Abstract
Juglans regia (walnut) is a species belonging to the family Juglandaceae. Broadly spread in diverse temperate and subtropical regions, walnut is primarily cultivated for its nuts. In France, Colletotrichum sp. on walnut was detected for the first time in 2007; in 2011 the disease led to 50–70% losses in nut production. A combined approach of metabarcoding analysis and multi-locus genetic characterization of isolated strains has been used for taxonomic designation and to study the genetic variability of this pathogen in France. Evidence indicates that four Colletotrichum species are associated with walnut in France: 3 belong to the C. acutatum species complex and 1 to the C. gloeosporioides species complex. Results also show that C. godetiae is the most abundant species followed by C. fioriniae; while C. nymphaeae and another Colletotrichum sp. belonging to the C. gloeosporioides complex are found rarely. Representative isolates of detected species were also used to confirm pathogenicity on walnut fruits. The results show a high variability of lesion’s dimensions among isolates tested. This study highlights the genetic and pathogenic heterogeneity of Colletotrichum species associated with walnut anthracnose in France providing useful information for targeted treatments or selection of resistant cultivars, in order to better control the disease.
Similar content being viewed by others
Introduction
The English/Persian walnut (Juglans regia L., 1753), or common walnut, is a species that is native to Central Asia and belongs to the Juglandaceae family. The genus Juglans includes approximately 21 species; all species produce nuts but only Juglans regia is extensively cultivated for commercial production1. The common walnut is a tree broadly spread in diverse temperate and subtropical regions of North and South America, Asia, Australia, New Zealand, South Africa and Europe, where it grows widely or semi-cultivated. In Europe, common walnut was most likely introduced from Iran and eastern Turkey by Greek commerce a thousand years ago2. Common walnut is primarily cultivated for its nuts, which are harvested from wild stands, backyard gardens or commercial orchards. Nuts are collected for home consumption or sold on the market for their nutritional values and their high polyunsaturated fats content, including omega-3, consumed either as a snack or in baked foods. Furthermore, walnut trees are utilized for their high quality wood to make a wide array of products3. The total world production of J. regia is estimated to be about 3.4 million tonnes; China is the world’s largest producer of walnuts with a total production of about 1.7 million tonnes4. In 2014, European Union produced about 169,621 tonnes of walnuts with France the largest producer with about 34,767 tonnes of walnuts yielded, followed by Romania (31,514 tonnes) and Greece (22,310 tonnes)4. In France walnut cultivation occupies an area of about 19,712 ha4; orchards are the main production sites whereas harvest on isolated trees has strongly decreased in the last decades. In France, the establishment of new orchards, mainly localised in two large areas, balanced this reduction: South-East (Auvergne-Rhône-Alpes region) and the South-West (mainly Dordogne, Lot, Corrèze and Gironde departments). In French walnut orchards, the two main historical diseases were bacterial wilt (caused by Xanthomonas campestris pv. juglandis, walnut blight) causing yield losses of up to 50%. and anthracnose caused by Ophiognomonia leptostyla (formerly Gnomonia juglandis, Ascomycota, Sordariomycetes). Since 2007, a new fungal disease associated to the Colletotrichum genus has appeared in French walnut trees causing fruits browning (anthracnose symptoms) which then become unmarketable5.
Colletotrichum is a globally distributed plant-associated fungal genus able to cause disease on a wide variety of woody and herbaceous plants6, including walnut, on which the pathogen causes a new form of walnut anthracnose. Colletotrichum acutatum species complex is a diverse yet relatively closely related group of plant pathogenic fungi within the genus, recently suggested as a model system to study evolution and host specialization in plant pathogens7. In 2005, Sreenivasaprasad and Talhinhas reported C. acutatum sensu lato associated with J. regia8, however no information about the geographic origin and the pathogenicity were reported. The same year Juhasova et al. reported the presence of C. gloeosporioides on walnut fruits in Slovakia, but the importance of the disease was not indicated9. Later Damm et al. described two C. godetiae strains associated with walnut: one isolated in Austria and another one of unknown origin10. The walnut anthracnose disease caused by Colletotrichum spp. is not only restricted to Europe. Recently, 3 reports described C. gloeosporioides sensu lato as the causal agent of anthracnose on J. regia in Shandong province, China11,12,13. Zhu et al. 2015 also reported leaf spot disease caused by C. fioriniae on walnut trees in Hechi, Guangxi region, China, which led to severe reductions in nut production14. Symptoms are described as water-soaked circular to semi-circular leaf spots, later becoming tan bordered, greyish-white in the centre and dark brown to the margins; lesions are 3 to 4 mm in diameter. Morphological and molecular characterization confirmed the presence of C. fioriniae. Artificial inoculations and re-isolation of the pathogen from the leaves demonstrated that the causal agent of the disease was C. fioriniae. Efforts to contain the pathogen spread were made. To date, chemical control has been the main approach to control the disease, although it may lead to environmental concerns and drug resistance in the pathogen15. Therefore, identification of resistant cultivars is required.
In France, Colletotrichum sp. on walnut has been detected for the first time in 2007 as part of a study regarding the bacteriosis of walnut5. Later, in 2011, symptoms of anthracnose appeared on walnut leading to 50–70% losses in nut production; the causal agent was identified as belonging to the Colletotrichum genus5. To our knowledge, this is the first report of an epidemic event of walnut anthracnose caused by Colletotrichum spp. in Europe. The disease mainly affects the surface of the fruit in June and is characterized by small brown or black dry spots. These spots tend to become circular and dark in colour. Orange conidial masses can appear (i.e., acervuli) on the necrotic spots during the season (depending on meteorological conditions). Eventually, the nut becomes completely necrotic and falls prematurely (Fig. 1).
Development of anthracnose symptoms on a walnut fruit. Left: in June small brown to black necrosis, here taking also the aspect of a run-out, appear on young fruit. Centre: around August orange conidial masses can usually be observed. The necrosis has a dry aspect and deforms the husk. Right: The nut can become completely necrotic and deformed, with conidial masses, and falls of the tree.
These symptoms sometimes may be misleading: in the early stages of the disease, necrotic areas can be confused with those caused by Xanthomonas campestris pv. juglandis; symptoms may also be confused with those caused by Ophiognomonia leptostyla, although the spots caused by O. leptostyla present a typical light-green colouration in the centre5.
Considering the severity of the disease on walnut, the focus of the present study was to assess the extent of the genetic and pathogenic diversity of Colletotrichum spp. populations associated with walnut anthracnose in France. We used two different approaches: 1. Metabarcoding analysis of Colletotrichum spp. diversity in plant tissues; 2. Multi-locus phylogenetic analysis of a collection of Colletotrichum spp. isolates established through the work. We selected the most disease-affected area as our sampling zone. Pathogenicity was confirmed by inoculation tests on walnut (cultivar Lara) grown in France.
Results
Metabarcoding data
A total of 1,993,250 ITS sequences (from 53,197 to 190,494 per sample) were obtained for the 17 samples collected. A total of 52,663 (2.64%) ITS sequences for the genus Colletotrichum were obtained. The overall percentage of Colletotrichum species varied from 0.001 in the sample collected in parcel FP38 to 20.12 for sample collected in parcel FP24 (Fig. 2). Only 3 samples had a proportion of Colletotrichum ITS sequences greater than 5% (FP24, FP18 and FP36), while 9 samples had abundances below 1% (FP20, FP21, FP9 FP38, FP26, FP37, FP32, FP35 and FP31). Among all the Colletotrichum sequences, 3 C. acutatum sensu lato ITS genetic groups8 were detected by metabarcoding approach. C. acutatum sensu lato was present in all the samples analysed. C. acutatum group A4, corresponding to C. godetiae10, was present in each sample, with abundances between 61.94 and 100% of the total Colletotrichum sequences obtained. Results shown C. godetiae to be the most abundant species in all samples except FP37, which has C. acutatum group A3, corresponding to C. fioriniae10, as the most abundant Colletotrichum species (40.89% and 59.11% respectively). C. fioriniae was the second most abundant species found, which is present on 11/17 samples, with abundances between 0.39 and 59.11%. In 5 samples the proportion of C. fioriniae was above 10%, and in 2 samples was below 1%. A third genetic group belonging to the C. acutatum species complex, and identified as group A28, was detected. C. acutatum group A2 was present only in one sample analysed (FP31), representing an 8.25% of all the Colletotrichum sequences. Due to the low resolution of the ITS locus in the C. acutatum species complex and the presence of multiple species in the same genetic group, a correct identification at species level was not possible for this set of sequences.
Percentage of occurrence of Colletotrichum spp. sequences in the overall number of ITS sequences obtained by metabarcoding (grey bars on the left) and relative percent abundances of Colletotrichum acutatum sensu lato ITS groups described by Sreenivasaprasad and Talhinhas8, (red, blue and green bars on the right). Post codes and parcel codes are reported in the centre of the figure. Samples are ordered according to geographical position from east to west.
Isolate collection
In the present study, a total of 116 samples were obtained (Table 1). Isolate 2015-4-1 was obtained from a scale insect belonging to the Coccoidea superfamily (order Hemiptera), while the other isolates were collected from fruits, buds, leaves and stems of five cultivars and several hybrids of walnut. Eighty-four strains (~72%) were isolated in the South-Eastern (SE) region, while 32 strains (~28%) were isolated in the South-Western (SW) region (Fig. 3).
Geographic distribution, postcode and number of samples used to characterize Colletotrichum species associated with walnut anthracnose in France. MB corresponding to the metabarcoding samples analysed. Red circles correspond to sites where only classic fungal isolations have been carried out while purple circles correspond to sites where classic isolation and metabarcoding sample have been collected. Geographical information about parcels sampled are reported in the table.
On PDA plates incubated at room temperature (~20 °C), cultures have two main morphological types.
The first morphotype was light grey, with cottony aerial mycelium becoming darker with age and with reverse colours ranging from brownish orange to dark grey with black spots (Fig. 4A1,A2). The majority of isolates with this morphology were later characterized as C. godetiae. The second morphotype was white to light grey on the upper side and brownish pink to vinaceous with black spots on reverse (Fig. 4B1,B2). All isolates with this morphology were later characterized as C. fioriniae. In our study two other species were isolated from walnuts, one isolate (2016-1-3) belongs to C. gloeosporioides species complex, and one isolate (2016-5-1) was identified as C. nymphaeae; the morphotypes of these two isolates are quite similar to those of the first type, but the isolate 2016-5-1 has a more orange reverse (Fig. 4C1,C2,D1,D2). When cultivated under daylight conditions the colonies showed diurnal zonation sometimes visible on the reverse side as concentric dark circles (Fig. 4A2,B2). Whatever their morphology, all the cultures have dark melanised structures similar to acervuli that oozed orange-coloured conidia. Conidia were hyaline and unicellular, cylindrical to fusiform, pointed at one or both ends (except for those from isolate 2016-5-1 which show both ends rounded), and measured 10.0 to 14.0 μm × 3.0 to 4 μm (Fig. 4A3,B3,C3 and D3) (at least 20 conidia were measured for each isolate). Both cultural and morphological characteristics were similar to those described for C. acutatum sensu lato8 with the exception of isolate 2016-5-1, for which conidial morphology is similar to that of C. gloeosporioides sensu lato16.
Ten-days Colletotrichum spp. cultures grown on PDA and isolated from nuts lesions. 1: upper side, 2: reverse, 3: conidia of A: C. godetiae (2015-24-3); B: C. fioriniae (2015-41-1); C: C. gloeosporioides sensu lato (2016-1-3); D: C. nymphaeae (2016-5-1). Conidia have been stained by cotton blue (scale bar: 20 µm).
Species identification and genetic diversity
In order to identify the species complex of each isolate obtained during this study, a phylogenetic tree of the Colletotrichum genus was built. The multi-locus analysis using the ITS, GAPDH and TUB2 performed on the 116 isolates of Colletotrichum spp. associated with walnut-growing regions revealed that 115 isolates belonged to the C. acutatum species complex and 1 isolate to the C. gloeosporioides species complex. For C. acutatum species, the phylogenetic analysis of 115 isolates and 39 reference isolates, using C. orchidophilum as outgroup, was performed. The multi-locus sequence alignment obtained concatenating ITS, CHS-1, TUB2, ACT, HIS3 and GAPDH loci, consisted of 2124 characters, of which 1591 were conserved, 303 were parsimony-informative and 208 were singleton (Supplementary Table 1).
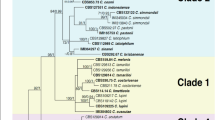
Based on the multi-locus phylogenetic analysis (Fig. 5), the 115 C. acutatum sensu lato isolates belong to three different species: C. godetiae (C. acutatum group A4), C. fioriniae (C. acutatum group A3) and C. nymphaeae (C. acutatum group A2). C. godetiae, with 80 isolates (69% of the samples), was the most abundant species, including the isolate 2015-4-1, isolated from an insect in 38160. Considering all the isolates, C. godetiae was identified in 14 out of 16 geographical sites with 100% isolates of C. godetiae identified in 26380 (SE) and 46200 (SW). C. fioriniae was the second most abundant species with 34 isolates (29.3% of the samples) found in 14 out of 16 sites, among which 24250 (SW) and 28840 (SE) resulted in 100% samples of C. fioriniae. Finally, one isolate (2016-5-1), which resulted from 46600 (SW), was identified as C. nymphaeae (Fig. 5). Except for the sites where C. godetiae was not present, and excluding the ones with 100% abundance, the presence of C. godetiae in the sites varied from 20% (26470, SE) to 90% (38160, SE), while the abundance of C. fioriniae varied from 10% in 38160 (SE) to 80% in 26470 (SW). Considering the two main regions, C. godetiae was the most abundant species in both SE and SW areas with 56.25% and 73.81% abundance, respectively. The haplotype network analysis performed over the 115 isolates of C. acutatum sensu lato resulted in 4 different haplotypes of C. fioriniae, 7 different haplotypes of C. godetiae and 1 haplotype of C. nymphaeae (Fig. 6). Their geographical distribution revealed 7 haplotypes in SW regions, covering all the three species, and 9 haplotypes in SE regions, covering C. fioriniae and C. godetiae. Three haplotypes were exclusively present in the SW regions and covered all the three species, while five haplotypes were present in the SE regions only, covering the C. fioriniae and C. godetiae species. A total of 17 nucleotide variations were counted in both populations of C. fioriniae and C. godetiae. The AMOVA results (Table 2) showed that more than 82% of molecular variation is contained within the populations (isolates from each field), and a significant (P < 0.01) differentiation was detected among the populations relative to the total population (FST = 0.179) and among populations within groups (FSC = 0.121). Even showing different haplotypes structure (Fig. 6), differentiation was not significant (P = 0.072, FCT = 0.066) among groups (geographical regions), which indicates that these regions must be connected by some mechanism of dispersion.
Bayesian inference phylogenetic tree reconstructed from a combined ITS, HIS3, GAPDH, CHS-1, TUB2 and ACT sequence alignment of 154 isolates of the C. acutatum species complex including the outgroup. Bayesian posterior probability (BPP) values (above 0.50) are shown at the nodes. The thickened nodes represent BPP of 1. Isolates obtained in this study are emphasized in bold font. C. orchidophilum CBS 632.8 is used as outgroup. Main clades within the C. acutatum species complex from Damm et al. (2012) are indicated in red. The scale bar represents the number of expected substitutions per site. Information such as tissue sampled, cultivar and geographic information (in brackets) for the isolates obtained in this work are reported.
Median-joining network of 12 Colletotrichum acutatum species haplotypes based on concatenation of ITS, HIS3, GAPDH, CHS-1, TUB2 and ACT sequences alignments. Circles areas are proportional to the number of strains with a specific haplotype. Segments reported in the connecting lines represent number of mutations between haplotypes. Circles slices area is proportional to the number of strains isolates from a specific geographic area whereas colours indicate the geographic origin according to legend (from yellow to red indicate south west (SW) of France while from green to blue indicate south east (SE) of France).
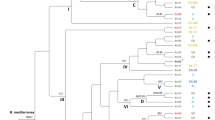
For C. gloeosporioides sensu lato, 1 isolate and 39 reference isolates, with C. sydowii as outgroup, were analysed. Phylogenetic analysis was performed on a multi-locus concatenated sequence alignment (ITS, CHS-1, CAL, ACT, SOD2, TUB2, GS, GAPDH and ApMAT locus) resulting in 5716 characters, of which 3658 were conserved, 768 parsimony-informative and 1051 singletons (Supplementary Table 1). Based on the multi-locus phylogenetic analysis, the C. gloeosporioides sensu lato isolate (2016-1-3) deriving from site 38470, in the SE region, does not belong to any accepted species and is closely related to C. rhexiae and C. fructivorum (Fig. 7).
Bayesian inference phylogenetic tree reconstructed from a combined ITS, GAPDH, CHS-1, ACT, TUB2, GS, SOD2, ApMAT and CAL sequence alignment of 40 isolates of the C. gloeosporioides species complex including the outgroup. Bayesian posterior probability (BPP) values (above 0.50) are shown at the nodes. The thickened nodes represent BPP of 1. Isolates obtained in this study are emphasized in bold font. Colletotrichum sydowii CBS 135819 is used as outgroup. The scale bar represents the number of expected substitutions per site. Information such as tissue sampled, cultivar and geographic information (in brackets) for the isolates obtained in this work are reported.
Pathogenicity tests
Nineteen days after inoculation, all fruits clearly showed necrotic lesions, all strains tested were pathogenic on walnuts fruits; Koch’s postulates, therefore, were verified.
When diameters of necrotic lesions were submitted to ANOVA, all isolates produced lesions whose diameter was significantly bigger than those on control (P = 0.0001).
Data were then submitted to post hoc Tukey’s test whose results are showed in Fig. 8. Generally, isolates could be divided into two groups: the first including C. fioriniae 2015-26-1, C. godetiae 2015-24-3, C. fioriniae 2015-41-1, C. nymphaeae 2016-5-1, C. fioriniae 2015-19-2 and C. gloeosporioides sensu lato 2016-1-3 that showed no significant intra-grouping differences among them; the second included two C. godetiae strains (2015-39-2 and 2015-19-1) that caused lesions significantly smaller than those produced by the other isolates but significantly larger than controls.
Histograms showing average lesions size of 8 Colletotrichum reference isolates on walnut fruits (cultivar Lara). Bars indicate the average diameters of the lesion in cm. Standard deviations are reported as lines at the end of each bar. Letters at the extreme of each bar indicate significant differences based on ANOVA Tukey post hoc test results.
Discussion
In 2011, an epidemic of anthracnose on walnut was observed in France. This was shown to be caused by members of the genus Colletotrichum5, leading to 50–70% of losses with some orchards experiencing 100% losses. In the past decade, anthracnose on walnut caused by Colletotrichum spp. was also reported in the Shandong province and in the Guangxi region, in China12,13,14. However, Colletotrichum species causing epidemic infections of walnut anthracnose in Europe have never been characterized. Information regarding the presence of Colletotrichum spp. on walnut in Europe is scarce; however one strain of C. godetiae and one of C. gloeosporioides have been associated with this plant in Austria10 and Slovakia9 respectively. Hence, there was a need to characterize the species of Colletotrichum associated with walnut, which was the basis of the present study. The current study represents the first identification of Colletotrichum species associated with anthracnose of walnut in France using a metabarcoding and a multi-locus phylogenetic combined approach.
Molecular identification of the pathogenic species associated with walnut provides a useful tool to help to understand the distribution and the interactions between the host and its pathogens. In this study, a total of 116 isolates were obtained from infected walnuts tissues. In France, walnut is mainly cultivated in the Auvergne-Rhône-Alpes region in SE and in the Occitanie and Nouvelle-Aquitaine regions in SW. Samples were collected where the disease incidence was higher, mainly in the former Rhône-Alpes region for SE samples and between Aquitaine, Midi-Pyrénées and Limousin former regions for SW samples. Moreover, parts of these areas were sampled and used for metabarcoding analysis.
The multi-locus characterization method led to the identification of four different species: 80 isolates of C. godetiae (69%), 34 isolates of C. fioriniae (29.3%), 1 isolate of C. nymphaeae (0.86%) and 1 isolate of C. gloeosporioides sensu lato (2016-1-3, 0.86%). These results are coherent with data obtained from the metabarcoding analysis where the most abundant sequences belong to C. acutatum group A4 (C. godetiae, 17/17 of the samples), corresponding to 89.88% of the total Colletotrichum sequences, followed by C. acutatum group A3 (C. fioriniae, 11/17 of the samples), corresponding to 9.64% of the Colletotrichum sequences obtained.
Metabarcoding analysis is a powerful DNA sequencing technique that provides a realistic approximation of the quantitative presence of species in a sample. It is also a useful tool to characterize the species recovered in a sample17.
However, it is important to highlight that metabarcoding analysis, due to the presence of chimeric sequences or differences in template DNA copy number, can suffer from biases which may lead to an overestimation or underestimation of the species present in a sample17. Moreover a metabarcoding approach can detect false positive due to the persistence of DNA in the environment after cells have lost viability18. On the other hand, fungal isolation methods are suitable to characterize the species of a sample and to cover its variability, since they are based on phenotypic characters that may be highly selective. Therefore, in order to correctly identify the cultivable pathogenic species associated to a specific host, metabarcoding analysis should always be coupled with isolation methods.
Whilst being accepted widely as the universal fungal barcode region, the ITS region is not able to delimit species with the genus Colletotrichum, and especially not within its species complexes such as C. acutatum sensu lato. In contrast, the use of fungal isolation methods coupled with the multilocus genetic characterization enabled the definition of the C. acutatum A2 genetic group as C. nymphaeae. Furthermore, fungal isolations allowed the recovery of a fourth Colletotrichum species belonging to the C. gloeosporioides species complex and closely related to C. rhexiae and C. fructivorum.
Samples derived from the southern part of France, were mapped and divided on the basis of their geographical origin. The two most representative species, C. godetiae and C. fioriniae, do not show a uniform distribution between the two areas, and no significant differentiation was found at the haplotype level between the two areas. All things considered, on the basis of the samples we had and the results we obtained, we could not find any correlation that could indicate a common origin of the haplotypes where the disease initially originated. Moreover, based on the data obtained in this study, no correlation can be observed considering the cultivar or the matrix from which the samples were isolated. However, further investigations covering a more extended sample area, a wider temporal distribution and sampling a higher number of isolates, may contribute to clarify whether species, geographical areas and cultivars are correlated.
The study also highlighted a high genetic variation between the two most abundant species, C. godetiae and C. fioriniae. Particularly, C. godetiae presented in seven distinct haplotypes while C. fioriniae resulted in four haplotypes, although a higher number of samples were obtained during the study. Proportionally, the number of haplotypes over the number of isolates resulted similar in both species, with isolates differing from each other for only one to seventeen nucleotide variations.
Interestingly, one isolate of C. godetiae was isolated from an insect body (2015-4-1). A scale insect, which did not present any symptom of disease, alive at the time of sampling, was caught and assessed for the presence of Colletotrichum sp. The insect was sampled because in 2010, one year before the epidemic event occurred, some areas suffered a big attack of cochineals. Although the capacity of this C. godetiae isolate to cause disease on the insect was not investigated, the ability of this fungus to colonize and infect insects is documented19,20. Similarly, Gaffuri et al. 201521 reported the presence of Colletotrichum acutatum sensu lato on the Asian chestnut gall wasp (Dryocosmus kuriphilus) affecting chestnut (Castanea sativa); authors speculate about the ecological role of the insect in the spread of this fungus on other chestnut plants. Undoubtedly, the presence of C. godetiae on the body of the insect should be investigated considering the ability of the insect to act as a pathogen vector, especially because adult male insects are winged and able to fly and certain stadia of the nymph, called crawlers, are able to move and are considered the main dispersal agents for Coccoidea22. Scale insects are also a considerable inoculum source, since female insects heavily feed on different parts of the plant causing important injuries on the tissues, thus facilitating the pathogen penetration23.
Pathogenicity tests revealed that two isolates of C. godetiae (2015-39-2 and 2015-19-1), one of the most abundant species isolated from walnuts affected by anthracnose, produced smaller lesions compared to the other strains when artificially inoculated on fruit. Similar situations have been reported in other pathosystems; for example C. gloeosporioides species are found only occasionally on strawberry in the UK, though in vitro assays reported those as the most aggressive species24. The large presence of C. godetiae on anthracnose lesions may be related to environmental factors, which promote the pathogen diffusion causing a population burst. Further studies, using a more consistent set of isolates and cultivars, are needed to obtain additional data about the aggressiveness of the isolates and the susceptibility of the tested cultivars to Colletotrichum spp.
Characterization of the Colletotrichum species associated with walnut anthracnose provides considerable knowledge and allows targeted treatments to be implemented. This is of particular concern considering that distinct Colletotrichum species respond differently to specific groups of chemical compounds25,26. Moreover, the knowledge of the etiological agents of a disease allows the development of diagnostic procedures that can help to monitor and limit the disease. Finally, in order to better elucidate the epidemiology and the pathogen behaviour, it is important to define those factors contributing to species abundance.
Material and Methods
Sampling
Plant tissues for metabarcoding analysis
Walnut buds were collected from 17 parcels during May-June 2016. In total, 10 parcels were surveyed in South-East (SE) of France (Two parcels in: Beaulieu, 38470; Cras, 38210. And one parcel in: Laissaud, 73800; La Buissière, 38530; Geyssans, 26750; Saint Romans, 38160; Peyrins, 26380; La Motte, 26470) and 7 in South-West (SW) of France (One parcel in: Toulenne, 33210; Terrasson La Villedieu, 24120; Puybrun, 46130; Saint Cybranet, 24250; and three parcels in: Montvalent, 46600) (Fig. 3).
For each parcel, twenty walnut buds from 10 different plants were cut with a sterilized scalpel, mixed and ground with liquid nitrogen in an autoclaved mortar and pestle. DNA was extracted from plant tissues using FastDNA® SPIN kit (MP Biomedicals, Santa Ana, CA, USA) following the manufacturer’s instructions. Quality and concentration of purified DNA were determined using a UV spectrophotometer (NanoDrop1000, Thermo Scientific, USA), and dilutions of at least 10 ng/μL were prepared for each DNA sample using nuclease-free water (Promega, Madison, WI, USA).
Colletotrichum spp. isolation and morphological description
From July 2015 to May 2016, plant tissue samples were collected from 36 parcels in 16 locations of southern France as shown in Fig. 3. Isolation was performed on fruits, buds, leaves and stems of walnut trees affected by walnut anthracnose.
Collected plant material was cut in small pieces, washed three times (the first one by using a 1% (v/v) NaClO water solution for 1 min, then twice for 2 min using sterile water) and dried on a paper sheet in sterile conditions. Samples were placed in Petri dishes (90 mm) containing Potato Dextrose Agar medium (PDA, Difco Laboratories, USA) and 100 ppm of streptomycin sulphate (Sigma-Aldrich, St Louis, MO, USA), then incubated for at least four days at room temperature. After four/seven days, three to five small agar plugs containing fungal mycelium, identified as Colletotrichum sp. by macroscopic and microscopic observations, were transferred to a fresh PDA plate and incubated in the dark at 25 °C for 10 days. One sample (2015-4-1) was obtained from an asymptomatic insect (Hemiptera: Sternorrhyncha: Coccidae) isolated from the branch of a walnut tree.
Cultures were maintained at 25 °C on PDA for up to a week under a 12 h light/dark cycle. Long-term storage involved cryoconservation of spores in liquid nitrogen.
Morphological observations (mycelium colour, texture, zonation, growing margin, and colour of the reverse side) of all isolates were made on cultures grown on PDA plates incubated at room temperature (~20 °C) under natural daylight27.
Observations and measurements of conidial size and shape have been made by microscopic observation at ×1000 on spores (20 randomly chosen) harvested after 10 to 14 days incubation and mounted in cotton blue27.
Metabarcoding analysis of Colletotrichum spp. diversity in walnut buds
A total of 17 samples were used for amplicon PCRs and Illumina Miseq PE300 sequencing, which was performed at the McGill University and Génome Québec Innovation Centre, Montréal, Canada. Primers ITS1F and ITS428 were used to amplify the internal transcribed spacer.
Data Analysis and Statistics
Although expected, a low level of joined pair reads for the analysis of ITS sequences were obtained, leading us to choose an alternative approach with QIIME29. The forward and reversed reads were merged in both multiple fasta files independently, using multiple_split_libraries_fastq.py.
ITS1 and ITS2 regions were first extracted separately from read1 and read2 nonchimera-fasta files respectively, using ITSx30 before being concatenated in a new fasta file. Chimera detection was made in the new fasta file, with ITS1 and ITS2 concatenated and lacking in 5.8 region sequence, using the UCHIME algorithm31 with vsearch v1.1.3 (https://github.com/torognes/vsearch) and the UNITE/INSDC representative/reference sequences version 7.032 as reference database. Only non-chimeric sequences were used for OTU picking using the QIIME script pick_open_reference_otus.py, with BLAST33 as taxonomic assignment method and a modified database from UNITE plus INSD non-redundant ITS database version 7.134. The modified database was obtained by extracting, using ITSx software, and concatenating ITS1 and ITS2 region sequences from UNITE v7.1 database. To minimize the overestimation of rare OTUs in the community analysis, we include only OTUs with sequence count greater than 1035,36. OTUs with “No blast hit” were also discarded to determine the total number of ITS sequences obtained per sample.
For taxonomic assignment at Colletotrichum species complex level, the same approach and parameters were used for OTU selection with a home-made ITS-Colletotrichum database. The database was obtained selecting entire ITS sequences from representative strains according to currently accepted species of Colletotrichum37. Species were selected based on phylogenetic distribution in order to cover the diversity of the genus. ITS1 and ITS2 region sequences were extracted using ITSx software, and concatenated. Only OTUs with e-value = 0 and 97% of similarity based on blastn results against ITS-Colletotrichum database were selected. All the ITS raw reads files have been deposited at NCBI and are available under Bioproject ID SRP126756, with the BioSample accession numbers from SRS2758044 to SRS2758060.
Multi-locus phylogenetic analysis of Colletotrichum species associated with walnut anthracnose
Genomic DNA extraction and PCR amplification
10-day-old fungal mycelium was scraped from the surface of a PDA plate using a sterile scalpel and transferred into a sterile 2 mL tube. Genomic DNA was then extracted using the FastDNA® SPIN kit (MP Biomedicals, Santa Ana, CA, USA) following the manufacturer’s instructions with an initial homogenization step using the Retsch MM400 instrument (Retsch GmbH) at 30 Hz for 30 sec, for two times. The DNA was resuspended in 100 µL of sterile nuclease-free water, quantified and checked in quality using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, DE, USA). DNA aliquots were stored at a temperature of −20 °C for further use.
In order to establish the species complex designation, for each isolate, the internal transcribed spacer (ITS) region, partial sequence of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene and partial sequence of the beta-tubulin 2 gene (TUB2) (exons 3 through 6, including introns 2 through 4), regions were initially sequenced and compared with reference sequences38. Other loci were subsequently amplified to determine the species designation according to Damm et al.10 for the C. acutatum species complex and to Weir et al.16 for the C. gloeosporioides species complex.
For isolates belonging to the C. acutatum species complex, partial sequences of the chitin synthase 1 gene (CHS-1), actin gene (ACT) and histone H3 gene (HIS3) were amplified and sequenced. For isolates identified as belonging to the C. gloeosporioides species complex, partial sequence of the chitin synthase 1 gene (CHS-1), actin gene (ACT), glutamine synthetase (GS), calmodulin (CAL) and Apn2/Mat1-2-1 intergenic spacer (ApMAT) were amplified and sequenced.
Amplification reactions were performed in 25 μL volume using 0.025 U/μL of GoTaq Flexi DNA polymerase (Promega) and 1 × GoTaq Flexi buffer (Promega), 25–50 ng of template DNA, 0.08 μM of each primer, 2 mM of MgCl2 and 0.2 mM of 10 mM dNTP mix (Promega). For GAPDH and TUB2 genes, primer concentration was increased to 0.2 μM while dNTP mix concentration was decreased to 0.08 mM. A list of the primers and conditions used in this study is reported in Table 3.
Amplification products were analysed by electrophoresis in 1 × TAE buffer (40 mM Tris-acetate, 1 mM EDTA) with 1% (w/v) agarose gel (LE, analytical grade agarose; Promega) prepared using 1 × TAE buffer and detected by UV fluorescence after GelRed™ (Biotium Inc., CA) staining, according to manufacturer’s instructions. The BenchTop 100-bp DNA ladder (Promega) was used as molecular size marker. PCR products were sent to Eurofins MWG (Ebersberg, Germany) for purification and sequencing in forward and reverse, using the same primers used for PCR. ABI trace files were analysed and consensus sequences were generated using Geneious® 10.0.6 (Biomatters, http://www.geneious.com).
Phylogenetic analysis and species identification
To establish the species complex of each isolate, a phylogenetic tree of the Colletotrichum genus was constructed using a concatenated alignment of ITS, TUB2 and GAPDH39. For the isolates belonging to the acutatum complex, phylogenetic analysis was conducted using a sequence dataset enriched with 39 ex-type and other reference strains of species belonging to the C. acutatum complex, C. orchidophilum was used as outgroup. For the isolate belonging to the gloeosporioides complex, sequences of 39 reference strains were used and C. sydowii was used as outgroup. All reference sequences based on Marin-Felix et al.38 are available and listed in Supplementary Table 2.
The sequences obtained were aligned using MAFFT v. 7.30440. Multiple sequence alignments were exported to MEGA741 and the best-fit substitution model was calculated for each separate sequence dataset. The multi-locus concatenated alignment was performed using Geneious 10.0.6. Using MrBayes 3.2.642, the Markov chain Monte Carlo (MCMC) algorithm was performed to generate phylogenetic trees with Bayesian posterior probabilities for combined sequence datasets using, for each locus, the nucleotide substitution models determined by MEGA7. Four MCMC chains were run simultaneously for random trees for 5,000,000 generations. Samples were taken every 1,000 generations. The first 25% of trees were discarded as burn-in phase of each analysis and posterior probabilities were determined from the remaining trees.
To visualize intraspecific evolutionary and geographic relationships between isolates the Median-joining network algorithm43 was used to build a haplotypes network using the software PopART v1.744. Analysis of molecular variance (AMOVA) was performed with Arlequin 3.545 to compare the genetic structure of 2 groups: samples from South East (SE; haplotypes = 6, isolates = 31), samples from South West (SW; haplotypes = 10, isolates = 83). For this purpose, conventional F-statistics and 10,000 permutations to test significance were used with haplotype frequencies.
Pathogenicity tests
Eight representative Colletotrichum strains (C. godetiae 2015-19-1, 2015-24-3 and 2015-39-2; C. fioriniae 2015-19-2, 2015-26-1 and 2015-41-1; C. nymphaeae 2016-5-1; C. gloeosporioides sensu lato 2016-1-3; Table 1), selected among the isolates obtained during this study, were used to perform pathogenicity tests on artificially wounded fruits (cultivar Lara).
Fruits, harvested 100 days after the beginning of fruit enlargement, were first washed with distilled water and then surface sterilized using a 70% (v/v) ethanol solution for 1 min, rinsed twice with distilled water and dried on a paper sheet. Surface sterilized fruits were wounded on the pericarp using a 2 mL pipette tip and an agar plug (0.2 cm in diameter) containing the fungal mycelium, was placed in the wound. 5 Wounded fruits inoculated with agar without mycelium were used as control. For each strain 5 fruits were inoculated. The test was independently replicated twice. Inoculated fruits were then incubated in a moist chamber at 24 °C.
The development of the necrosis was daily monitored and the two perpendicular necrosis diameters were recorded 4, 8 and 14 days after the first symptoms appeared, corresponding to 9, 13 and 19 days post inoculation. Data from the final measurements were submitted to analysis of variance (ANOVA and Tukey’s multiple post hoc range test), with isolate as independent variable, by using Systat 11 (Systat Software, USA) and assuming P < 0.05 as significant level.
At the end of the experiment, each strain was re-isolated from the affected fruits and cultured on PDA and streptomycin sulphate in order to confirm the identity (based on morphological characters) of the causal agent.
References
Martínez-García, P. J. et al. The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. Cell Mol. Biol. 87, 507–532 (2016).
McGranahan, G. & Leslie, C. Breeding Walnuts (Juglans Regia). In Breeding Plantation Tree Crops: Temperate Species 249–273 (Springer, 2009).
Molnar, T. J. et al. Persian walnuts (Juglans regia L.) in Central Asia. In Northern Nut Growers Association 101st Annual Report 56–69 (2011).
Statistical Databases. World and France total production, Walnut with shell. FAO, FAOSTAT, Available at: http://www.fao.org/faostat/ (2014).
Giraud, M. & Verhaeghe, A. Fiche bio-agresseur: Anthracnose à Colletotrichum sp. en verger de noyers. Arboric. Fruitière 2 (2015).
Cannon, P. F., Damm, U., Johnston, P. R. & Weir, B. S. Colletotrichum – current status and future directions. Stud. Mycol. 73, 181–213 (2012).
Baroncelli, R. et al. The Colletotrichum acutatum species complex as a model system to study evolution and host specialization in plant pathogens. Front. Microbiol. 8, 2001 (2017).
Sreenivasaprasad, S. & Talhinhas, P. Genotypic and phenotypic diversity in Colletotrichum acutatum, a cosmopolitan pathogen causing anthracnose on a wide range of hosts. Mol. Plant Pathol. 6, 361–378 (2005).
Juhásová, G., Ivanová, H. & Spišák, J. Occurrence and spread of the parasitic microscopic fungi on walnut (Juglans regia L.) on various localities of Slovakia. Trak. Univ. J. Nat. Sci. 6, 19–27 (2015).
Damm, U., Cannon, P. F., Woudenberg, J. H. C. & Crous, P. W. The Colletotrichum acutatum species complex. Stud. Mycol. 73, 37–113 (2012).
Xiaa, L. I. U., Keqianga, Y. A. N. G., Yufenga, Z. H. U. & Yanfeia, Y. I. N. Laboratory toxicity of eight fungicides against Colletotrichum gloeosporioides causing walnut anthracnose. Chin. J. Pestic. Sci. 15, 412–420 (2013).
Zhu, Y. F., Yin, Y. F., Qu, W. W. & Yang, K. Q. Occurrence and spread of the pathogens on walnut (Juglans regia) in Shandong province, China. Acta Hortic. 1050, 347–351 (2014).
Wang, Q. H. et al. First Report of Walnut Anthracnose Caused by Colletotrichum fructicola in China. Plant Dis. 102, 247 (2018).
Zhu, Y. Z., Liao, W. J., Zou, D. X., Wu, Y. J. & Zhou, Y. First report of leaf spot disease on walnut caused by Colletotrichum fioriniae in China. Plant Dis. 99, 289 (2015).
Zhu, Y. et al. Construction of a high-density genetic map using specific length amplified fragment markers and identification of a quantitative trait locus for anthracnose resistance in walnut (Juglans regia L.). BMC Genomics 16, 614 (2015).
Weir, B. S., Johnston, P. R. & Damm, U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 73, 115–180 (2012).
Thomas, A. C., Deagle, B. E., Eveson, J. P., Harsch, C. H. & Trites, A. W. Quantitative DNA metabarcoding: improved estimates of species proportional biomass using correction factors derived from control material. Mol. Ecol. Resour. 16, 714–726 (2016).
Pochon, X., Zaiko, A., Fletcher, L. M., Laroche, O. & Wood, S. A. Wanted dead or alive? Using metabarcoding of environmental DNA and RNA to distinguish living assemblages for biosecurity applications. PLOS ONE 12, e0187636 (2017).
Marcelino, J. et al. Colletotrichum acutatum var. fioriniae (teleomorph: Glomerella acutata var. fioriniae var. nov.) infection of a scale insect. Mycologia 100, 353–374 (2008).
Gasparoto, M. C. G. et al. Honeybees can spread Colletotrichum acutatum and C. gloeosporioides among citrus plants. Plant Pathol. 66, 777–782 (2017).
Gaffuri, F. et al. Colletotrichum acutatum associated with Dryocosmus kuriphilus galls on Castanea sativa. For. Pathol. 45, 169–171 (2015).
Gullan, P. J. & Martin, J. H. Sternorrhyncha: (Jumping Plant-Lice, Whiteflies, Aphids, and ScaleInsects). in Encyclopedia of Insects (Second Edition) (eds. Resh, V. H. & Cardé, R. T.) 957–967 (2009).
Mansour, R., Griss-Lebdi, K., Suma, P., Mazzeo, G. & Russo, A. Key scale insects (Hemiptera: Coccoidea) of high economic importance in a Mediterranean area: host plants, bio-ecological characteristics, natural enemies and pest management strategies - a review. Plant Prot. Sci. 53, 1–14 (2017).
Baroncelli, R. et al. Molecular diversity of anthracnose pathogen populations associated with UK strawberry production suggests multiple introductions of three different Colletotrichum species. PLoS ONE 10, e0129140 (2015).
Freeman, S., Katan, T. & Shabi, E. Characterization of Colletotrichum species responsible for anthracnose diseases of various fruits. Plant Dis. 82, 596–605 (1998).
Bragança, C. A. D., Damm, U., Baroncelli, R., Massola Júnior, N. S. & Crous, P. W. Species of the Colletotrichum acutatum complex associated with anthracnose diseases of fruit in Brazil. Fungal Biol. 120, 547–561 (2016).
Cannon, P. F., Buddie, A. G. & Bridge, P. D. The typification of Colletotrichum gloeosporioides. Mycotaxon 104, 189–204 (2008).
Anderson, I. C. & Cairney, J. W. G. Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environ. Microbiol. 6, 769–779 (2004).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Bengtsson-Palme, J. et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 4, 914–919 (2013).
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011).
Nilsson, R. H. et al. A Comprehensive, Automatically Updated Fungal ITS Sequence Dataset for Reference-Based Chimera Control in Environmental Sequencing Efforts. Microbes Environ. 30, 145–150 (2015).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Kõljalg, U. et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277 (2013).
Oliver, A. K., Brown, S. P., Callaham, M. A. Jr. & Jumpponen, A. Polymerase matters: non-proofreading enzymes inflate fungal community richness estimates by up to 15%. Fungal Ecol. 15, 86–89 (2015).
Brown, S. P. et al. Scraping the bottom of the barrel: are rare high throughput sequences artifacts? Fungal Ecol. 13, 221–225 (2015).
Jayawardena, R. S. et al. Notes on currently accepted species of Colletotrichum. Mycosphere 7, 1192–1260 (2016).
Marin-Felix, Y. et al. Genera of phytopathogenic fungi: GOPHY 1. Stud. Mycol. 86, 99–216 (2017).
Baroncelli, R. et al. Gene family expansions and contractions are associated with host range in plant pathogens of the genus Colletotrichum. BMC Genomics 17, 555 (2016).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Bandelt, H. J., Forster, P. & Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Leigh Jessica, W., David, B. & Shinichi, Nakagawa PopART: full‐feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116 (2015).
Excoffier, L. & Lischer, H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010).
White, T., Bruns, T., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. in PCR Protocols: A Guide toMethods and Applications(eds. Innis, M., Gelfand, D., Shinsky, J. & White, T.)315–322 (1990).
Guerber, J. C., Liu, B., Correll, J. C. & Johnston, P. R. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia 95, 872–895 (2003).
Woudenberg, J. H. C., Aveskamp, M. M., de Gruyter, J., Spiers, A. G. & Crous, P. W. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia - Mol. Phylogeny Evol. Fungi 22, 56–62 (2009).
Carbone, I. & Kohn, L. M. A Method for Designing Primer Sets for Speciation Studies in Filamentous Ascomycetes. Mycologia 91, 553–556 (1999).
Crous, P. W., Groenewald, J. Z., Risède, J.-M., Simoneau, P. & Hyde, K. D. Calonectria species and their Cylindrocladium anamorphs: species with clavate vesicles. Stud. Mycol. 55, 213–226 (2006).
Silva, D. N. et al. Application of the Apn2/MAT locus to improve the systematics of the Colletotrichum gloeosporioides complex: an example from coffee (Coffea spp.) hosts. Mycologia 104, 396–409 (2012).
Acknowledgements
This work was supported by FranceAgriMer and French Ministry of Agriculture (CASDAR #29-2015-03- Maladies émergentes) coordinated by G.L.F. The authors gratefully acknowledge the producers of walnuts for the use of the data from their orchards.
Author information
Authors and Affiliations
Contributions
R.B. conceived and designed the experiments; D.D.L. and R.B. performed PCRs and phylogenetic analyses; J.F.C.D. performed the metabarcoding analysis; C.M., M.G., D.K. and A.V. provided plant samples and performed pathogenicity tests; P.N. and M.C. performed fungal isolations observations and DNA extractions; S.S. performed statistical analyses; G.L.F. supervised the project; R.B., D.D.L. and J.F.C.D. wrote the first draft of the manuscript. All authors read, corrected and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Da Lio, D., Cobo-Díaz, J.F., Masson, C. et al. Combined Metabarcoding and Multi-locus approach for Genetic characterization of Colletotrichum species associated with common walnut (Juglans regia) anthracnose in France. Sci Rep 8, 10765 (2018). https://doi.org/10.1038/s41598-018-29027-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-29027-z
This article is cited by
-
Multilocus phylogenetic analysis of Colletotrichum gloeosporioides causing anthracnose disease of Murraya koenigii in India
Indian Phytopathology (2023)
-
Development of species-specific primers and highly sensitive duplex ddPCR assay for the identification and detection of chili anthracnose
European Journal of Plant Pathology (2022)
-
Genome-wide identification and characterization of long non-coding RNAs conferring resistance to Colletotrichum gloeosporioides in walnut (Juglans regia)
BMC Genomics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.