Abstract
One of the most important factors driving amphibian declines worldwide is the infectious disease, chytridiomycosis. Two fungi have been associated with this disease, Batrachochytrium dendrobatidis and B. salamandrivorans (Bsal). The latter has recently driven Salamandra salamandra populations to extirpation in parts of the Netherlands, and Belgium, and potentially also in Germany. Bsal has been detected in the pet trade, which has been hypothesized to be the pathway by which it reached Europe, and which may continuously contribute to its spread. In the present study, 918 amphibians belonging to 20 captive collections in Germany and Sweden were sampled to explore the extent of Bsal presence in captivity. The fungus was detected by quantitative Polymerase Chain Reaction (qPCR) in ten collections, nine of which lacked clinical symptoms. 23 positives were confirmed by independent processing of duplicate swabs, which were analysed in a separate laboratory, and/or by sequencing ITS and 28 S gene segments. These asymptomatic positives highlight the possibility of Bsal being widespread in captive collections, and is of high conservation concern. This finding may increase the likelihood of the pathogen being introduced from captivity into the wild, and calls for according biosecurity measures. The detection of Bsal-positive alive specimens of the hyper-susceptible fire salamander could indicate the existence of a less aggressive Bsal variant or the importance of environmental conditions for infection progression.
Similar content being viewed by others
Introduction
Modern amphibians (Lissamphibia), the oldest class of extant tetrapods1, have inhabited Earth since the Triassic and have survived several of the major mass extinction events2. However, they are presently one of the animal groups most threatened with extinction3,4. In 2017, the Global Amphibian Assessment revealed that 41% of the 6,609 species reviewed, showed evidence of decline, and 35 were already extinct5. While habitat degradation and destruction still appear to be the main culprits6, an emerging infectious disease, chytridiomycosis, is currently considered another major factor7, having led to numerous declines and extinctions worldwide8,9,10,11. Chytridiomycosis is the manifestation of an infection with one of two pathogens: Batrachochytrium dendrobatidis (Bd;12,13), a fungus that lethally infects many frogs (Anura)12,14,15 but also salamanders (Caudata)16 and caecilians (Gymnophiona)17; and B. salamandrivorans (Bsal;10) that appears to lethally affect only salamanders18,19. The clinical symptoms caused by these pathogens involve excessive skin shedding, and reddened or discoloured skin which, in Bd, leads to erythema and malfunctions in respiration and osmoregulation12,20,21; and, in Bsal, to deep epidermal ulcerations which are subsequently colonized by other microorganisms10. Several studies have explored methods to clear chytrid infections, with heat treatments and antifungals among the viable options for both pathogens22,23.
The chytrid fungus Bd has been detected in several parts of Europe24,25,26, inducing population declines in some cases but not leading to massive declines in the majority of species (with the exception of Alytes midwife toads in Spain27). Bsal was first detected as the cause of a mass-mortality event in fire salamanders (Salamandra salamandra) in the Netherlands10,28, but has now been detected also in Belgium and western Germany, causing mass mortality in fire salamanders but also infecting various newt species29. Currently, the potential spread of Bsal30 is one of the most pressing conservation challenges not only for European, but also for the conservation of the global salamander diversity19.
The import of newt and salamander species from Asia as pets, for research, or for zoological collections, is the hypothesized route of introduction of Bsal into European wildlife18,31. Therefore, captive collections of amphibians have the potential to be particularly relevant and potentially dangerous pathways for the further spread of Bsal into the wild. Currently, Bsal has been detected in captive collections in the United Kingdom and in Germany32,33, but not in the United States34.
After Bsal was demonstrated to occur in captive salamanders32, we carried out an additional study to assess the extent of occurrence of this pathogen in captive amphibian collections mainly within Germany. Here, we present the results of this study, based on 918 skin swabs taken from individual amphibians from captive collections screened for Bd and Bsal using quantitative PCR (qPCR), with numerous Bsal positives confirmed by direct sequencing and/or independent analysis of duplicate swabs in a second laboratory.
Methods
Our study is based on samples taken by the private collection owners for routine health screenings. The owners of the collections check the individuals regularly for disease signs, such as skin ulcerations, lethargy, abnormal skin shedding and death. With the exception of the collection that experienced mass dying to a Bsal infection (Collection 1) at an earlier time point32, none of the clinical signs were observed in any other collection. In order to maintain the anonymity of collection owners, precise locations and species composition of the collections are not provided.
A total of 918 samples belonging to 111 species (11 Anura and 100 Caudata) and 46 subspecies (all Caudata) from 20 private captive amphibian collections were sampled between May of 2015 and May of 2016 (Supplementary Table 1 and Table 1). Eighteen of the collections were located in Germany (across nine federal states) and two in Sweden. All collection owners are members of the AG Urodela (‘Arbeitsgemeinschaft’ der Deutschen Gesellschaft für Herpetologie und Terrarienkunde e.V.), a working group of the German Society of Herpetology and Herpetoculture (DGHT). Members of the DGHT receive formation and guidelines on several aspects related to amphibians, and more specifically salamanders, including housing and how to carry out health checks. Housing conditions of amphibians varied among collections, with most terrestrial species being kept in groups of three or four individuals within glass terraria, while aquatic species were kept at higher densities, up to approximately 30 individuals. Enclosures ranged from 20 × 25 × 40 cm to 40 × 40 × 80 cm. Terrestrial and aquatic enclosures were equipped with access to hiding places.
From all captive collections, two individuals from every enclosure were sampled. Each individual was handled with clean nitrile gloves and its ventral surface was rubbed 10 times, simultaneously with two sterile rayon swabs (MW113; Medical Wire & Equipment, Corsham, UK). Each of these two swabs of an individual was kept separately in a sterile 1.5 ml centrifuge tube and stored at −20 °C until DNA extraction. A specific protocol for sampling that has been followed by breeders can be found in the supplementary materials. Additionally, one breeder (Collection 2) provided a clean swab as a sampling control, unfortunately other breeders failed to do so. The negative control was collected half way through the sampling event by opening a clean swab and placing it in a centrifuge tube. Three collections – also including Collection 1 - in which Bsal was detected were heat treated following established methods23,35. Individuals from this collection were re-sampled after the heat treatment following the same protocol as described above.
One swab per sampled individual was analysed in the laboratory of the Technical University Braunschweig (Braunschweig, Germany). Genomic DNA was extracted from the swabs using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s Animal Tissues protocol with Pretreatment for Gram-Positive Bacteria. Incubation for the initial enzymatic lysis was extended to 1 h, and the temperature of the proteinase K lysis was increased to 70 °C to increase DNA yield36. One sterile swab was included in every third round of extraction as a negative control, with approximately 50 rounds being run.
For the samples that were found to be positive, DNA from the duplicate swab was extracted and analyzed using the same method but by a different person in the laboratory of Trier University (Trier, Germany), a laboratory accredited for the European Bsal initiative37. For nine samples for which no duplicate swab was available, such a double check was not possible. As an additional control, nine negatives were also analyzed in Trier, all of which were negative.
In both laboratories, qPCRs amplified a region of the ITS rRNA (120 bp), following a standard protocol38 with one alteration: the use of KlearKall Master Mix (LGC genomics, Middlesex, UK). Quantitative PCRs were performed on a CFX96 Real-Time System (Bio-Rad Laboratories Inc., Hercules, CA) in Braunschweig, and on a StepOnePlus (Applied Biosystems, Foster City, CA) in Trier. Each sample was run in duplicate in Braunschweig; when replicates of a sample showed contradictory results, a third replicate was run. Samples analyzed at Trier University were each run in triplicate. Each qPCR plate in Braunschweig had two replicates of Bd and Bsal standards (0.1–1,000 zoospores) and two negative controls, while in Trier each plate had Bsal standards in triplicate (0.1–1,000 zoospores) and three negative controls. Quantitative PCR amplification signals were only considered as positives when the signal was between the highest (1,000 zoospores) and the lowest standard (0.1 zoospores) and when the amplification curve was logarithmic. The estimated zoospore equivalents were converted to number of zoospores per swab (hereafter z/s) based on the extraction volume used. Loads were not normally distributed, so median and 75% inter quartile range (IQR) are reported.
Because our sampling approach involves two different swabs being taken from the same individual, one to be analysed initially in a laboratory, and the second one being analysed in a different laboratory to confirm the observations when the first one is positive, it is particularly robust against contamination during DNA extraction and subsequent manipulation of DNA templates (see Discussion). However, due to the lack of negative controls from the majority of breeders we cannot exclude contamination during sampling process. It must be emphasized that our approach also implies that differences among the results (but not confirmed by qPCR replication in a second laboratory) in the two laboratories can be expected, as with low infection loads, the amount of pathogen DNA might differ among the swabs. We considered samples as positive for Bsal, if they were positive in both laboratories; unconfirmed positive when they were only positive in one laboratory, and negative when they were negative in the first laboratory (only nine negative samples were repeated in the second laboratory, all of which were negative). For Bd, samples found positive in the first laboratory were not further tested, as our focus was on Bsal; thus, all Bd results are reported as unconfirmed positives.
From all Bsal-positive samples, stretches of DNA were sequenced to confirm its genetic identity with the Bsal type strain (Supplementary Table 3). Two regions were amplified, the ITS (Short: forward: 5′–TGC TCC ATC TCC CCC TCT TCA–3′, and reverse: 5′–TGA ACG CAC ATT GCA CTC TAC–3′; Long: forward: 5′–CAA CGG ATC TCT TGG CTC TC–3′, and reverse: 5′–GGT TTG CCT TAA TTT CAT AAT GG–3′) and the 28 S (forward: 5′-ACG CTT GAA ACC AGT ATT GAG TG–3′, and reverse: 5′–TAC AGC TGC GTT CCT CAG TC–3′) regions. The ITS short38 and the 28 S39 primers were as described in literature, while the ITS long was designed by us. Samples were enzymatically cleaned with ExoSap and sequenced using standard approaches on an ABI 3730xl capillary sequencer. Additionally negative controls were included to control for sequencing contamination. Sequences were controlled for quality by checking the chromatograms and were manually corrected when necessary in CodonCode Aligner. Subsequently sequences were blasted in the National Center for Biotechnology Information platform (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The majority of samples yielded PCR products and could be successfully sequenced for at least one of these gene segments.
Results
Of the 918 samples collected, 13 (1.4%) were unconfirmed positive for Bd, 23 (2.5%) were positive for Bsal, and 18 (2.0%) were unconfirmed positive for Bsal, with no co-occurrence of both pathogens in the same individual. Out of 28 of the positives (68%), 16 remained positive (39.0%) and 12 as unconfirmed positive (29.3%) after double checked at the Trier University. For all of these samples, DNA sequences matching Bsal were obtained (see below). The sampling control (a clean swab provided by breeder of Collection 2), the extraction controls, the qPCR controls and the sequencing controls were all negative.
None of the collections showed mortality or clinical signs of Bd or Bsal infection, including Collection 1 that had a Bsal-induced mortality event six months before for its Salamandra species32.
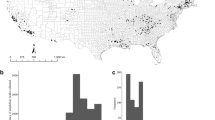
Both pathogens (Bd and Bsal) were detected in Germany (Fig. 1). The Bsal positives in Sweden could not be confirmed since no duplicate swab was available. Out of 20 collections sampled, six (30%) showed unconfirmed positive signals for Bd and nine (45%) showed positive and unconfirmed positive signals for Bsal (positive - 6 (30%); unconfirmed positive - 3 (15%)), with three private collections showing unconfirmed positive qPCR signals for both Bd and Bsal (Table 1). The median (IQR) loads (excluding the zeros of samples with three replicates) for Bd and Bsal were 101.1 (19.2) and 105.0 (965.8) (positive - 211.2 (1094.4); unconfirmed positive - 5.7 (283.3)) z/s, respectively.
Distribution of positives and unconfirmed positive samples for both chytrid fungi, Batrachochytrium dendrobatidis (Bd) and B. salamandrivorans (Bsal), in captive collections from Germany and Sweden. Europe map (“leere Grundkarte von Europa”) was created by Ktrinko, is under Creative Commons Attribution-Share Alike 3.0 Unported license (https://creativecommons.org/licenses/by-sa/3.0/deed.en), can be found on https://commons.wikimedia.org/wiki/File:EuropaleereKarte.svg. Germany map (“Map of States of Germany (Area States & City States)”) was created by Roman Poulvas and David Liuzzo, is under Creative Commons Attribution-Share Alike 2.0 Germany license (https://creativecommons.org/licenses/by-sa/2.0/de/deed.en), can be found on https://en.wikipedia.org/wiki/File:Map_of_Lands_of_Germany_(Area_States_and_City_States).svg. Both maps were re-colored and elements (such as pie charts, legend and scale) added.
Out of the 157 species and subspecies sampled, 11 (7.0%, three Anura and eight Caudata) showed unconfirmed positive signals for Bd, and 20 (12.7%, all Caudata) showed positive and unconfirmed positive signals for Bsal (positive − 12 (7.6%); unconfirmed positive – 8 (5.1%)) (Table 2). Batrachochytrium dendrobatidis was found (but not confirmed by qPCR replication in a second laboratory) in the following genera: Alytes, Bombina and Gastrotheca (Anura), and Ichthyosaura, Neurergus, Salamandra, Salamandrella, Siren, Taricha and Triturus (Caudata). Salamanders with Bsal-positives or unconfirmed positives belonged to the genera: Ambystoma (Ambytomatidae), Cynops, Laotriton, Paramesotriton, Pleurodeles and Salamandra (Salamandridae) (Laotriton, Paramesotriton, Pleurodeles only with unconfirmed positives). With the exception of the samples from the Bsal outbreak in Collection 1, the loads were below 400 z/s for both pathogens across the sampled species (below 200 when excluding the unconfirmed positives) (Supplementary Table 1).
From all the Bsal positives and unconfirmed positives, 15 (65.2%) and 10 (55.5%) yielded sequences of good quality for the ITS region; and 14 (60.9%) and 4 (22.2%) for the 28 S region, respectively. All but three ITS sequences obtained fully matched the homologous sequence of the Bsal type strain (Genbank accession number KC762295) with 100% similarity, and all 28 S sequences fully matched the Bsal type strain sequence. Of the three deviant ITS sequences, one (from a skin swab of A. maculatum) had three nucleotide substitutions (positions 281, 283 and 314), one (from a skin swab of Pleurodeles nebulosus) had three heterozygous positions (positions 234, 235 and 312), and one (from a skin swab of S. salamandra) had one heterozygous position (position 383), compared to the Bsal type strain (Supplementary Table 3).
To three private collections (Collections 1, 3 and 5) on which Bsal was detected, a heat treatment was applied. These collections were sampled a second time afterwards. Collection 1 previously had high loads of Bsal (~1,000 zoospores) and suffered a mass mortality event (Table 132), and Collections 3 and 5 had lower loads ( < 100 z/s), but individuals showed no clinical signs. All samples from the second sampling round of these three collections were negative for Bsal (Table 1). No further mortality events or clinical signs of Bsal infection were detected in these or any of the other sampled collections in the following ca. 18–24 months after sampling took place.
Discussion
The chytrid fungus Bd is known to be widespread both in the wild and in captivity15,40,41,42. For Bsal, its distribution in the wild has been explored recently29, but its presence in captivity is restricted to only to two reports so far32,33. Our study indicates that Bsal might be more widespread in captivity than expected, although it appears to be absent in the USA34.
At present, and with the exception of Collection 1 at an earlier time point32, no clinical signs of chytridiomycosis have been reported from the captive collections studied herein, yet Bsal was detected in approximately one third of them, and approximately half, if also considering unconfirmed positives (Table 1). While interpretation of zoospore loads is difficult due to the semi-quantitative nature of qPCR and the potential for copy number variation43,44, it is worth noting that despite half of the loads being below 100 zoospores (lowest zoospore quantity known to cause mortality in amphibians19), some of the Bsal-positives were above that threshold in species such as the common fire salamander (S. salamandra) that are known to succumb to chytridiomycosis with lower loads19. One explanation for this lack of clinical signs is that some of the positives might be from a novel, less virulent strain of Bsal, or a different species of chytrid fungus that co-amplifies with the applied Bsal probe. This explanation might apply to some samples as the sequences obtained from the A. maculatum, Pleurodeles nebulosus and S. salamandra Bsal-positive samples yielded sequences slightly differing from the ITS rRNA sequence of the type strain (Supplementary Table 3). The possible existence of such genetically deviant strains requires further confirmation because due to insufficient DNA template it was not possible to repeat the respective PCRs, and accordingly artefacts through polymerase synthesis and/or subsequent sequencing errors thus cannot be fully excluded at this point. With that said, the remaining sequences obtained from Bsal-positive individuals yielded sequences 100% identical to the type strain of Bsal (Supplementary Table 3). The presence of asymptomatic positives in species that are susceptible to Bsal, such as Salamandra and Pleurodeles18, might also indicate that under certain conditions in captivity, these susceptible salamanders are able to tolerate and exist with the pathogen. This may be explained by adverse environmental conditions (such as higher temperatures and less water pools) that are not optimal for the pathogen, potentially making it difficult for Bsal to develop and disseminate rapidly and minimizing the escalation of the infection to clinical disease.
From the unconfirmed positives identified in this screening, 12 showed contradictory results (Supplementary Table 2). Two of these, amplified in the laboratory of the University of Trier had an amplification signal above the highest standard as in the laboratory of the Technical University of Braunschweig. It is expected that with an appropriate dilution these samples would have tested also positive in Braunschweig as well. Eight of the unconfirmed positives (five amplified in Trier, and three amplified in Braunschweig) presented loads below 7 zoospores. This variation in low-load positives is likely related to the standards varying slightly in the actual DNA present in them, and so samples that did not make the cut off in one laboratory, did so in the other. To minimize such issues, negative sampling controls are necessary, i.e. swabs taken out of their sterile envelope and placed into a vial, by the same person and at the same time the sampling is performed. Unfortunately, in this study the standardized sampling procedure as suggested by the DGHT missed this point. We would therefore like to point out that for future studies such negative controls should be taken. Given that gloves were changed between individuals and all involved breeders guaranteed to have followed the instructions, we believe that the possibility of cross contamination between individuals does not impact our main findings, i.e. our conclusion of Bsal being present in the respective collections.
We hypothesize these widespread qPCR positives indicate asymptomatic Bsal infections, but alternative interpretations require discussion. Although we cannot exclude sample cross-contamination during sampling, this factor should not affect our main conclusions as such contamination would only alter the species identification of the positive individuals, not the presence of Bsal in the respective collection. The possibility that our results are exclusively explained by false positives is unlikely for several reasons. First, none of the negative controls showed amplification for Bsal or Bd. Second, most replicated swabs showed the same results in both laboratories. Third, sequences from both regions (ITS and 28 S) of the positive and unconfirmed positive samples sequenced (positive – ITS: 15, 28 S: 14; and unconfirmed positives – ITS: 10, 28S: 4) matched to the Bsal type strain. Both regions were amplified as to control for the potential of false positives39. And finally, samples were processed simultaneously with approximately 1,000 other samples from the wild (partly included in Spitzen-van der Sluijs et al. and Sabino-Pinto et al.29,45), and none of these was found to be Bsal-positive, with the exception of 26 samples from one geographical region29 of a recent Bsal spread, also confirmed by the laboratory in Trier. Additionally, as discussed above, our results might reflect detection of less virulent strains of Bsal than those known to cause mass mortality in fire salamanders. It is important to emphasize again that our approach, taking two swabs simultaneously from the same individual, and separately analysing those in two different laboratories (including separate DNA extractions) differs from most qPCR replicate approaches in which merely the qPCR is repeated in a second laboratory often from the same extracted individual DNA template. Hence, our duplicate swabbing approach is particularly robust against contamination as it does not only exclude qPCR artefacts and contamination of reagents, but also contamination of the DNA template with Bsal DNA during DNA extraction and subsequent manipulation in the laboratory. Still, we also highlight once more that the lack of sampling controls, with the exception of the one provided by the breeder of Collection 2, does not completely exclude the potential of within-collection contamination.
In 2015, a private collection (Collection 1) showed high mortality as a result of Bsal infection32. Heat treatment is known to clear the fungus in laboratorial set-ups23,35, but their efficiency when applied under less strict conditions was until now not known. The private keeper of this collection increased the temperature of the rooms in which the salamanders were kept to ~25 °C for 10 days as suggested by Blooi et al.35. During the first five days of the treatment mortality decreased and stopped completely after that point. The same collection was re-sampled six months after the treatment after temperature had been reset to original conditions. Our results show that no sign of Bsal (or Bd) was detected in any of the sampled individuals, and no additional mortality in this collection has been reported to date. The same heat treatment was applied also to two other collections (Collections 3 and 5) in which asymptomatic Bsal infections were detected. Again, no positives were detected after the heat treatment (Table 1), confirming the effectiveness of this mitigation method in captivity.
Overall, approximately half of the collections sampled in this project were positive or unconfirmed positive for Bd and Bsal, with three collections having both pathogens (although not in the same individuals) (Table 1). This is a crucial information for the development of further mitigation strategies46. Raising awareness to the necessity of screening and quarantine is of paramount importance, as is the implementation of biosecurity measures. The apparent occurrence of asymptomatic infections poses a new challenge as infections might go undetected and thereby increase the likelihood of further transmission and spread of Bsal both in captivity (from exchanged salamanders and newts between collections) and into the wild.
The introduction of Bsal and possibly of other, discovered or yet undiscovered, pathogens via animal imports or existing captive collections into the wild can have devastating effects to native amphibian populations10,18,28,29. Additionally, its spread within collections can lead to mass mortality events32. We argue that Bsal should be declared and treated as an epizootic disease by the European Union and national countries, and recommend that quarantine and biosafety rules should be adapted and developed to limit the spread of Bsal through Europe. As a first step, the unregulated import of salamanders and newts potentially carrying Bsal into European countries should be stopped immediately – as it has been done in the case of the USA, Hungary and Switzerland already – until proper measures and safety rules can be applied.
Availability of data and material
All data generated or analysed during this study are included in this published article and its supplementary information files.
References
Benton, M. J. Phylogeny of the major tetrapod groups: Morphological data and divergence dates. J. Mol. Evol. 30, 409–424 (1990).
Whittaker, K., Koo, M., Wake, D. & Vredenburg, V. Global Declines of Amphibians. Encyclopedia of Biodiversity (Elsevier, 2013).
IUCN. The IUCN red list of threatned species. Version 2016-3 http://www.iucnredlist.org (2017).
Wake, D. B. & Vredenburg, V. T. Colloquium paper: are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc. Natl. Acad. Sci. USA 105, 11466–11473 (2008).
IUCN. The IUCN Red List Of Threatened Species. Version 2017-3, Available at: http://www.iucnredlist.org (2017).
Baillie, J., Griffiths, J., Turvey, S., Loh, J. & Collen, B. Evolution Lost: Status and Trends of the World’s Vertebrates. Zoological Society of London (2010).
Fisher, M. C., Garner, T. W. J. & Walker, S. F. Global emergence of Batrachochytrium dendrobatidis and amphibian chytridiomycosis in space, time, and host. Annu. Rev. Microbiol. 63, 291–310 (2009).
Rachowicz, L. J. et al. Emerging infectious disease as a proximate cause of amphibian mass mortality. Ecology 87, 1671–1683 (2006).
Crawford, A., Lips, K. & Bermingham, E. Epidemic disease decimates amphibian abundance, species diversity, and evolutionary history in the highlands of central Panama. Proc. Natl. Acad. Sci. USA 107, 13777–13782 (2010).
Martel, A. et al. Batrachochytrium salamandrivorans sp. nov. causes lethal chytridiomycosis in amphibians. Proc. Natl. Acad. Sci. USA 110, 15325–15329 (2013).
Carvalho, T., Becker, C. & Toledo, L. Historical amphibian declines and extinctions in Brazil linked to chytridiomycosis. Proc. R. Soc. London B Biol. Sci. 284, 20162254 (2017).
Berger, L. et al. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc. Natl. Acad. Sci. USA 95, 9031–9036 (1998).
Longcore, J., Pessier, A. & Nichols, D. Batrachochytrium dendrobatidis gen. et sp. nov., a chytrid pathogenic to amphibians. Mycologia 91, 219–227 (1999).
Preuss, J. F. et al. Batrachochytrium dendrobatidis in near threatened and endangered amphibians in the southern Brazilian Atlantic Forest. North. West. J. Zool. 11, 360–362 (2015).
Group, G. B.-M. The Global Bd-Mapping Project: Bd-Maps. Imperial College of London and US Forest Service Available at: www.bd-maps.net (2017).
Sharifi, M., Farasat, H., Vaissi, S., Parto, P. & Haghighi, Z. M. S. Prevalence of the amphibian pathogen Batrachochytrium dendrobatidis in endangered Neurergus microspilotus (Caudata: Salamandridae) in Kavat stream, western Iran. Glob. Vet. 12, 45–52 (2014).
Gower, D. et al. Batrachochytrium dendrobatidis infection and lethal chytridiomycosis in caecilian amphibians (Gymnophiona). Ecohealth 10, 173–183 (2013).
Martel, A. et al. Recent introduction of a chytrid fungus endangers Western Palearctic salamanders. Science 346, 630–631 (2014).
Stegen, G. et al. Drivers of salamander extirpation mediated by Batrachochytrium salamandrivorans. Nature 544, 353–356 (2017).
Berger, L., Speare, R. & Skerratt, L. Distribution of Batrachochytrium dendrobatidis and pathology in the skin of green tree frogs Litoria caerulea with severe chytridiomycosis. Dis. Aquat. Organ. 68, 65–70 (2005).
Parker, J., Mikaelian, I., Hahn, N. & Diggs, H. Clinical diagnosis and treatment of epidermal chytridiomycosis in African clawed frogs (Xenopus tropicalis). Comp. Med. 52, 265–268 (2002).
Woodhams, D. et al. Treatment of amphibians infected with chytrid fungus: learning from failed trials with itraconazole, antimicrobial peptides, bacteria, and heat therapy. Dis. Aquat. Organ. 98, 11–25 (2012).
Blooi, M. et al. Successful treatment of Batrachochytrium salamandrivorans infections in salamanders requires synergy between voriconazole, polymyxin E and temperature. Sci. Rep. 5, 117788 (2015).
Garner, T. et al. Chytrid fungus in Europe. Emerg. Infect. Dis. 11, 1639–1641 (2005).
Ohst, T., Gräser, Y., Mutschmann, F. & Plötner, J. Neue Erkenntnisse zur Gefährdung europäischer Amphibien durch den Hautpilz Batrachochytrium dendrobatidis. Zeitschrift für Feldherpetologie 18, 1–17 (2011).
Tobler, U., Borgula, A. & Schmidt, B. Populations of a susceptible amphibian species can grow despite the presence of a pathogenic chytrid fungus. PLoS One 7, e34667 (2012).
Bosch, J. Martı́nez-Solano, I. & Garcı́a-Parı́s, M. Evidence of a chytrid fungus infection involved in the decline of the common midwife toad (Alytes obstetricans) in protected areas of central Spain. Biol. Conserv. 97, 331–337 (2001).
Spitzen-van der Sluijs, A. et al. Rapid enigmatic decline drives the fire salamander (Salamandra salamandra) to the edge of extinction in the Netherlands. Amphibia-Reptilia 34, 233–239 (2013).
Spitzen-van der Sluijs, A. et al. Expanding distribution of lethal amphibian fungus Batrachochytrium salamandrivorans in Europe. Emerg. Infect. Dis. 22, 1286–1288 (2016).
Schmidt, B., Bozzuto, C., Lötters, S. & Steinfartz, S. Dynamics of host populations affected by the emerging fungal pathogen Batrachochytrium salamandrivorans. R. Soc. Open Sci. 4, 160801 (2017).
Laking, A., Ngo, H., Pasmans, F., Martel, A. & Nguyen, T. Batrachochytrium salamandrivorans is the predominant chytrid fungus in Vietnamese salamanders. Sci. Rep. 7, 44443 (2017).
Sabino-Pinto, J. et al. First detection of the emerging fungal pathogen Batrachochytrium salamandrivorans in Germany. Amphibia-Reptilia 36, 411–416 (2015).
Cunningham, A. et al. Emerging disease in UK amphibians. Vet. Rec. 176, 468 (2015).
Klocke, B. et al. Batrachochytrium salamandrivorans not detected in U.S. survey of pet salamanders. Sci. Rep. 7, 1–5 (2017).
Blooi, M. et al. Treatment of urodelans based on temperature dependent infection dynamics of Batrachochytrium salamandrivorans. Sci. Rep. 5, 8037 (2015).
Belden, L. K. et al. Panamanian frog species host unique skin bacterial communities. Front. Microbiol. 6, 1171 (2015).
Project partners. Mitigating Batrachochytrium salamandrivorans in Europe Available at: https://bsalinfoeurope.wixsite.com/eubsalmitigation2017/partners (2017).
Blooi, M., Pasmans, F., Spitzen-van der Sluijs, A., Vercammen, F. & Martel, A. Duplex real-time PCR for rapid simultaneous detection of Batrachochytrium dendrobatidis and Batrachochytrium salamandrivorans in amphibian samples. J. Clin. Microbiol. 51, 4173–4177 (2013).
Iwanowicz, D. et al. Potential concerns with analytical methods used for the detection of Batrachochytrium salamandrivorans from archived DNA of amphibian swab samples, Oregon, USA. Herpetol. Rev. 48, 352–355 (2017).
Annis, S., Dastoor, F., Ziel, H., Daszak, P. & Longcore, J. A DNA-based assay identifies Batrachochytrium dendrobatidis in amphibians. J. Wildl. 40, 420–428 (2004).
Boyle, D., Boyle, D., Olsen, V., Morgan, J. & Hyatt, A. Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis. Aquat. Organ. 60, 141–148 (2004).
Havlíková, B., Baláž, V. & Vojar, J. First systematic monitoring of Batrachochytrium dendrobatidis in collections of captive amphibians in the Czech Republic. Amphibia-Reptilia 36, 27–35 (2015).
Rebollar, E., Woodhams, D., LaBumbard, B., Kielgast, J. & Harris, R. Prevalence and pathogen load estimates for the fungus Batrachochytrium dendrobatidis are impacted by ITS DNA copy number variation. Dis. Aquat. Organ. 123, 213–226 (2017).
Longo, A. et al. ITS1 copy number varies among Batrachochytrium dendrobatidis strains: implications for qPCR estimates of infection intensity from field-collected amphibian. PLoS 8, e59499 (2013).
Sabino-Pinto, J., Bletz, M., Iturriaga, M., Vences, M. & Rodríguez, A. Low infection prevalence of the amphibian chytrid fungus Batrachochytrium dendrobatidis (Chytridiomycetes: Rhizophydiales) in Cuba. Amphibia-Reptilia 38, 1–7 (2017).
Canessa, S. et al. Decision-making for mitigating wildlife diseases: From theory to practice for an emerging fungal pathogen of amphibians. J. Appl. Ecol. https://doi.org/10.1111/1365-2664.13089 (2018).
Acknowledgements
We thank the owners of the amphibian collections for their interest and participation in this project. We thank Karin Fischer for the help in the lab in Trier. This work was financially supported by a grant of the Deutsche Bundesstiftung Umwelt (DBU). M. Vences was supported by a grant of the Deutsche Forschungsgemeinschaft (VE247/9-1).
Author information
Authors and Affiliations
Contributions
J.S.P. and S.S. designed the study. J.S.P. analysed and interpreted the data, and wrote the manuscript. M. Veith contributed to the confirmation of the results. M. Vences and M. Veith funded the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sabino-Pinto, J., Veith, M., Vences, M. et al. Asymptomatic infection of the fungal pathogen Batrachochytrium salamandrivorans in captivity. Sci Rep 8, 11767 (2018). https://doi.org/10.1038/s41598-018-30240-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-30240-z
This article is cited by
-
United States amphibian imports pose a disease risk to salamanders despite Lacey Act regulations
Communications Earth & Environment (2023)
-
Batrachochytrium salamandrivorans’ Amphibian Host Species and Invasion Range
EcoHealth (2022)
-
Pooled samples and eDNA-based detection can facilitate the “clean trade” of aquatic animals
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.