Abstract
During mammalian pre-implantation embryonic development, dramatic and orchestrated changes occur in gene transcription. Pregnancy rates were low when yak females were crossbred with cattle breeds, but few studies exist to describe the unique molecular network regulation behind the pre-implantation development of these embryos. We determined the transcriptomes of crossbred embryos derived from yak oocytes in vitro fertilized with Jersey sperm using Illumina RNA-seq for the first time in this study. Embryos were sampled at the 2-, 4-, and 8-cell, morula and blastocyst stages. The results showed that in total, 291.9 million short reads were generated from the five libraries of 2-, 4-, and 8-cell, morula and blastocyst stages, with 276.2 million high-quality reads selected for further analysis. Eighty to 91% of the clean reads were aligned against the yak reference genome. A total of 19,072 transcripts were identified in five libraries, of which 7,785 transcripts were co-expressed in each stage and 2,013 transcripts were stage-specific. When a |log2 ratio| ≥1 and q-value ≤ 0.05 were set as thresholds for identifying differentially expressed genes (DEGs), we detected a total of 3,690 to 10,298 DEGs between any two consecutive stages. Based on the results of GO and KEGG enrichment, some of these DEGs potentially play an important role in regulating pre-implantation development, but they are most likely stage-specific. There were 2,960, 7,287, 6,420, 7,724 and 10,417 DEGs in 2-, 4-, 8-cell, morula and blastocyst stages between the crossbred embryos and purebred embryos of the yak, respectively, leading to a large difference in GO terms and pathways. In conclusion, we sequenced transcriptomes of in vitro-produced crossbred embryos of yak and cattle during pre-implantation and provided comprehensive examinations of gene activities. These will be helpful for development of assisted reproductive technology and better understanding the early maternal-fetal or maternal-embryonic dialog in inter-species crossbreeding.
Similar content being viewed by others
Introduction
The yak (Bos grunniens) is one of the world’s most remarkable domestic animals ‒ an herbivore living in and around the Himalayas and further north at altitudes ranging from 2,500 to 5,500 m with no frost-free period. They are very important to local people for providing milk and meat, as few other domestic animals can survive in such harsh conditions. However, the production performance of yak is inferior to that of improved cattle breeds1,2.
The economic traits of F1 hybrids derived from yak females crossbred with improved cattle breeds either by natural mating or artificial insemination are greatly improved. The F1 hybrids derived from dairy cattle breeds produce 100‒300% more milk than the yak, while those derived from beef cattle breeds grow faster and produce 50‒100% more meat than the yak1. However, there is a marked difference between the pregnancy rates of purebred service (>70%) compared with crossbred service (<30%)3, and the underlying causes of this difference have not been well studied. Mammalian pre-implantation embryonic development is a complex process including fertilization, cleavage divisions, compaction, and blastulation, governed by dramatic and orchestrated changes occurring in gene transcription. Our previous study indicated that the fertilization stage was normal when yak oocytes were fertilized with cattle sperm, but the cleavage rates and blastocyst rates were lower4. The development of RNA sequencing technologies permits the study of gene regulation at an unprecedented level. Such studies have been successfully conducted in mouse5, pig6, cattle7,8, human9 and yak10. However, these data have limited utility in crossbred embryogenesis of the yak due to the large differences in gene expression and genome sequences between different species11,12,13. Here, we provide the first comprehensive description of gene activities during the in vitro development of crossbred embryos of yak and cattle.
Results
Illumina HiSeq mRNA sequencing
The average cleavage rates and blastocyst rates after yak oocytes were in vitro fertilized (IVF) with Jersey sperm were 78.4% and 36.3%, respectively. In total, 291.9 million short reads were generated from the five libraries of yak crossbred embryos, i.e., the 2-, 4-, and 8-cell, morula and blastocyst stages throughout pre-implantation, with 276.2 million high-quality reads selected for further analysis. Eighty to 91% of the clean reads were aligned against the yak reference genome. A total of 19,072 transcripts were identified in five libraries, of which 7,785 transcripts were co-expressed in each stage. A total of 370, 737, 251, 120 and 535 transcripts were really stage-specific at the 2-, 4-, and 8-cell, morula and blastocyst stages, respectively, and the rest of the genes are common between two, three of four stages (Fig. 1). In total, 9,600 to 15,400 transcripts were detected in each in vitro stage, of which, 496, 564, 747, 441 and 519 novel transcripts were detected at the 2-, 4-, and 8-cell, morula and blastocyst stages, respectively. As embryo development proceeded, BMP15, ZP3, 4, PPARG, SLBP, FRZB and KIT (the maternal expression profiles) were decreased (Fig. 2A), whereas ATP5B, UBE3A, SNURF, ZO3, CLDN4, MAPK13, JUP, PCGF4, RRAD and NANOG (the embryonic expression profiles) were increased at specific stages (Fig. 2B).
Differentially expressed genes (DEGs) during pre-implantation development
Differences in gene expression at five stages during the pre-implantation development of crossbred embryos were examined, and DEGs were identified by pairwise comparisons of any two consecutive embryonic stages (Fig. 3, Supplementary Table 1). We detected a total of 3,690 to 10,298 DEGs between two consecutive stages. The number of DEGs identified in the comparison of 2- vs. 4-cell, 4- vs. 8-cell, 8-cell vs. morula, and morula vs. blastocyst stages increased with increasing the developmental stages. Four-cell vs. 8-cell stage of development presents the lower number of up-regulated genes, but it increases in the following stages of development. The number of down-regulated DEGs increased from the 2-cell stage to the morula stage but decreased from the morula stage to the blastocyst stage. The number of up-regulated DEGs was higher across the 2- vs. 4-cell stage than the 4- vs. 8-cell stage, but thereafter, it increased with increasing developmental stages.
Number of differentially expressed genes during pre-implantation development of yak crossbred embryos. The X axis represents four different stage comparisons of pre-implantation development, including from 2-cell to 4-cell (2 v 4), from 4-cell to 8-cell (4 v 8), from 8-cell to morula (8 v M), and from morula to blastocyst (M v B).
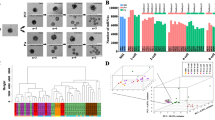
We performed hierarchical clustering of all DEGs using the Euclidean distance method associated with complete linkage (Fig. 4a,b). We used the SOTA function in the clValid package to classify DEGs into 12 clusters. Finally, we chose the six clusters with the most significant variations, as shown in Fig. 4b. Six clusters were plotted from the expression patterns. The genes in clusters K1 (1,120 genes) and K2 (1,467 genes) contained most of the DEGs, but the genes in the two clusters had different expression patterns. The genes in K1 were expressed at a relatively steady level throughout the developmental stages, whereas the ones in K2 dramatically decreased at the blastocyst stage. The expression patterns of genes in clusters K3 (521 genes) and K4 (518 genes) were similar, but genes in K4 were expressed at higher levels than those in K3. They were down-regulated as the embryonic developmental stage increased from the 2-cell stage to the morula stage but were dramatically up-regulated at the blastocyst stage. Genes in cluster K5 (579 genes) were up-regulated between the 2-cell stage and the morula stage but dramatically down-regulated at the blastocyst stage. Genes in cluster K6 (771 genes) were slightly down-regulated at the 8-cell stage and up-regulated at the morula stage, then dramatically down-regulated at the blastocyst stage.
Overview of DEG analysis during the five consecutive stages of yak crossbred pre-implantation development. (a) Heat map of DEGs across five developmental stages, including the 2-cell (2), 4-cell (4), 8-cell (8), morula (M), and blastocyst (B) stages. (b) Expression patterns of the genes in the six main clusters, namely, K1-K6, corresponding to the heatmap. (c) GO-term function enrichment analysis of the different clusters.
We further analysed the overrepresented GO functions within each cluster. The enriched GO terms of biological process are shown in Fig. 4c. The K3 cluster contained the most overrepresented GO terms among all of the clusters. Some gene GO terms were enriched in a particular cluster, such as the structural constituents of ribosomes and protein serine/threonine/tyrosine kinase activities, with protein kinase activities enriched specifically in K3.
Significantly related GO terms and pathways
We used GO assignments to classify the functions of DEGs in pairwise comparisons of cDNA libraries between different developmental stages of the yak crossbred embryos. In the Biological Process (BP) category, there were 29, 10 and 34 GO terms significantly enriched in the comparisons of 2- vs. 4-cell, 4- vs. 8-cell, and 8-cell vs. morula stages, respectively, but there was no GO term significantly enriched in the morula vs. blastocyst stage (Table 1, Supplementary Table 2). In the 2- vs. 4-cell stage and 4- vs. 8-cell stage, the most significant GO terms were the same, i.e., detection of chemical stimulus and sensory perception of chemical stimulus, but in 8-cell vs. blastocyst stage, the most significant GO terms were multicellular organismal process and single-multicellular organism process. In the Cellular Component (CC) category, there were 19 GO terms significantly enriched in the comparison of 2- vs. 4-cell stage, and the most significant GO terms were extracellular space, intrinsic to membrane, and extracellular region. In the comparison of 4- vs. 8-cell stage, there were 10 GO terms significantly enriched, and the most significant GO terms were plasma membrane, cell periphery and intrinsic to plasma membrane. In the comparison of 8-cell vs. morula stage, there were 12 GO terms significantly enriched, and the most significant GO terms were intrinsic to membrane, integral to membrane and extracellular space. In the morula vs. blastocyst stage, there were 9 GO terms significantly enriched, and the most significant GO terms were cytosolic ribosome, plasma membrane and cell periphery (Table 2, Supplementary Table 3). In the Molecular Function (MF) category, there were 43 GO terms significantly enriched in the comparison of 2- vs. 4-cell stage, and the most significant GO terms were receptor activity, signalling receptor activity, and G-protein coupled receptor activity. In the comparison of 4- vs. 8-cell stage, there were 7 GO terms significantly enriched, and the most significant GO terms were G-protein coupled receptor activity, signalling receptor activity and transmembrane signalling receptor activity. In the comparison of 8-cell vs. morula stage, there were 7 GO terms significantly enriched, and the most significant GO terms were receptor activity, signalling receptor activity and transmembrane signalling receptor activity. In the comparison of morula vs. blastocyst stages, there was only one GO terms significantly enriched, i.e., signalling receptor activity (Table 3, Supplementary Table 4).
Pathways enriched for differentially expressed genes are shown in Table 4. There were 15 significantly enriched pathways in the 2- vs. 4-cell stage. The most significantly enriched pathways were olfactory transduction, ubiquitin-mediated proteolysis, and protein processing in the endoplasmic reticulum. There were no significantly enriched pathways in the 4- vs. 8-cell stage. There were 18 significantly enriched pathways in the 8-cell vs. morula stage. The most significantly enriched pathways were associated with ribosomes, Parkinson’s disease, and oxidative phosphorylation. There was only one significantly enriched pathway in the morula vs. blastocyst stage, i.e., RNA transport.
Validation of RNA-seq results by qRT-PCR
To validate the findings of Smart-seq2 by qRT-PCR, we randomly selected four gene transcripts (SKP1, CD63, ZAR1 and H3) that were expressed differentially during embryonic development as candidate genes. The qRT-PCR results revealed that the expression patterns of the studied transcripts were in agreement with those observed in the RNA-seq analysis (Fig. 5)
Comparative analysis of transcriptome between the crossbred embryos and purebred embryos of the yak
There were 2,960, 7,287, 6,420, 7,724 and 10,417 DEGs in 2-, 4-, 8-cell, morula and blastocyst stages between the crossbred embryos and purebred embryos of the yak, respectively (Fig. 6a). There were a number of significantly enriched GO terms (Fig. 6b) and KEGG pathways (Table 5) in each developmental stage between the two types of embryos. In BP category, the most significantly enriched GO term was cell surface receptor signalling pathway, defense response, detection of chemical stimulus, and locomotion in 2-, 4-, 8-cell and blastocyst stages, respectively, but there was no significantly enriched GO term in morula stage. In CC category, the most significantly enriched GO term was intrinsic to membrane in all stages compared. In MF category, the most significantly enriched GO term was receptor activity in 2- and 4-cell stages, but it was G-protein coupled receptor activity in 8-cell and morula stages, and transmembrane signalling receptor activity in blastocyst stage (Supplementary Table 5). The most significantly pathway was RNA transport in 2-cell and morula stages, but it was spliceosome, olfactory transduction, and ribosome biogenesis in eukaryote in 4-, 8-cell and blastocyst stages, respectively (Table 5).
Discussion
We used high-throughput sequencing to generate comprehensive transcriptome profiles of crossbred embryos of yak and cattle at the 2-, 4-, and 8-cell embryo, morula and blastocyst stages for the first time. This offers valuable information for investigating the causes of low pregnancy rates when yak females are mated with Bos taurus and the mechanism that regulates embryonic development of these crossbred embryos. However, in vitro-produced embryos are known to be developmentally less competent than in vivo-derived embryos, and the expression of genes involved in transcription and translation in in vitro cases is different from in in vivo-cultured bovine embryos14,15,16,17 and mouse embryos18,19. Therefore, future studies comparing the transcriptomes of in vitro vs. in vivo crossbred embryos of yak and cattle will provide essential information for improving assisted reproductive technology.
In the various developmental stages of the crossbred embryos of yak and cattle analysed in the present study, transcripts from 9,600 to 15,400 different genes per developmental stage were identified (Fig. 1). This was similar to the number of expressed genes detected in bovine embryos and human embryos7,8,16,20, except for the lower number of expressed genes in the morula stage detected in the present study. The lower number of transcripts detected at the morula stage may indicate that the current in vitro culture system is inferior. To provide information about the expression changes of individual genes over time, we further performed hierarchical clustering of all DEGs using the Euclidean distance method. Although the dynamics of actual gene expression changes in individual genes was very complex, the k-means clustering provided a good overview of the expression trends and formed a wave-like expression pattern. As a result, six clusters were identified during pre-implantation development (Fig. 4). Several regulated genes specific to each cluster were different from those of other mammals9,10,11,12,13,16,20, although early mammalian development encompasses dynamic cellular, molecular and epigenetic events that are largely conserved from mouse to man21.
Early embryonic development is controlled by maternal RNA and proteins accumulated during oogenesis and oocyte maturation22,23. As development proceeds, these maternally derived substances are degraded, while embryonic genome activation (EGA) gradually occurs7,24,25. The EGA is initiated at a species-specific time point. It occurs at the 2-cell stage in mice26, between the 4- and 8-cell stages in humans27 and pigs6,26, and between the 8- and 16-cell stages in bovines23,26. Previous reports have alluded to minor bovine EGA between the zygote and 4-cell stage10,16,28,29,30,31. However, our previous report in the yak10 and this data show that a high number of genes were significantly up-regulated between 2-cell and 4-cell stage, indicating that EGA occurs before the 8-cell stage in the yak and its crossbred embryos. BMP15, ZP3, 4, PPARG, SLBP, FRZB and KIT are oocyte markers27,32,33,34,35,36,37, and they were down-regulated with the development of the embryos (Fig. 2A). The number of down-regulated DEGs increased from the 2-cell stage to the morula stage of these crossbred embryos (Fig. 3), also indicating that maternally derived transcripts and proteins are gradually degraded during this period. ATP5B, UBE3A, SNURF, ZO3, CLDN4, MAPK13, JUP, PCGF4, RRAD and NANOG are previously known to be embryonically expressed7,23,36,38,39. They were up-regulated with the development of the embryos or expressed at specific stages (Fig. 2B). They are important in regulating embryonic development. For example, NANOG transcripts were first observed at the eight-cell stage in the bovine embryo7, and its expression is required for the bovine embryonic development40. Mouse embryos cultured in the presence of Clostridium perfringens enterotoxin inhibitory to CLDN4 failed to form a mature blastocele cavity, demonstrating the importance of CLDN4 in the normal formation of blastocysts41. In addition, CLDN4 could be potentially involved in uterine implantation42.
Our previous study10 and qRT-PCR analyses of the amplified RNA in the present study (Fig. 5) verified that the Smart-seq2 accurately reflect the relative abundance of selected amplified transcripts (SKP1, CD63, ZAR1 and H3) in the samples. SKP1 mRNA synthesis was activated at early bovine embryonic stages, which suggests that these transcripts are necessary to prepare the embryo for EGA. The level of SKP1 protein significantly increased from MII oocytes to 4-cell embryos but then significantly decreased again43. Increasing importance for all aspects of inter-cell communications is attributed to extracellular vesicles (EVs) released by eukaryotic and prokaryotic cells44. Giacomini et al.45 showed EVs to be CD63, CD9 and ALIX, suggesting their predominant exosomal nature. The proteins mediate signal transduction events that play a role in the regulation of cell development, activation, growth and motility. Zar1 was the first oocyte-specific maternal-effect gene identified to play an essential role during the oocyte-to-embryo transition in humans and mice, as elucidated by knockout experiments in mice46. H3.3-mediated paternal chromatin remodelling is essential for the development of pre-implantation embryos and the activation of the paternal genome during embryogenesis47. The results of both RNA-seq and qRT-PCR showed that SKP1, CD63, ZAR1 and H3 were expressed in the pre-implantation development of crossbred embryos of yak and cattle, and their expression patterns were similar to previous observations in other mammals43,44,45,46,47.
In the present study, we found that there were a large number of DEGs (Fig. 6a), different enriched GO terms (Fig. 6b) and pathways (Table 5) between each stage of crossbred embryos and purebred embryos of the yak. The most significantly enriched pathways for differentially expressed genes were olfactory transduction, ubiquitin-mediated proteolysis, and protein processing in the endoplasmic reticulum before the 4-cell stage, while ribosome, Parkinson’s disease, and oxidative phosphorylation pathways were highly represented after the 8-cell stage in crossbred embryos of yak and cattle (Table 5). This is different from the findings in bovine embryos reported by Jiang et al.8, except those for the ribosome pathway. The oxidative phosphorylation pathway is one of the obligatory energy metabolism pathways in most species throughout pre-implantation development48, and we also identified this pathway as enriched from the 8-cell to the morula stage of crossbred embryo of the yak. We identified ubiquitin-mediated proteolysis as enriched from the 2- to 4-cell and from the 8-cell to morula stages, and the most common pathway was RNA transport throughout the pre-implantation development of crossbred embryos of yak and cattle. In general, the pathways enriched by DEGs were not completely the same as those found in the bovine8,16,49, yak10, pig6, mouse and human11,37,42. Embryonic expression profiles across these mammalian species are different. Therefore, the regulatory pathways involved in pre-implantation development appear to be species-specific.
Our preliminary study indicated that the cleavage rates and blastocyst rates were lower when yak oocytes were in vitro fertilized with cattle sperm4. However, the report by Sun et al.50 and the present study showed that the developmental competence of the crossbred embryos is comparable to that of bovine embryos in the optimal in vitro culture conditions. Fertility defect is multifactorial origin. Among the possible origins of recurrent pregnancy loss are uterine structural defaults, defective immunological dialog between the embryo (or the fetus) and the uterus sometimes in relation with immunological disorders (such as autoimmune diseases), thrombophilia, and free radical metabolism imbalance. Numerous variants of genes are supposed to be intervening in the different facets of the early maternal-fetal or maternal-embryonic dialog, and eventually modify the outcome of fertilization, leading to success or failure of post-implantation development51,52,53. From the foregoing, there were a number of DEGs, and different GO terms and pathways between the crossbred embryos and the purebred embryos of the yak. It is worthy to further study on whether these differences effect on the early maternal (yak) - fetal (crossbred) or maternal (yak) - embryonic (crossbred) dialog, and eventually on pregnancy rate of female yaks borne crossbred embryos or fetal.
Conclusions
We sequenced the transcriptomes of in vitro-produced crossbred embryos of yak and cattle during pre-implantation with the Illumina 2500 sequencing platform and provided a comprehensive examination of gene activities. This is the first report to investigate the mechanism that regulates embryonic development in crossbred embryos of yak and cattle using high-throughput sequencing, which will be helpful for development of assisted reproductive technology in yak crossbreeding. There were a number of DEGs, different GO terms and pathways between the crossbred embryos and the purebred embryos of the yak, however, there is a need to study whether these differences effect on the early maternal-fetal or maternal-embryonic dialog, and eventually on pregnancy rate of female yaks borne crossbred embryos or fetal.
Methods
IVM, IVF, IVC and embryo sample collection
All animal procedures were approved by the Institutional Animal Care and Use Committee of the Southwest Minzu University and all methods were performed in accordance with the relevant guidelines and regulations. Yak ovaries were collected at local slaughterhouses in October. IVM was performed as previously described by Xiao et al.54, and IVF and IVC were performed as previously described by Yao et al.55 with some modifications. Briefly, cumulus-oocyte complexes (COCs) were collected in Dulbecco’s phosphate buffered saline (DPBS) supplemented with 6 mg/ml BSA using a low-power (20×) stereomicroscope (Leica MZ75, Germany). The COCs were rinsed three times in DPBS containing 5% (v/v) foetal calf serum (FCS) and twice in TCM 199 (Gibco, Grand Island, NY, USA) supplemented with 20% (v/v) FCS, 5 μg/ml FSH, 5 μg/ml LH (Bioniche Inc, Belleville, Canada), 1 μg/ml oestradiol-17β, and 100 U/ml penicillin and 100 g/ml streptomycin (Sigma Chemical Co, St. Louis, MO, USA) (maturation medium). Oocytes with homogenous cytoplasm and several layers of cumulus cells were selected for IVM. Up to 30 COCs were placed in each culture well (Nunc Inc, Naperville IL, USA) containing 600 μl of maturation medium covered with 300 μl mineral oil (Sigma Chemical Co, St. Louis, MO, USA) and were matured at 38.5 °C for 24 h in a humidified incubator with 5% CO2. Motile spermatozoa were selected using a swim-up technique. Briefly, 200 μl of Jersey frozen/thawed semen was overlaid with 1.0 ml of SpermRinse™ (Vitrolife, Sweden) in a 1.5-ml EP tube and incubated at 38.5 °C. After 50 min, 600 μl of the supernatant was collected and centrifuged at 500 × g for 5 min, and then 50 μl of the concentrated sperm fraction was removed for IVF.
After IVM, the COCs were further washed twice in IVFTM (Vitrolife, Sweden) before being transferred into 4-well plates (up to 30 per well) containing 500 μl IVFTM covered with mineral oil per well. Motile spermatozoa were added to produce a final concentration of 2 × 106 sperms/ml, and after a period of 22 h post-insemination (hpi), cumulus cells were removed by 0.2% hyaluronidase. Presumptive zygotes were cultured in 50-μl drops of G1TM for 72 h and then cultured in G2TM (Vitrolife, Sweden) covered with mineral oil in a humidified incubator with 90% N2 and 5% CO2 in air. Each culture droplet contained 10‒15 embryos. All embryos were carefully evaluated under a stereomicroscope, and only morphologically intact embryos scored as grade 1 according to the Manual of the International Embryo Transfer Society (IETS)56 were used. Embryos were pooled in groups of 10 in the case of 2-cell (42‒46 hpi), 4-cell (68‒72 hpi), and 8-cell (88‒96 hpi) stages or in singles for morula (120 hpi) and blastocyst (168 hpi) stages and were washed three times in saline, then immediately frozen and stored at −80 °C until use.
Library preparation and sequencing
The collected embryos were lysed to release all RNA using cell lysis buffer (Sigma-Aldrich), and RNA was then reverse-transcribed by Smart-Seq2 into first cDNA. Smart-seq2 was carried out by Annoroad Gene Technology (Beijing, China) according to the method previously described57,58,59. Briefly, first-strand cDNA was synthesized from the RNA using oligo-dT, superscript II reverse transcriptase (Invitrogen), first-strand buffer (Invitrogen), RNase inhibitor (Clontech), and template-switching oligonucleotides (TSO) primers. Second-strand cDNAs were synthesized using IS PCR primers and KAPA HiFi HotStart ReadyMix (KAPA Biosystems) on a thermal cycler (S1000, Bio-Rad). cDNAs were consequently pre-amplified, purified and recovered. After purification, the distribution of fragments and the quality of amplified products were examined with the Agilent high-sensitivity DNA chip (Agilent Technologies, Palo Alto, CA, USA) using a Bioanalyzer 2100 (Agilent). The qualified library contained no fragment <500 bp, with peak value at 1.5–2 kb.
The cDNA samples were fragmented by Tagmentation (Tn5) to obtain cDNA fragments, ligated with a sequencing adapter, and then subjected to PCR amplification with KAPA HiFi DNA polymerase on a thermal cycler (S1000, Bio-Rad). The amplified cDNA was purified with AMPure XP beads (Beckman-Coulter). The cDNA concentration was determined on a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA), and the distributions of the fragments of amplified products were examined with the Agilent high-sensitivity DNA chip (Agilent Technologies, Palo Alto, CA, USA) using a Bioanalyzer 2100 (Agilent). The peak value of DNA fragments was within the range of 300–800 bp. The qualified cDNA libraries were sequenced on the Illumina X-Ten platform (Illumina, San Diego, CA, USA) with read lengths of paired-ends at 125 bp.
Analysis of sequencing results: mapping and differential expression
Raw reads obtained from the RNA-seq were cleaned by removing adapter sequences, reads containing ploy-N, and low-quality sequences (Q < 20) as previously described60. Clean reads were then aligned to the yak reference genome (BosGru_v2.0)61 using Tophat2 (v2.0.12). Novel transcripts were identified from TopHat alignment results using the Cufflinks (v2.2.1) (http://cufflinks.cbcb.umd.edu/howitworks.html) reference annotation-based transcript (RABT) assembly method, with lengths ≥180 bp, sequence depths ≥2, and distances within 200 bp of the annotated gene62.
Gene expression levels were normalized by considering the RPKM value (reads per kilobase of the exon model per million mapped reads)63. Differentially expressed gene (DEG) analysis was conducted using the R package DEGseq (v1.18.0). The P values were adjusted using the Benjamini & Hochberg method64. The significant DEGs between subsequent stages were identified with corrected ‘p < 0.05 and |log2 fold change| >1’ in this study. The heat maps were drawn using the R packages (v3.1.1) as follows: function ‘heatmap.2’ of the ‘gplots’ package and the ‘sota’ function in the ‘clValid’ package, with default Euclidean distance and the hierarchical clustering method.
Gene ontology and pathway enrichment analysis of DEGs
The main functions of the differentially expressed genes were determined using GO analysis (http://www.geneontology.org/) provided by the NCBI, which can discover gene regulatory networks on the basis of biological processes, molecular functions and cellular components. A hypergeometric test was used to find significantly enriched GO terms or pathways in DEGs by comparison with the genome background. Multiple tests were adjusted by the FDR method64. The adjusted p-value ≤0.05 was set as the significant threshold. Pathway annotations of DEGs were performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.enome.jp/kegg/).
Validation of the Smart-seq2 results by qRT-PCR
The total RNA from each pool of crossbred yak embryos (n = 3) was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Then, RNA was dissolved in sterile water and stored at −80 °C until use. Quantitative real-time PCR (qRT-PCR) was performed to validate the expression of four selected genes (SKP1, CD63, ZAR1 and H3). They were amplified with specific primers (Supplementary Table 6) on an ABI 7500 Fast instrument (Applied Biosystems). All values were normalized to the internal control, H2A. The efficiency of each primer pair was calculated over a 3.5 log dilution range, and the relative gene expression values were calculated using the 2−△△Ct method65.
Comparative analysis of transcriptome between the crossbred embryos and purebred embryos of the yak
In vitro production and mRNA sequencing of yak embryos were exactly as same as the above described. Raw sequencing data of yak embryos were stored in the NCBI Sequence Read Archive (Accession number SRP127024)10. The DEGs in each stage between these two types of embryos, and GO and KEGG pathway analysis were the same with the above described.
References
Wiener, G., Han, J. L. & Long, R. J. The Yak. 2nd ed. Bangkok: The Regional Office for Asia and the Pacific of the Food and Agriculture Organization of the United Nations; (2003).
Zi, X. D. et al. Growth performance, carcass composition and meat quality of Jiulong-yak (Bos grunniens). Asian-Aust J Anim Sci. 17, 410–414 (2004).
Zi, X. D. et al. Induction of estrus in suckled female yaks (Bos grunniens) and synchronization of ovulation in the non-sucklers for timed artificial insemination using progesterone treatments and Co-Synch regimens. Anim Reprod Sci. 92, 183–192 (2006).
Zi, X. D. et al. Developmental competence of embryos derived from reciprocal in vitro fertilization between yak (Bos grunniens) and cattle (Bos taurus). J Reprod Dev. 55, 480–483 (2009).
Li, L., Zheng, P. & Dean, J. Maternal control of early mouse development. Development 137, 859–870 (2010).
Cao, S. et al. Specific gene-regulation networks during the pre-implantation development of the pig embryo as revealed by deep sequencing. BMC Genomics 15, 4 (2014).
Graf, A. et al. Fine mapping of genome activation in bovine embryos by RNA sequencing. Proc Natl Acad Sci USA 111, 4139–4144 (2014).
Jiang, Z. et al. Transcriptional profiles of bovine in vivo preimplantation development. BMC Genomics 15, 756 (2014).
Yan, L. et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 20, 1131–1139 (2013).
Zi, X. D. et al. Characterization of transcriptional complexity during preimplatation development of the yak (Bos grunniens) using RNA-Seq. Reprod Domest Anim. https://doi.org/10.1111/rda.13167 (2018).
He, K., Zhao, H., Wang, Q. & Pan, Y. A comparative genome analysis of gene expression reveals different regulatory mechanisms between mouse and human embryo pre-implantation development. Reprod Biol Endocrinol. 8, 41 (2010).
Xie, et al. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 20, 804–815 (2010).
Jiang, Z. et al. mRNA levels of imprinted genes in bovine in vivo oocytes, embryos and cross species comparisons with humans, mice and pigs. Sci Rep. 5, 17898 (2015).
Lonergan, P. et al. Temporal divergence in the pattern of messenger RNA expression in bovine embryos cultured from the zygote to blastocyst stage in vitro or in vivo. Biol Reprod. 69, 1424–1431 (2003).
Corcoran, D. et al. Suppressed expression of genes involved in transcription and translation in in vitro compared with in vivo cultured bovine embryos. Reproduction 131, 651–660 (2006).
Kues, W. A. et al. Genome-wide expression profiling reveals distinct clusters of transcriptional regulation during bovine preimplantation development in vivo. Proc Natl Acad Sci USA 105, 19768–19773 (2008).
Gad, A. et al. Molecular mechanisms and pathways involved in bovine embryonic genome activation and their regulation by alternative in vivo and in vitro culture conditions. Biol Reprod. 87, 100 (2012).
Wang, S., Cowan, C. A., Chipperfield, H. & Powers, R. D. Gene expression in the preimplantation embryo: in-vitro developmental changes. Reprod Biomed Online 10, 607–616 (2005).
Giritharan, G. et al. Effect of in vitro fertilization on gene expression and development of mouse preimplantation embryos. Reproduction 134, 63–72 (2007).
Xue, Z. et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597 (2013).
Cui, W. et al. Towards functional annotation of the preimplantation transcriptome: an RNAi screen in mammalian embryos. Sci Rep. 6, 37396 (2016).
Tadros, W. & Lipshitz, H. D. The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042 (2009).
Lavagi, I. et al. Single-cell RNA sequencing reveals developmental heterogeneity of blastomeres during major genome activation in bovine embryos. Sci Rep. 8, 4071 (2018).
Schultz, R. M. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update 8, 323–331 (2002).
Misirlioglu, M. et al. Dynamics of global transcriptome in bovine matured oocytes and preimplantation embryos. Proc Natl Acad Sci USA 103, 18905–18910 (2006).
Sirard, M. A. Factors affecting oocyte and embryo transcriptomes. Reprod Domest Anim. 47(Suppl 4), 148–155 (2012).
Braude, P., Bolton, V. & Moore, S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332, 459–461 (1988).
Barnes, F. L. & Eyestone, W. H. Early cleavage and the maternal to zygotic transition in bovine embryos. Theriogenology 33, 141–152 (1990).
Marcucio, R. S., Hopwood, R. M., Ignotz, G. G. & Currie, W. B. Translation of zygotically derived mRNA in a cell cycle specific manner in 2-cell cattle embryos. J Reprod Fert Abstr Series 15, 46 (1995).
Hyttel, P., Viuff, D., Avery, B., Laurincik, J. & Greve, T. Transcription and cell cycle dependent development of intranuclear bodies and granules in two-cell bovine embryos. J Reprod Fert. 108, 263–270 (1996).
Memili, E., Dominko, T. & First, N. L. Onset of transcription in bovine oocytes and early embryos. Mol Reprod Dev. 51, 36–41 (1998).
Bilodeau-Goeseels, S. & Schultz, G. A. Changes in ribosomal ribonucleic acid content within in vitro-produced bovine embryos. Biol Reprod. 56, 1323–1329 (1997).
Brambrink, T. et al. Application of cDNA arrays to monitor mRNA profiles in single preimplantation mouse embryos. Biotechniques 33, 376–385 (2002).
Cabot, R. A., Hannink, M. & Prather, R. S. CRM1-mediated nuclear export is present during porcine embryogenesis. Biol Reprod. 67, 814–819 (2002).
Campbell, L. & Hardwick, K. G. Analysis of Bub3 spindle checkpoint function in Xenopus egg extracts. J Cell Sci. 116, 617–628 (2003).
Adjaye, J. et al. Conserved molecular portraits of bovine and human blastocysts as a consequence of the transition from maternal to embryonic control of gene expression. Physiol Genomics 31, 315–327 (2007).
Sudheer, S. & Adjaye, J. Functional genomics of human pre-implantation development. Brief Funct Genomic Proteomic 6, 120–132 (2007).
Adjaye, J. et al. Cross-species hybridisation of human and bovine orthologous genes on high density cDNA microarrays. BMC Genomics 5, 83 (2004).
Lee, M. T. et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364 (2013).
Simmet, K. et al. Bovine OCT4 (POU5F1) knockout embryos fail during the second lineage differentiation due to loss of NANOG. Reprod, Fertil Dev. 29, 138 (2016).
Moriwaki, K., Tsukita, S. & Furuse, M. Tight junctions containing claudin 4 and 6 are essential for blastocyst formation in preimplantation mouse embryos. Dev Biol. 312, 509–522 (2007).
Munch, E. M. et al. Differentially expressed genes in preimplantation human embryos: potential candidate genes for blastocyst formation and implantation. J Assist Reprod Genet. 33, 1017–1025 (2016).
Benesova, V., Kinterova, V., Kanka, J. & Toralova, T. Characterization of SCF-complex during bovine preimplantation development. PLoS One 11, e0147096 (2016).
Willms, E. et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep. 6, 22519 (2016).
Giacomini, E. et al. Secretome of in vitro cultured human embryos contains extracellular vesicles that are uptaken by the maternal side. Sci Rep. 7, 5210 (2017).
Wu, X. et al. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet. 33, 187–191 (2003).
Kong, Q. et al. Histone variant H3.3-mediated chromatin remodeling is essential for paternal genome activation in mouse preimplantation embryos. J Biol Chem. 293, 3829–3838 (2018).
Harvey, A. J., Kind, K. L. & Thompson, J. G. REDOX regulation of early embryo development. Reproduction 123, 479–486 (2002).
Zuo, Y. et al. Exploring timing activation of functional pathway based on differential co-expression analysis in preimplantation embryogenesis. Oncotarget 7, 74120–74131 (2016).
Sun, Y. G., Xu, J. T., Caizi, D. Z. & Ma, Z. J. The study on cattle × yak in vitro fertilization and embryo transfer. Acta Veterinaria Zootechnica Sinica 44, 719–726 (2013).
Saadeldin, I. M., Oh, H. J. & Lee, B. C. Embryonic-maternal cross-talk via exosomes: potential implications. Stem Cells Cloning 8, 103–107 (2015).
Vaiman, D. Genetic regulation of recurrent spontaneous abortion in humans. Biomed J. 38, 11–24 (2015).
Quintero-Ronderos, P. et al. Novel genes and mutations in patients affected by recurrent pregnancy loss. PLoS One 12, e0186149 (2017).
Xiao, X. et al. Effect of addition of FSH, LH and proteasome inhibitor MG132 to in vitro maturation medium on the developmental competence of yak (Bos grunniens) oocytes. Reprod Biol Endocrinol. 12, 30 (2014).
Yao, G. et al. Developmental potential of clinically discarded human embryos and associated chromosomal analysis. Sci Rep. 6, 23995 (2016).
International Embryo Transfer Society. Manual of the International Embryo Transfer Society (International Embryo Transfer Society, Savoy, IL), 3rd ed.; 1998.
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 10, 1096–1098 (2013).
Chen, Q. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400 (2016).
Wang, N. et al. Effect of vitrification on the mRNA transcriptome of bovine oocytes. Reprod Domest Anim. 52, 531–541 (2017).
Guo, Q. et al. De novo transcriptome sequencing and digital gene expression analysis predict biosynthetic pathway of rhynchophylline and isorhynchophylline from Uncaria rhynchophylla, a non-model plant with potent anti-alzheimer’s properties. BMC Genomics 15, 676 (2014).
Qiu, Q. et al. The yak genome and adaptation to life at high altitude. Nat Genet. 44, 946–949 (2012).
Roberts, A., Pimentel, H., Trapnell, C. & Pachter, L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27, 2325–2329 (2011).
Wanger, G. P., Kin, K. & Lynch, V. J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285 (2012).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Statist Soc B. 57, 289–300 (1995).
Schmitgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 3, 1101–1108 (2008).
Acknowledgements
This study was funded by the Fundamental Research Funds for the Central Universities, Southwest Minzu University (no. 2015NZYTD02).
Author information
Authors and Affiliations
Contributions
X.D.Z. performed all aspects of the study and wrote the manuscript. S.L. and B.L. performed IVF and analysed data. W.X. and X.R.X. performed the IVF and the experimental design.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zi, Xd., Liu, S., Xia, W. et al. Transcriptional profiles of crossbred embryos derived from yak oocytes in vitro fertilized with cattle sperm. Sci Rep 8, 11571 (2018). https://doi.org/10.1038/s41598-018-29912-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-29912-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.