was approved by the Institutional Ethics Boards of the Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College. The human materials in the study were stool samples collected from eight healthy children aged 3–6 years.
Sample collection and virus isolation
A total of 96 stool samples were collected from three kindergartens from June to August in 2013 in Kunming, China. The three kindergartens are located in the east, north and south of Kunming. The children were asymptomatic and did not present a fever, cough, or rash and did not have contact with patients recently infected by EVs in the 2 weeks before sample collection. The children’s ages ranged from 3 to 6 years31. The cell line, KMB17, was used to isolate viruses from the stool specimens according to the standard procedures32. The samples that induced a cytopathic effect (CPE) were considered positive. All of the positive isolates were stored at −80 °C.
VP1 RT-PCR, sequencing, and typing
Viral RNA was extracted from the cell culture supernatants with a QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA, USA) in accordance with the manufacturer’s instructions. The enterovirus universal primer pairs 222 and 224 were used to amplify the partial VP1 gene sequence33 using a PrimeScriptTM One-Step RT-PCR Kit Ver.2 (Takara, Dalian, China). The positive PCR products were sequenced using an ABI 3730XL automatic sequencer (Applied Biosystems, Foster City, CA, USA) at the Tsingke Sequencing Company (Kunming, China).
The partial VP1 nucleotide sequence (nucleotide position 2617–3273) was analysed using the Enterovirus Genotyping Tool for serotyping34. Virus strains showing >75% nucleotide sequence identity with a known enterovirus serotype were designated the relative serotype of EVs. Estimates of average evolutionary divergence of the sequence pairs within and between E-12 genotypes were performed using MEGA6 (Table 1). The analyses were conducted using the Tamura-Nei model. The rate variation among the sites was modelled with a gamma distribution (shape parameter = 1).
Whole genomic sequencing
Primers for amplifying the complete genome were designed using the published sequence of the Travis strain and internal primers were designed for the ‘primer walking’ strategy as previously reported35. The positive PCR products were sequenced using an ABI 3730XL automatic sequencer (Applied Biosystems, Foster City, CA, USA) at the Tsingke Sequencing Company. The primers for PCR amplification and sequencing are listed in Table 4. The 5′ extremity of the three genomes was not sequenced by RACE RT-PCR or circularization.
Sequence analysis and recombination analysis
BioEdit (version 7.2.3) was used for nucleotide and amino acid sequence alignment. The phylogeny of the E-12 strains and other members of the species EV-B strains was conducted via MEGA (version 7.0)36 and generated by the neighbour-joining method with a Kimura two-parameter model. Bootstrapping was performed with 1000 duplicates and the bootstrap values greater than 75% were considered statistically significant for grouping. The Simplot 3.5.1 software, which was programmed with a 200-nt window moving in 20-nt steps and a Jukes-Cantor correction37, was used for the similarity plot and bootscanning analysis. The highest similarity of the nucleotide sequences of enteroviruses in all of the sequenced genomic regions of the K624/YN/CHN/2103 strain using BLAST online is shown in Table 3.
Nucleotide sequence accession number
The accession numbers of the complete VP1 nucleotide sequences and full-length genome sequences of the E-12 strains identified in this study were as follows: MF083144-MF083151 and MF083152-MF083154, respectively.
References
Song, Y. et al. Phylogenetic Characterizations of Highly Mutated EV-B106 Recombinants Showing Extensive Genetic Exchanges with Other EV-B in Xinjiang, China. Sci Rep-Uk. 7, 43080, https://doi.org/10.1038/srep43080 (2017).
Nasri, D. et al. Basic Rationale, Current Methods and Future Directions for Molecular Typing of Human Enterovirus. Expert Rev Mol Diagn. 7, 419–434, https://doi.org/10.1586/14737159.7.4.419 (2007).
Zhou, F. et al. Use of the 5′ Untranslated Region and VP1 Region to Examine the Molecular Diversity in Enterovirus B Species. J Med Microbiol. 63, 1339–1355, https://doi.org/10.1099/jmm.0.074682-0 (2014).
Trallero, G. et al. Enteroviruses in Spain Over the Decade 1998–2007: Virological and Epidemiological Studies. J Clin Virol. 47, 170–176, https://doi.org/10.1016/j.jcv.2009.11.013 (2010).
Cabrerizo, M. et al. Molecular Characterization of Enteroviruses Associated with Neurological Infections in Spain, 2008. J Med Virol. 85, 1975–1977, https://doi.org/10.1002/jmv.23693 (2013).
Li, W. et al. Molecular Epidemiological Study of Enteroviruses Associated with Encephalitis in Children From Hangzhou, China. Medicine. 95, e4870, https://doi.org/10.1097/MD.0000000000004870 (2016).
Odoom, J. K. et al. Human Enteroviruses Isolated During Acute Flaccid Paralysis Surveillance in Ghana: Implications for the Post Eradication Era. Pan Afr Med J. 12, 74 (2012).
Tang, J. et al. Molecular Epidemiology and Recombination of Human Enteroviruses From AFP Surveillance in Yunnan, China From 2006 to 2010. Sci Rep-Uk. 4, 6058, https://doi.org/10.1038/srep06058 (2015).
Kraus, W., Zimmermann, H., Zimmermann, A., Eggers, H. J. & Nelsen-Salz, B. Infectious cDNA Clones of Echovirus 12 and a Variant Resistant Against the Uncoating Inhibitor Rhodanine Differ in Seven Amino Acids. J Virol. 69, 5853–5858 (1995).
Abdelkhalek, I. et al. Molecular Epidemiology of Coxsackievirus Type B1. Arch Virol. 160, 2815–2821, https://doi.org/10.1007/s00705-015-2561-5 (2015).
Junttila, N. et al. New Enteroviruses, EV-93 and EV-94, Associated with Acute Flaccid Paralysis in the Democratic Republic of the Congo. J Med Virol. 79, 393–400, https://doi.org/10.1002/jmv.20825 (2007).
Shaukat, S. et al. Characterization of a Novel Enterovirus Serotype and an Enterovirus EV-B93 Isolated from Acute Flaccid Paralysis Patients. Plos One. 8, e80040, https://doi.org/10.1371/journal.pone.0080040 (2013).
Bingjun, T. et al. Molecular Typing and Epidemiology of Non-Polio Enteroviruses Isolated From Yunnan Province, the People’s Republic of China. J Med Virol. 80, 670–679, https://doi.org/10.1002/jmv.21122 (2008).
Fernandez-Garcia, M. D., Kebe, O., Fall, A. D. & Ndiaye, K. Identification and Molecular Characterization of Non-Polio Enteroviruses From Children with Acute Flaccid Paralysis in West Africa, 2013-2014. Sci Rep. 7, 3808, https://doi.org/10.1038/s41598-017-03835-1 (2017).
Rao, D. C. et al. Non-Polio Enteroviruses and their Association with Acute Diarrhea in Children in India. Infect Genet Evol. 17, 153–161, https://doi.org/10.1016/j.meegid.2013.04.011 (2013).
Zhang, J. et al. Molecular Characterization of a New Human Coxsackievirus B2 Associated with Severe Hand-Foot-Mouth Disease in Yunnan Province of China in 2012. Arch Virol. 162, 307–311, https://doi.org/10.1007/s00705-016-3075-5 (2017).
Zheng, H. et al. Isolation and Characterization of a Highly Mutated Chinese Isolate of Enterovirus B84 from a Patient with Acute Flaccid Paralysis. Sci Rep-Uk. 6, 31059, https://doi.org/10.1038/srep31059 (2016).
Zhou, Y. et al. Metagenomics Study of Viral Pathogens in Undiagnosed Respiratory Specimens and Identification of Human Enteroviruses at a Thailand Hospital. Am J Trop Med Hyg. 95, 663–669, https://doi.org/10.4269/ajtmh.16-0062 (2016).
De, W. et al. A Large Outbreak of Hand, Foot, and Mouth Disease Caused by EV71 and CAV16 in Guangdong, China, 2009. Arch Virol. 156, 945–953, https://doi.org/10.1007/s00705-011-0929-8 (2011).
Tang, J. et al. Complete Genome Analysis of an Enterovirus EV-B83 Isolated in China. Sci Rep-Uk. 6, 29432, https://doi.org/10.1038/srep29432 (2016).
Zhang, T. et al. Epidemics and Frequent Recombination within Species in Outbreaks of Human Enterovirus B-Associated Hand, Foot and Mouth Disease in Shandong China in 2010 and 2011. Plos One. 8, e67157, https://doi.org/10.1371/journal.pone.0067157 (2013).
Ma, H. et al. Recombination in Human Coxsackievirus B5 Strains that Caused an Outbreak of Viral Encephalitis in Henan, China. Arch Virol. 158, 2169–2173, https://doi.org/10.1007/s00705-013-1709-4 (2013).
Li, Y. et al. Epidemiological and Genetic Analysis Concerning the Non-Enterovirus 71 and Non-Coxsackievirus A16 Causative Agents Related to Hand, Foot and Mouth Disease in Anyang City, Henan Province, China, From 2011 to 2015. J Med Virol. 89, 1749–1758, https://doi.org/10.1002/jmv.24847 (2017).
McMinn, P. C. An Overview of the Evolution of Enterovirus 71 and its Clinical and Public Health Significance. Fems Microbiol Rev. 26, 91–107 (2002).
Tian, X. et al. New Coxsackievirus B4 Genotype Circulating in Inner Mongolia Autonomous Region, China. Plos One. 9, e90379, https://doi.org/10.1371/journal.pone.0090379 (2014).
Chen, P. et al. A Coxsackievirus B5-associated Aseptic Meningitis Outbreak in Shandong Province, China in 2009. J Med Virol. 85, 483–489, https://doi.org/10.1002/jmv.23478 (2013).
Liu, N. et al. An Outbreak of Aseptic Meningitis Caused by a Distinct Lineage of Coxsackievirus B5 in China. Int J Infect Dis. 23, 101–104, https://doi.org/10.1016/j.ijid.2014.02.005 (2014).
Milia, M. G. et al. Recent Outbreak of Aseptic Meningitis in Italy Due to Echovirus 30 and Phylogenetic Relationship with Other European Circulating Strains. J Clin Virol. 58, 579–583, https://doi.org/10.1016/j.jcv.2013.08.023 (2013).
Zhao, Y. N., Jiang, Q. W., Jiang, R. J., Chen, L. & Perlin, D. S. Echovirus 30, Jiangsu Province, China. Emerg Infect Dis. 11, 562–567, https://doi.org/10.3201/eid1104.040995 (2005).
Park, S. K. et al. Transmission of Seasonal Outbreak of Childhood Enteroviral Aseptic Meningitis and Hand-Foot-Mouth Disease. J Korean Med Sci. 25, 677, https://doi.org/10.3346/jkms.2010.25.5.677 (2010).
Sun, H. et al. Molecular Evolution of Two Asymptomatic Echovirus 6 Strains that Constitute a Novel Branch of Recently Epidemic Echovirus 6 in China. Virol J. 14, 140–149, https://doi.org/10.1186/s12985-017-0809-2 (2017).
Prim, N. et al. Combining Cell Lines to Optimize Isolation of Human Enterovirus From Clinical Specimens: Report of 25 Years of Experience. J Med Virol. 85, 116–120, https://doi.org/10.1002/jmv.23426 (2013).
Nix, W. A., Oberste, M. S. & Pallansch, M. A. Sensitive, Seminested PCR Amplification of VP1 Sequences for Direct Identification of All Enterovirus Serotypes From Original Clinical Specimens. J Clin Microbiol. 44, 2698–2704, https://doi.org/10.1128/JCM.00542-06 (2006).
Kroneman, A. et al. An Automated Genotyping Tool for Enteroviruses and Noroviruses. J Clin Virol. 51, 121–125, https://doi.org/10.1016/j.jcv.2011.03.006 (2011).
Kaczorowski, T. & Szybalski, W. Genomic DNA Sequencing by SPEL-6 Primer Walking Using Hexamer Ligation. Gene. 223, 83–91 (1998).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33, 1870–1874, https://doi.org/10.1093/molbev/msw054 (2016).
Lole, K. S. et al. Full-Length Human Immunodeficiency Virus Type 1 Genomes From Subtype C-infected Seroconverters in India, with Evidence of Intersubtype Recombination. J Virol. 73, 152–160 (1999).
Acknowledgements
This work was supported by the Basic Research Projects of Yunnan Province, China (2017FA006) and the Chinese Academy of Medical Sciences Initiative for Innovative Medicine (2016-I2M-3-026).
Author information
Authors and Affiliations
Contributions
H.L., J.Z., Z.Y. and S.M. conceived the study and drafted the paper. Y.Z., H.Z. and X.H. performed the experiments. K.L. and H.S. helped to interpret results and contributed to the writing. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, H., Zhang, J., Zhao, Y. et al. Molecular characterization of echovirus 12 strains isolated from healthy children in China. Sci Rep 8, 11716 (2018). https://doi.org/10.1038/s41598-018-30250-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-30250-x
This article is cited by
-
Genetic Characteristics and Phylogeographic Dynamics of Echovirus
Journal of Microbiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.