Abstract
Understanding the general principles governing the functioning of biological networks is a major challenge of the current era. Functionality of biological networks can be observed from drug and target interaction perspective. All possible modes of operations of biological networks are confined by the interaction analysis. Several of the existing approaches in this direction, however, are data-driven and thus lack potential to be generalized and extrapolated to different species. In this paper, we demonstrate a systems pharmacology pipeline and discuss how the network theory, along with gene ontology (GO) analysis, co-expression analysis, module re-construction, pathway mapping and structure level analysis can be used to decipher important properties of biological networks with the aim to propose lead molecule for the therapeutic interventions of various diseases.
Similar content being viewed by others
Introduction
Analysis of protein interaction network for targets of FDA approved drugs and genes related to disease in OMIM revealed that most drug targets are not even closer to the genes specifically involved in the disease and hence reflects the lack of selectivity in traditional drugs towards the genetic cause1. Besides, biasness of literature-mined interaction sets towards well-known proteins, dependence of current approach on target profile similarity or identification of shortest path between drug targets in the interactome has proved to be less efficient in the analysis of relationship between drugs and disease2,3.
However, an interdisciplinary approach like the ones used by Albert-László Barabási Group has reflected its efficiency to predict novel targets and other uses of the existing drugs through network-driven knowledge4,5,6,7,8. In addition, recent findings have demonstrated that genes associated with a disease, tend to cluster in a disease oriented module and represent a connected sub-network within the interactome9,10,11. Many online databases and network approaches have been developed to handle drug-target interaction such as prediction of drug target interaction by integrating protein sequences and drug chemical strcutures12, network construction on the basis of heterogeneous biological data13, non-coding RNAs and drug targets based networks14, drug target interaction prediction models15,16, rotation forest-based drug target prediction17,18, and drug target prediction using deep neural networks19. This led us to think, that for a drug to be efficient enough to cure a disease, it must target proteins within or in the immediate vicinity of the disease module formed by the well-associated genes20,21. Hence, to understand therapeutic action of drugs at different levels of biological organization, we developed an unsupervised and unbiased network-driven framework to come-up with a drug-disease proximity measure that would help us to quantify the therapeutic effect of drugs.
In this study, we selected Picroliv to understand of the context within which drug-target interactions at molecular level can lead to distal effectors in a process that result in adverse phenotypes at the organ and organismal levels. Picroliv, is one of the active compounds yielded by underground parts of Picrorhiza kurroa, growing at elevations of 3,000–5,000 meters. It is usually a mixture of kutkoside and picroside-I in 1:1.5 ratio22. While the other major product synthesized by the underground part is kutkin composed of picroside-I and picroside-II23.
The active principal component of Picrorhiza kurroa is kutkin comprehend kutkoside and the iridoid glycoside picrosides I, II. Picroside I, also known as 6′-O-cinnamoylcatalpol, forms a stable mixture with kutkoside to form kutkin24. Another isolated catalpol derivative, identified as 6-O-vanilloylcatalpol, was named Picroside II23. Traditionally picrorhiza has been used to treat disorders of the liver and upper respiratory tract, dyspepsia, chronic diarrhea, and scorpion sting. Studies on Picrorhiza show its crucial role in restoring the depleted glutathione levels in rats infected with malaria22,24. Further studies on picrorhiza reveal its anti-lipid peroxidative effect25. Recent studies show that Picroside II plays a critical role in preventing the alterations that take place in I/R injury26,27. Although the anticancerous activity of picrorhiza has been exploited, its exact molecular mechanisms of actions and related pathways and targets remains poorly understood28.
To achieve the desired therapeutic effect while reducing the risk of unpropitious conditions, with a known drug, it is imperative to identify the neighborhood of these targets within which they have their action29. Consequently, using information from known drug target and creating networks of associated target proteins; we can understand how drugs can have beneficial as well as pernicious consequences10. Based upon these observations, relevant drugs for specific disease could be filtered out to provide only the beneficial population of drugs to the patients.
To decipher the regulatory interactions and underlying mechanistic behavior of picrorhiza, a target-pathway network re-construction was performed to discover the relationship between the drug and its relevant targets and pathways. Construction and analysis of such intricate network not only requires the basic concepts of network biology but also an understanding of how the interaction between drug and its relevant target determines regulation of various phenotypic characters in a diseased state. Besides the direct consequences of the interaction between drug and its target, drug action also depends on the consequences within the physiological system. Therefore a holistic approach is required to deal with drug-target interaction network for the selection of putative drug candidates.
As stated earlier, integration of concepts from various fields can help to reach the best solution for a given problem. Hence, we integrated advanced application of computational and experimental information through literature based support in our work to build networks for analyzing drug action and to develop poly-pharmacology for complex diseases and predict therapeutic efficacy and adverse event risk for individuals prior to commencement of therapy. In this study, we demonstrate a systems pharmacology pipeline and discuss how the network theory in combination with gene ontology (GO) analysis, co-expression analysis, module re-construction, pathway mapping and structural analysis can be used to decipher important properties of biological networks.
Results and Discussion
To acquire holistic view through empirical data, literature mining was performed to identify known targets for Picroside I, II, III, and IV. Unlike P-I and P-II, no target was identified for P-III and P-IV which led us to drop them for further analysis. The reason for such outcome can be imparted to its inability to cross the blood-brain-barrier30. Further, to uncover unknown drug targets those are yet to be verified experimentally, mapping of P-I and P-II structures was done against protein/receptor library through PharmMapper with threshold limited to 3031. Combined results from literature mining and PharmMapper, showed the presence of targets common to both and hence were categorized as primary targets while the ones found only in PharmMapper were categorized as secondary targets for downstream analysis.
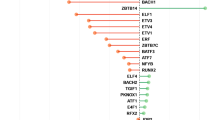
To further address the question that whether targets taken as secondary are appropriate or not, we retrieved top ten co-expressed genes by considering primary targets as query dataset based on confidence score. Selected nodes were then considered for degree distribution with betweenness centrality, which would state the importance of genes with respect to their association with involved pathways. In this analysis, prioritization of node was done based on k and Bc correlation. The node size reflects the association score, i.e., more the association score bigger will be the node and vice-versa.
Genes selected through co-expression analysis were combined together for P-I and II separately and gene ontology analysis was performed using GORILLA to identify the role of target association genes in Biological processes, Molecular functions and Cellular components32. Based on p-value, association type weak or strong was indicated, which further referred based on well-compiled GO databases. To further understand the role of primary targets we performed the docking analysis using PatchDock server, which checks various conformations and suggests the best one (Table 1). Additionally, we performed the PatchDock analysis with other FDA-approved drugs available in the market to find out the best drug based on molecular interactions in selected targets against P-I, P-II and other drugs33. Prioritized targets were cross-checked with literature and found to be key player in carcinogenesis and therefore their role in various malignancies was found to be crucial.
Primary targets were considered for module definition and tried to converge on pathways on the basis of co-expression based association score. Genes were highlighted using different colors and size, where red color represents the association between degree and betweenness centrality of nodes and node size shows the co-expression association between the interacting nodes (Fig. 1). Mapping of these modules on pathways was performed through pathway reconstruction (Fig. 2).
Pathway Analysis
For drug-target interaction network we have considered literature mining techniques, scoring functions on the basis of co-expression modules derived from cancer and KEGG database as reference for giving a support factor for holistic network visualization by using Reactome Pathway Database and Pathway Interaction Database. The pipeline presented is based upon working modules where we have compiled the information through the stepwise procedures and outcome of one step can be used as an input for the next step. At few points cross validation is also being applied to present the refined information to the further steps. Pipeline is being verified through all the available data sets for the analysis and finally the robust one is being proposed. We have broadly explored the possible routes and diversion points on the basis of node involvement in networks and data is being generated from standard pathways available. Pathway analysis revealed that genes are distributed in pathways associated with various diseases such as, Cancer associated signaling, Hepatitis–B, Human T-cell leukemia virus type 1 (HTLV-I) Infection, Tuberculosis, Influenza A, Thyroid Hormonal Signaling Pathway and many more. But careful evaluation and mapping on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using combined score resulted association of maximum number of genes with cancer associated signaling, viz. receptor based Death-Associated Protein Kinase 1 (DAPK1) activation, Transforming Growth Factor Beta (TGFβ) signaling, Interleukin (IL) 2, 4 signaling and cytokine signaling. All these signaling cascade results in transcriptional activation and leads to carcinogenesis (Fig. 2).
Death-Associated Protein Kinase mediated signaling
Calcium/calmodulin-dependent serine/threonine kinases (CDK) play a major role in activation of various signaling via regulating apoptotic pathways34. Such kind of activation helps in stimulation of induction of autophagy through JNK regulation as shown in Fig. 2. BRAF1 targeted by both picroside derivatives for inhibiting pathway module as shown in Table 1.
Transforming growth factor beta mediated signaling
TGFβ signaling acts as crucial regulator in various apoptotic and proliferative pathways. The signaling includes binding to Transforming Growth Factor Beta Receptor TGFBβ-II which further initiates formation of SMAD complex and its phosphorylation ultimately resulting in transcriptional activation of various genes in nucleus35. FKB1A, CASP1, CASP3 and TGFB2 targeted by P-I and P-II for blocking TGFβ mediated signaling (Table 1).
Interleukin Mediated Signaling
IL-2 and IL-4 one of the types of promotes differentiation and proliferation of T helper 2 (TH2) cells and the synthesis of immunoglobulin E (IgE)36. Generally, it activates mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K), signal transducers and activators of transcription (STAT), and mammalian target of rapamycin (mTOR) signaling modules, leading to both mitogenic and anti-apoptotic signals37. IL2 considered as target for inhibition for inhibiting interleukin mediated singaling (Table 1).
Cytokine Mediated Signaling
Cytokines plays critical role in the regulation of a various normal functions ranging from cellular proliferation, differentiation and survival to specialized cellular functions enabling host resistance against pathogens. Also, release of cytokines in response to inflammation, immunity or infection can supress cancer development and progression38. The JAK-STAT pathway trigerred by cytokines to achieve their ultimate goal can be thought of promising way for cancer therapy in humans38. MAPKs acts as central points for target inhibition due to hyperphosphorylation events. Hence, considered as inhibition of proliferative pathways.
On the basis of reconstructed pathway, key nodes were selected to perform structural study using PatchDock server. Docking of P-I and P-II was performed and found that picrosides can be used as active inhibitory molecule for cancer treatment as it targets at multiple level which is evident from Fig. 3 and data presented in Table 1. But, there is need to prioritize contender on the basis of personalized gene expression of candidate targets in patients. Picroside derivatives combinely plays a crucial role in inhibition of BRAF, FKB1A, CASP1, CASP3, TGFB2, IL2 and MAPKs through various signaling routes and therefore, can be considered as potent inhibitory molecule for further experimental analysis.
Structural representation of docking result for (A) Picroside-I and targets listed in Table 1A and B) Picroside-II and targets listed in Table 1B. The structural information given as output from PatchDock helps in deciphering the binding site of our ligand with the targets. With the help of parameters listed out in Table 1 we can filter out the best targets and infer their structural interactions.
Conclusively, our study presents a novel path to trace down the potential targets and propose them either for treating multiple diseases or for combinatorial therapy by identifying the exact course of disease transmission. It is anticipated that our network based drug-target interaction analysis protocol will assist computational biologists to look for similar patterns in other disease targets and biomedical scientists to design new therapeutic interventions based upon these findings.
Conclusions
Understanding of regulatory mechanism and subsequent effect on phenotypic level can not be dechiphered through individual genes only, but needs to include coordination of set of genes or gene groups. Hence, there is a need to study combinatorial effects of drugs by targeting multiple triggring points at same instance. With the aid of presented pipeline, biologists can infer key points involved in dysregulation of a particular mechanism given a medicinally important molecule using network-based perspective. For instance Picroside derivatives thought as medicinally important yet have not been broadly investigated for cancer treatment. Our study reveals key markers targeted by picroside derivatives through integration of data mining and network based approaches. The same revealation was found through computational molecular interactions and selected targets can act as potential markers for experimental validation.
Methods
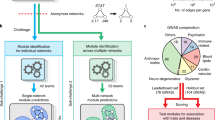
Complex chemical composition of the metabolic compounds found in medicinal herbs makes the understanding of therapeutic mechanism of action arduous. However, to clarify its mechanism of action at molecular level with an aim to know its usefulness in treating disorders, one has to have not only a deep insight into the molecular mechanism but also should opt a systematic approach to aid precise identification of therapeutic target. To achieve the same in this pipeline, literature mining of metabolites along with target network analysis was performed under systems pharmacology framework. Schematic workflow is shown in Fig. 4.
Systems Pharmacology framework for the identification and analysis of biomarkers through various modules viz. medicinal plant selection, metabolite screening, literature mining, pharmMapper analysis, coexpression analysis, gene ontology analysis, module construction, module-pathway mapping and docking study to screen out potential drug targets.
Literature Mining
With the advancement in scientific era, the information generated in the form of research articles being published in number of journals, is increasing at rapid rate and hence becomes a cumbersome task for a researcher to keep track of relevant literatures from MEDLINE manually. To make the task of information retrieval (IR) much easier, PubMed search engine was used to find all the hits with the query keywords like Picrorhiza (224 hits), Picroside (103 hits) Picroside-I (45 hits) and Picroside-II (78 hits). Screening for Picroside-III and Picroside-IV was also performed in similar fashion. Compiled scoping document of literature mining is available in Supplementary File 1.
Target Prediction
With the purpose of cross checking the P-I, P-II, P-III, and P-IV interaction with Homo sapiens known targets, we downloaded the 2D structure of picroside derivatives from the PubChem library. Further, the downloaded structures were given as an input for PharmMapper31. PharmMapper is a web server to predict therapeutic candidate drug targets for small molecule provided as query. To dug out the possible picroside interaction, score for candidate targets was performed by setting the parametric values of 2241 for human targets and a maximum number of 300 reserved matched targets were considered and all other parameters with default values.
Common Target Identification
A comparative analysis was performed between the targets retrieved from literature and PharmMapper in order to predict the verified target for further consideration of the same as a potential biomarker for various diseases. Targets common to both analysis were considered to have direct interaction for inhibition and therefore are called Primary Targets (PT) in our study. However, the targets that were present in literature and found to be affected but not present in the PharmMapper analysis were considered as Secondary Targets (ST) since no direct interaction was found at in-silico level.
where, TS is Target Screening, LM represents Literature Mining, PMR denotes PharmMapper Results, PT stands for Primary Targets and ST for Secondary Targets.
Gene Ontology and Co-Expression Network
To capture comprehensive view of how targets form signaling cascade to inhibit or enhance the disease response and their role in various domains like biological process, molecular function and cellular component; we performed gene ontology analysis. The analysis also gave an idea about the inter-connecting component in which biomarker association with neighboring genes can be identified. Besides, BLAST2GO software was used to identify various interactions between predicted targets on the basis of node score.
where
-
desc(g) represents all the descendant terms for a given GO term g
-
dist(g, ga) represents the number of edges between the GO term g and the GO term ga
-
g represents the element of GO, where GO is the whole set of all GO terms
-
gp(g) represents the number of gene outcomes given to a given GO term g
Score is calculated in terms of Biological Processes Score (BPS), Molecular Functions Score (MFS), and Cellular Components Score (CCS). Overall Gene Ontology Score (GOS) is represented as:
To elaborate the network and to gain comprehensive knowledge about the targets and their associated partners we downloaded the interacting partners from Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database39, which contains information from several sources, like in-silico prediction methods, experimental data and scientific literatures. For network construction analysis both parametric (Pearson Correlation Coefficient (PCC)) and non-parametric test (Spearman Correlation Coefficient (SCC)). But, SCC has not shown significant correlation but PCC showed significant correlation and overlapping results with available literature. Following the collection from STRING database, data was weighted, integrated and a confidence score was calculated for all protein interactions using Pearson Correlation Coefficient (PCC) which measures the linear correlation between two variables.
where ji and ki are the degrees of targets at both the ends of the ith connection, and M represents total connections in the network.
Results of the various in-silico predictions were inspected from different designated views. Moreover, normalization and categorization based on the association score of co-expressed modules was performed. Module Construction (MC) was performed by combining all calculated scores given in the MC equation.
where, CES stands for Co-Expression Score, CLC for Clustering Coefficient, BWC for Betweenness Centrality, and DON for Degree of Nodes. Information of selected parameters is given below:
A co-expression network is an undirected graph, with every node representing a gene and every edge representing the connection between these nodes. In this study, we used an in-house Perl script to calculate gene co-expression; we calculated various scores, assigned weights to each score, and finally generated a combined score. Methodology was adopted from our previous study on miRNA regulatory network analysis3.
Gene Ontology Score deals with three components, namely Biological Processes (BPs), Molecular Functions (MFs), and Cellular Components (CCs). BLAST2GO was used to link selected genes to map with the GO database in terms of BPs, MFs, and CCs. The genes that belonged to the same category were clustered. A node score function was defined for all targeted genes. Genes that had the same score were clustered in the same cluster category. Interconnection from one cluster to another cluster was performed on the basis of their respective association based on the node score.
The degree of a node in an undirected graph is the number of connexions or edges a node has with other nodes, and it is defined as deg(i) = k(i) = |N(i)| where N(i) is the number of the neighbours of node i. The degree distribution p(k) reveals the fraction of vertices with degree k. DON gives the idea of association of nodes with node of interest.
Clustering Coefficient is the measurement that shows the tendency of a graph to be divided into clusters. A cluster is a subset of vertices that contains lots of edges connecting these vertices to each other. Assuming that i is a vertex with degree deg(i) = k in an undirected graph G and that there are e edges between the k neighbors of i in G, then the Clustering Coefficient of i in G is given by:
Thus, Ci measures the ratio of the number of edges between the neighbors of i to the total possible number of such edges, which is k(k − 1)/2. It takes values as 0 ≤ Ci ≤ 1.
Betweenness Centrality shows that nodes which are intermediate between neighbors rank higher. Without these nodes, there would be no way for two neighbors to communicate with each other. Thus, betweenness centrality shows important nodes that lie on a high proportion of paths between other nodes in the network. For distinct nodes i, j, w ∈ V(G), let σij be the total number of shortest paths between i and j and σij(w) be the number of shortest paths from i to j that pass through w. Moreover, for w ∈ V(G), let V (i) denote the set of all ordered pairs, (i, j) in V(G) × V(G) such that i, j, w are all distinct. Then, the Betweenness Centrality is calculated as:
Pathway Mapping of Co-Expressed Modules
After identifying co-expressed gene modules, a mapping of associated partners with designated pathway was performed by manual literature survey followed by constructing static map using KEGG, REACTOME and Pathway Interaction Database (PID) as a reference pathway maps, to aid proper understanding of molecular mechanism of action and target implication40,41,42.
Patch Dock Analysis
Also, to understand the inhibitory role of selected targets with small molecules (P-I and P-II), molecular docking was performed based on shape complimentary principles using PatchDock web server33; as we are interested to observe variations in target binding energy of picroside derivates with already known drug targets.
References
Zhou, H., Gao, M. & Skolnick, J. Comprehensive prediction of drug-protein interactions and side effects for the human proteome. Sci. Rep. 5 (2015).
Zhou, X., Menche, J., Barabási, A.-L. & Sharma, A. Human symptoms–disease network. Nat. Commun. 5, 4212 (2014).
Bansal, A., Singh, T. R. & Chauhan, R. S. A novel miRNA analysis framework to analyze differential biological networks. Sci. Rep. 7, 14604 (2017).
Menche, J. et al. Integrating personalized gene expression profiles into predictive disease-associated gene pools. Npj Syst. Biol. Appl. 3, 10 (2017).
Gomez-Cabrero, D. et al. From comorbidities of chronic obstructive pulmonary disease to identification of shared molecular mechanisms by data integration. BMC Bioinformatics 17, 441 (2016).
Kitsak, M. et al. Tissue Specificity of Human Disease Module. Sci. Rep. 6, 35241 (2016).
Basler, G., Nikoloski, Z., Larhlimi, A., Barabási, A.-L. & Liu, Y.-Y. Control of fluxes in metabolic networks. Genome Res. gr.202648.115, https://doi.org/10.1101/gr.202648.115 (2016).
Bansal, A. & Srivastava, P. A. Transcriptomics to Metabolomics: A Network Perspective for Big Data. Httpservicesigi-Glob. -1-5225-2607-0ch008, 188–206, https://doi.org/10.4018/978-1-5225-2607-0.ch008 (2018).
Ghiassian, S. D. et al. Endophenotype Network Models: Common Core of Complex Diseases. Sci. Rep. 6, 27414 (2016).
Guney, E., Menche, J., Vidal, M. & Barábasi, A.-L. Network-based in silico drug efficacy screening. Nat. Commun. 7, 10331 (2016).
Vinayagam, A. et al. Controllability analysis of the directed human protein interaction network identifies disease genes and drug targets. Proc. Natl. Acad. Sci. USA 113, 4976–4981 (2016).
Meng, F.-R., You, Z.-H., Chen, X., Zhou, Y. & An, J.-Y. Prediction of Drug-Target Interaction Networks from the Integration of Protein Sequences and Drug Chemical Structures. Mol. Basel Switz. 22 (2017).
Chen, X. Editorial: Identifying Drug-target Interactions Based on Heterogeneous Biological Data - PART 1. Curr. Protein Pept. Sci. 19, 428–429 (2018).
Chen, X. et al. NRDTD: a database for clinically or experimentally supported non-coding RNAs and drug targets associations. Database J. Biol. Databases Curation 2017 (2017).
Chen, X. et al. Drug-target interaction prediction: databases, web servers and computational models. Brief. Bioinform. 17, 696–712 (2016).
Chen, X., Liu, M.-X. & Yan, G.-Y. Drug-target interaction prediction by random walk on the heterogeneous network. Mol. Biosyst. 8, 1970–1978 (2012).
Wang, L. et al. RFDT: A Rotation Forest-based Predictor for Predicting Drug-Target Interactions Using Drug Structure and Protein Sequence Information. Curr. Protein Pept. Sci. 19, 445–454 (2018).
Huang, Y.-A., You, Z.-H. & Chen, X. A Systematic Prediction of Drug-Target Interactions Using Molecular Fingerprints and Protein Sequences. Curr. Protein Pept. Sci. 19, 468–478 (2018).
Wang, L. et al. A Computational-Based Method for Predicting Drug-Target Interactions by Using Stacked Autoencoder Deep Neural Network. J. Comput. Biol. J. Comput. Mol. Cell Biol. 25, 361–373 (2018).
Menche, J. et al. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science 347, 1257601 (2015).
Ghiassian, S. D., Menche, J. & Barabási, A.-L. A DIseAse MOdule Detection (DIAMOnD) Algorithm Derived from a Systematic Analysis of Connectivity Patterns of Disease Proteins in the Human Interactome. Plos Comput. Biol. 11, e1004120 (2015).
Chander, R., Kapoor, N. K. & Dhawan, B. N. Picroliv, picroside-I and kutkoside from Picrorhiza kurrooa are scavengers of superoxide anions. Biochem. Pharmacol. 44, 180–183 (1992).
Kumar, V., Bansal, A. & Chauhan, R. S. Modular Design of Picroside-II Biosynthesis Deciphered through NGS Transcriptomes and Metabolic Intermediates Analysis in Naturally Variant Chemotypes of a Medicinal Herb, Picrorhiza kurroa. Front. Plant Sci. 8 (2017).
Dwivedi, Y., Rastogi, R., Garg, N. K. & Dhawan, B. N. Picroliv and its Components Kutkoside and Picroside I Protect Liver Against Galactosamine-Induced Damage in Rats. Pharmacol. Toxicol. 71, 383–387 (1992).
Gao, H. & Zhou, Y.-W. Anti-lipid peroxidation and protection of liver mitochondria against injuries by picroside II. World J. Gastroenterol. 11, 3671–3674 (2005).
Pratheeshkumar, P., Son, Y.-O., Korangath, P., Manu, K. A. & Siveen, K. S. Phytochemicals in Cancer Prevention and Therapy. BioMed Res. Int. 2015 (2015).
Kılıç, Y. et al. Effect of picroside II on hind limb ischemia reperfusion injury in rats. Drug Des. Devel. Ther. 11, 1917–1925 (2017).
Rathee, D., Thanki, M., Bhuva, S., Anandjiwala, S. & Agrawal, R. Iridoid glycosides-Kutkin, Picroside I, and Kutkoside from Picrorrhiza kurroa Benth inhibits the invasion and migration of MCF-7 breast cancer cells through the down regulation of matrix metalloproteinases: 1st Cancer Update. Arab. J. Chem. 6, 49–58 (2013).
Rolland, T. et al. A proteome-scale map of the human interactome network. Cell 159, 1212–1226 (2014).
Zhu, J. et al. A pre-clinical pharmacokinetic study in rats of three naturally occurring iridoid glycosides, Picroside-I, II and III, using a validated simultaneous HPLC-MS/MS assay. J. Chromatogr. B Analyt. Technol. Biomed. Life. Sci. 993–994, 47–59 (2015).
Liu, X. et al. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 38, W609–614 (2010).
Eden, E., Navon, R., Steinfeld, I., Lipson, D. & Yakhini, Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009).
Schneidman-Duhovny, D., Inbar, Y., Nussinov, R. & Wolfson, H. J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363–W367 (2005).
Singh, P., Ravanan, P. & Talwar, P. Death Associated Protein Kinase 1 (DAPK1): A Regulator of Apoptosis and Autophagy. Front. Mol. Neurosci. 9 (2016).
Massagué, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 (2012).
Paul, W. E. & Zhu, J. How are TH2-type immune responses initiated and amplified? Nat. Rev. Immunol. 10, 225–235 (2010).
Burotto, M., Chiou, V. L., Lee, J.-M. & Kohn, E. C. The MAPK pathway across different malignancies: A new perspective. Cancer 120, 3446–3456 (2014).
Thomas, S. J., Snowden, J. A., Zeidler, M. P. & Danson, S. J. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br. J. Cancer 113, 365–371 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–462 (2016).
Tanabe, M. & Kanehisa, M. Using the KEGG database resource. Curr. Protoc. Bioinforma. Chapter 1, Unit1.12 (2012).
Joshi-Tope, G. et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 33, D428–432 (2005).
Acknowledgements
Authors thank Jaypee University of Information Technology for providing financial support and lab facilities to carry out the research work.
Author information
Authors and Affiliations
Contributions
T.R.S. conceived the idea; A.B., P.A.S. and T.R.S. designed the project. A.B. and P.A.S. have performed network reconstruction and further analysis. All authors read, improvised and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bansal, A., Srivastava, P.A. & Singh, T.R. An integrative approach to develop computational pipeline for drug-target interaction network analysis. Sci Rep 8, 10238 (2018). https://doi.org/10.1038/s41598-018-28577-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-28577-6
This article is cited by
-
DTiGEMS+: drug–target interaction prediction using graph embedding, graph mining, and similarity-based techniques
Journal of Cheminformatics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.