Abstract
Archaeological evidence suggests that dogs were introduced to the islands of Oceania via Island Southeast Asia around 3,300 years ago, and reached the eastern islands of Polynesia by the fourteenth century AD. This dispersal is intimately tied to human expansion, but the involvement of dogs in Pacific migrations is not well understood. Our analyses of seven new complete ancient mitogenomes and five partial mtDNA sequences from archaeological dog specimens from Mainland and Island Southeast Asia and the Pacific suggests at least three dog dispersal events into the region, in addition to the introduction of dingoes to Australia. We see an early introduction of dogs to Island Southeast Asia, which does not appear to extend into the islands of Oceania. A shared haplogroup identified between Iron Age Taiwanese dogs, terminal-Lapita and post-Lapita dogs suggests that at least one dog lineage was introduced to Near Oceania by or as the result of interactions with Austronesian language speakers associated with the Lapita Cultural Complex. We did not find any evidence that these dogs were successfully transported beyond New Guinea. Finally, we identify a widespread dog clade found across the Pacific, including the islands of Polynesia, which likely suggests a post-Lapita dog introduction from southern Island Southeast Asia.
Similar content being viewed by others
Introduction
When people spread from west to east across the Pacific Ocean they did not travel alone. Often dogs (Canis familiaris), pigs (Sus scrofa), chickens (Gallus gallus) and rats (Rattus exulans) were part of these migrations. The presence of dog, pig, chicken and rat bones in archaeological sites attest to their inclusion in migratory voyages. However, moving domesticated animals across increasingly wider ocean gaps and onto often smaller, and more environmentally depauperate, islands of the Pacific, with no established agricultural economies, may have posed considerable challenges. The archaeological evidence for the introduction of domestic and commensal animals into the region is patchy and discontinuous1,2, which may signal some of the difficulties inherent in the establishment of viable animal populations, or indicate human choices made in relation to their transportation and management.
Explanations for the late Holocene human colonisation of Remote Oceania draw on archaeological and linguistic data to model the movement of Austronesian language speakers out of Taiwan and into the Pacific, via the Philippines. According to this model, this movement preceded the emergence of the Lapita Cultural Complex (LCC) in the Bismarck Archipelago which then spread across much of Near and Remote Oceania3,4. Archaeological evidence for the LCC is found from New Guinea to West Polynesia. Various scenarios have been suggested, incorporating archaeological, linguistic and biological evidence, to account for the dispersal processes and the extent to which interactions occurred between incoming Austronesian language speakers and existing human groups in Near Oceania (for a summary see3). Further to the east across the Pacific Ocean, dogs were part of the last major human migration that resulted in the initial colonisation of the islands of East Polynesia4.
Although East Asia is a critical region for dog domestication5,6,7, the subsequent translocation of dogs across Mainland Southeast Asia (MSEA) and Island Southeast Asia (ISEA) and into the islands of the Pacific is not well understood. Molecular genetic studies using mitochondrial DNA (mtDNA) that have investigated the origins and dispersals of dogs throughout the Pacific indicate that the dingo of Australia, New Guinea Singing Dog (NGSD) and ancient Polynesian dogs are all descended from East Asian dogs8. Despite their long association with people, dogs appear relatively late in the archaeological record of Oceania, and the timing of their arrival and dispersal trajectories appear to differ from that of people. The earliest archaeological dog remains are found in Australia and date to c. 3,500 years ago9, although this age is based on archaeological context and is not a direct date. Over time, the descendants of these dogs reverted to a completely independent state, and the population continued over successive generations without human involvement. These dogs, known as dingoes, are considered to be one of the few truly feral dog populations found anywhere in the world10. Nevertheless, some dingoes have been incorporated into Aboriginal communities as companions and hunting aides11. NGSD are genetically closely related to dingoes8. The history of these dogs is however little known and highly debated12. Modern lowland dogs in New Guinea today are potentially the result of admixture with introduced European breeds and may not be representative of true NGSD lineages; however, it is possible that NGSDs may still be present in isolated areas of the highlands.
Dog remains from archaeological sites provide an important source of information, not only about the dynamic relationship between dogs and people, but also to reconstructions of their genetic history. Using a 582 base pair (bp) portion of the control region of the mitochondrial genome (mitogenome) from modern dogs, Savolainen and colleagues demonstrated that all dingoes sampled belonged to a lineage known as the A29 haplotype8. Although this is one of a number of dog mtDNA lineages that reached ISEA, it was the only one successfully established in Australia. In addition, two short haplotypes (Arc1 and Arc2), were observed in archaeological dog remains from East Polynesia (sampled from Hawai’i, the southern Cook Islands and New Zealand), but not found in modern dingoes8. A later study by Oskarsson and colleagues investigated the origins and routes of the introductions of dingoes, NGSDs, and Polynesian dogs in further detail, using the same control region fragments as the previous study by Savolainen and colleagues13. The Arc1 haplotype was found to be indistinguishable from a number of widespread control region haplotypes found in modern dogs from China, MSEA and ISEA, while Arc2 had a much more restricted distribution, and appears to belong to the A75 lineage found only in modern Indonesian dogs. Oskarsson and colleagues compared the frequency and distribution of modern haplotypes in MSEA and ISEA, including Taiwan and the Philippines, with those from the dingo and ancient Polynesian samples. Neither the ancient Polynesian short haplotypes nor the A29 haplotype carried by dingoes were present in modern dogs sampled from Taiwan and the Philippines, suggesting that dogs were not introduced into the Pacific region from or via this northeastern route.
Second generation sequencing technology is now making it feasible to sequence the complete mitochondrial genome, rather than just a portion, and has also contributed to the viability of sequencing of mitogenomes from archaeological specimens14. As expected, studies of mitogenomes of modern domesticated species, such as cattle15 and sheep16, and from archaeological dog samples17 have shown that the control region sequences provide only a partial picture of genetic diversity. A recent study of dingoes using complete mitogenomes and nuclear markers has shown that there are at least two geographically subdivided genetic populations18, rather than the single population suggested by control region studies8.
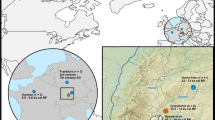
Here we present seven new complete and near-complete ancient mitogenomes and five partial ancient mitogenome sequences from archaeological dog specimens from Thailand, ISEA and the Pacific (Fig. 1, Supplementary Table S1), and mitogenomes from a sample of four modern dingoes from the Wellington Zoo in New Zealand (Supplementary Table S2). When combined with additional published mitogenomes from modern dogs from Southeast Asia, Australia and New Guinea (Supplementary Table S2), the aDNA data reveal a complex history of dog-human interactions in ISEA and the Pacific. Three temporally distinct dispersals are highlighted, and their possible origins and the sequence of introductions are revealed.
A map of Southeast Asia and the Pacific showing the source location of the specimens and associated haplogroups (assignment to haplogroup follows Duleba and colleagues19) and the median-joining network. The boundary between Near and Remote Oceania is also shown. Symbols identify the type of sequence: filled circle, ancient mitogenome; half circle, partial ancient sequence; hollow circle, modern mitogenome. Node colours represent the haplogroup, grey, A; red, A2b2, green, A2b3; yellow, A4’5; blue, B.
Results
Phylogenetic analyses of complete mitogenomes of modern dogs from across the world have shown that over 65% of the dogs sampled fall within Haplogroup (Hg) A19. There is geographical structure within this large haplogroup, with the greatest mitochondrial diversity found in East Asia5,19,20. Duleba and colleagues19 further identified several sub-clades that are present almost exclusively in East Asian modern dogs. These sub-clades were estimated to have the oldest evolutionary ages (between 15,000 and 38,700 years ago) and are hypothesised to represent the initial founding gene pool. Here we follow Duleba and colleagues’ phylogeny and nomenclature for dog haplogroups, which has the greatest resolution for sub-clade A2, the lineage carried by the majority of dogs we sequenced in our study.
All the specimens from which we obtained mitogenome sequences belong to Hg A, with the exception of one specimen from Taiwan which belongs to Hg B and two partial sequences from specimens from the south coast of Papua New Guinea which also possess single nucleotide mutations (SNPs) consistent with Hg B (Fig. 1, and see Supplementary Table S1 for haplogroup assignments and archaeological site locations, Supplementary Table S3 for variable positions relating to haplogroup assignment, and Supplementary Table S4 for defining SNPs for Haplogroup B observed in the ancient Papua New Guinea sequences). Within Hg A, the majority of sequences from the ancient Pacific Island dogs and two partial sequences from Thai dogs, belong to Dubela and colleagues’19 Hg A2b2. These share most, but not all, of the defining mutations with a modern specimen from Fiji (KM061586 [11 ws] in19) located on one of the Hg A2b2 terminal branches. The Pacific Island sequences that share this A2b2 lineage span a wide area from New Guinea to East Polynesia (Fig. 1). They include mitogenomes obtained from dog remains from the archaeological site of Taurama, near Port Moresby, in Papua New Guinea21; the Goemu site on the island of Mabuyag in the Torres Strait which separates New Guinea and Australia22; a partial sequence from an archaeological site in Graciosa Bay, Santa Cruz in the Solomon Islands23; and complete mitogenomes from samples from sites on Tokelau in West Polynesia24 and Hawai’i25 in East Polynesia. One previously published ancient mitogenome from New Zealand17, and two from the southern Cook Islands26 also share this A2b2 lineage. The dingoes belong to Hg A2b3, and share only a few defining mutations with the specimen from China on one of the two A2b3 terminal branches (EU789681 in19). One ancient specimen, from Timor-Leste, belongs to Hg A4’5, and has some of the defining mutations for sub-clade A5.
The two nearly complete ancient mitogenomes from dogs from the Iron Age archaeological site of Shisanhang in Taiwan (MS10333, MS10334) are highly divergent compared to the other ancient sequences, representing the A and B haplogroups. One of the specimens (MS10333) carries the A2b3 haplotype, which is observable in modern dingoes but not in any of the ancient Pacific samples reported here or previously published. Occupation of the Shisanhang site dates to between 2,300 and 500 BP, with most dates clustering around 1,500 to 1,000 BP, and contains evidence for long distance exchange with China27. The greatest genetic diversity of modern dogs is found in southeastern China5,7, and it is perhaps not unexpected to observe these divergent lineages in a Taiwanese site.
These ancient mitogenomes provide a series of individual insights into the history of dog dispersals from East Asia and their spread eastward across the Pacific, prior to the introgression of modern European dog breeds. Although the sample size is very small, these data from mtDNA sequences from only a few dogs, when combined with archaeological and linguistic data, demonstrate the potential of aDNA studies to differentiate between hypothesised dispersal events. A median-joining network (Fig. 1) shows the relationship between the complete or near complete ancient mitogenomes and modern mitogenomes from dingoes, NGSDs and Southeast Asian village dogs. Bayesian analysis to establish divergence times for the A and B haplogroups provides an estimate of between 3,000 to 21,000 years ago, which is broadly consistent with Duleba and colleagues19 findings but overall the tree lacks sufficient resolution to be very informative about such events (Supplementary Fig. S1). The topology of the tree remains the same whether or not dates from archaeological contexts are included, and is consistent with trees generated in MEGA7 using the maximum likelihood method28 (Supplementary Fig. S2). A median-joining network of the 582 bp control region haplotypes used previously to investigate dog domestication5 and dispersal in the Pacific8,13 also demonstrates the close relationship between the dingo haplotypes and the ancient Pacific samples (Supplementary Fig. S3).
Discussion
Pre-Lapita introductions
One of the earliest archaeological sites in ISEA with evidence of dogs is Matja Kuru 2 Cave in East Timor, with a dog burial that dates to about 3,000 years ago29. Stable isotope analyses indicate that the dog had a diet dominated by terrestrial plant foods, similar to that of Pacific pigs, rather than a diet high in hunted or marine foods. This suggests that the dog may have been associated with a food producing community, rather than one reliant on wild food sources29. The mitogenomic sequence we obtained from this specimen (MS10330) carries the A4’5 haplotype (after19), which is not shared by any other ancient specimen we sequenced. It is also clearly distinct from the sub-clade of Hg A2b3 where modern sequences from dingoes and NGSDs are located.
The introduction of dingoes to Australia remains enigmatic. Although molecular date estimates have raised the possibility of a late Pleistocene introduction of dogs to Australia13,18 there is, as yet, no archaeological evidence for dingoes prior to c. 3,500 years ago. In addition, divergence dates may relate to events that predate the arrival of dingoes to Australia, so have limited application to understanding the introduction timing and process. Recent genetic analyses of modern dingoes and NGSDs show a geographical distribution of dingo mtDNA lineages that distinguishes populations from the northwest and southeast of Australia18,30, which we also see in the modern dingo sequences we generated (Fig. 1). This population structure is also observable in results from nuclear and Y-chromosome analyses30,31,32,33. In addition, the two dingo groups, while more closely related to the NGSDs than to any other dogs, diverge from the NGSD sequence, which may indicate a period of isolation between the two groups of dogs. The only genetic data from NGSDs has been obtained from the captive population derived from only eight dogs34. Village dogs in New Guinea however possess many, if not all, of the behavioural and phenotypic characteristics ascribed to NGSDs35 and it is possible that sampling from isolated village populations on the New Guinea mainland, and highland NGSD populations if found to still exist, may also be informative about NGSD and dingo ancestral history.
The process of human settlement expansion across the western Torres Strait between New Guinea and Australia may have facilitated the dispersal of dogs/dingoes from New Guinea to Australia. Evidence of this human expansion appears in the archaeological record from around 3,800 years ago, but to date no dog remains have been found from this period36. Balme and O’Connor37 hypothesise that increasing numbers of small game animals observable in the archaeological record in the mid-Holocene may be indicative of the arrival and use of dingoes by people to assist with hunting. These chronometrically dated changes in faunal composition in archaeological sites, along with molecular evidence indicating at least two introductions to Australia and a close genetic relationship between dingoes and NGSDs, suggest that by the mid to late Holocene deliberate dog translocations were taking place between New Guinea and Australia. Securely dated archaeological material, along with further molecular data, would assist with the refining the date of introductions, routes and possible origins of the dingo.
Interestingly, one sequence we generated from a dog bone excavated from an Iron Age archaeological site in Taiwan, MS10133, is located on the network between these two groups of dingoes (Fig. 1). A similar link to modern Taiwanese dog lineages has been observed in dingo Y-chromosome markers32. Sacks and colleagues raise the possibility of a separate dispersal event from Taiwan, or the movement of dogs with Austronesian language speakers both out of China to the south through the Tonkin Gulf and to Taiwan, to explain this observation32.
The appearance of the Neolithic in ISEA and its extension out into the western Pacific, characterised by the presence of pottery in archaeological sites dated to between 3,800 and 3,300 years ago, involved the spread of new languages, ideas and identities alongside migration and recruitment from local communities38. Molecular analyses of dog remains from MSEA archaeological sites during this critical period of human expansion could greatly assist with tracking the translocations of dogs as part of this movement. What is known archaeologically is that dogs are absent from MSEA hunter gatherer sites. It was only with the expansion of rice and millet farmers into Southeast Asia from Chinese domestication centres that dogs made their appearance about 4,000 year ago, and from this point onwards, dog remains are commonly found. Archaeological excavation at An Song, in Southern Vietnam, has provided dog bones39 and at Khok Phanom Di, a central Thai site, dog bones and canid faeces containing rice chaff and fish bones were found40. The time period between 4,000 and 3,000 years ago in MSEA and ISEA is central to tracking the movement of dogs across Southeast Asia and out into the islands of Oceania. It is likely that as more samples of dogs from archaeological sites across the region become available, a more detailed picture of the history of ISEA dogs will be revealed.
Late Lapita dogs
Dogs are often argued to have been moved across the Pacific by people associated with the LCC4. However, there is only extremely limited archaeological evidence for dogs in early Lapita archaeological sites, and no dog bone has been reported from Lapita archaeological sites beyond the Bismarck Archipelago (for reviews see2,41). The presence of drilled dog teeth in Lapita archaeological sites of Near Oceania, along with the scarcity of other skeletal elements, also indicates that dog remains may have been incorporated into archaeological contexts by mechanisms unrelated to the transport of living dogs, such as the exchange of valuables. Although mtDNA control region sequences indicate a route through ISEA for the ancestors of dogs found in East Polynesian archaeological sites8,13, a possible link between Pacific dogs and modern Taiwanese dogs has also been suggested32.
Although we were unable to generate complete mitogenomes from the two archaeological specimens from the terminal Lapita and post-Lapita levels at archaeological sites at Edubu 1 (MS10031) and Bogi 1 (MS10023) from Caution Bay on the south coast of mainland Papua New Guinea, we were able to obtain limited coverage of part of their genomes (43% and 17% respectively). Compared to the rest of the ancient mitogenomes we sequenced, these specimens carry diagnostic mutations at several positions indicating that they fall within Hg B (Supplementary Table S3). Sample MS10031 comes from an archaeological context at Edubu 1 that is well dated to between 2,580 and 2,410 cal BP, and is probably closer to the start of this age range42,43. This archaeological site is associated with the very end of dentate-stamped ceramics at Caution Bay, that is, it dates to the terminal period of Lapita here as ceramics begin to lose all body decoration (i.e. as all ceramic vessel bodies become plain wares). This is a time of major change in Caution Bay ceramics. Bogi 1 is a multi-phase archaeological site representing an ancient village location. Sample MS10023 was securely embedded within an archaeological context that dates to within the narrow timeframe of 2,150 to 2,100 cal BP44. At Caution Bay, dentate-stamped ceramics that define Lapita pottery are commonly found in 2,600 cal BP sites but entirely cease by 2,550 cal BP, indicating that sample MS10023 post-dates Lapita in this area by 400 to 450 years. These two specimens are the first from the Pacific that have been identified with a Hg B lineage and suggest a common ancestor with one of our dog samples from an Iron Age archaeological site in Taiwan and hence a potential link with Taiwan. While this may fit with current linguistic models for the expansion of Austronesian languages from Taiwan, it is also possible, given the limited number of mtDNA sequences available to date, that the B haplogroup could be a shared common ancestral lineage throughout the broader ISEA gene pool. Only further genetic analyses of well-dated dog samples from ISEA will clarify the distribution of B lineages.
Later introduction of Remote Oceanic dogs
Despite the scarcity of dog remains in Lapita archaeological sites in the western Pacific, there is considerable archaeological evidence for people taking dogs with them during the colonisation of East Polynesia that began around 1,000 years ago45. Linguistic evidence, in addition to being used to model human migrations, is also informative about the nature of interactions between people and dogs during the colonisation of the Pacific46. Words for items that were a common part of Austronesian, Lapita and Polynesian cultures are transmitted consistently to daughter languages, and are often shared by languages across a wide area. Terms for dog however are not consistently shared across the Pacific and show considerable variability. There is no widely shared cognate for dog in either Proto-Austronesian or Proto-Oceanic (associated with the LCC), and in Oceanic languages the term for ‘dog’ is variable. By the time people were settling the islands of Polynesia the situation appears to have changed. There is a Proto-Polynesian term for dog, *kuli, which alongside the terms for chicken and pig, is a unique innovation. These terms first appear in Proto-Polynesian, and regular cognates consistently appear in daughter languages, including the Polynesian Outlier languages to the west47. This contrast suggests that, by the time that the colonisation of East Polynesia commenced, the Polynesians had developed linguistic and cultural innovations that included a highly successful method of transport and establishment of domestic animals not seen previously in the region46.
The complete and near-complete ancient mitogenomes we sequenced from dogs from archaeological sites from Atafu, an atoll of Tokelau in West Polynesia, and Hawai’i in East Polynesia, and three previously published sequences from New Zealand17 and the southern Cook Islands26, form a haplogroup A2b2 which is separate from dingoes and NGSDs (Fig. 1). A partial sequence from a dog burial from Graciosa Bay on Santa Cruz in the Solomon Islands also has markers consistent with this haplogroup. Ancient mitogenomes from dog specimens from the Taurama archaeological site near Port Moresby in Papua New Guinea and Goemu on Mabuyag island in the Torres Strait are also part of this haplogroup, which is distinct from the B lineage observed in the terminal Lapita and post-Lapita dogs from earlier archaeological sites along the south coast of mainland Papua New Guinea, or the dingo and NGSD group. This Hg A2b2 comprises two sub-clades, with haplotypes from Near Oceania and Polynesia found in both clades (Supplementary Table S1, Supplementary Figs S1 and S2).
The Taurama archaeological site dates to between 2,000 and 1,200 BP, where a dog burial was found in an archaeological context associated with Early Papuan Pottery (EPP)48. EPP archaeological sites begin to appear along the south coast of mainland Papua New Guinea around 2,000 BP and provide evidence of a period of cultural change, including new pottery designs49. The mtDNA sequence from the Taurama dog bone (MS10329) is the earliest known Oceanic occurrence of Hg A2b2* and may indicate the arrival of dogs from a different source population to those in the terminal and post-Lapita Caution Bay archaeological sites.
The appearance of this new A2b2* genetic lineage may be linked to a notable increase in the numbers of dog bone in Near Oceania and the sudden appearance of dog bones in West Polynesian archaeological sites beginning around 2,000 years ago46, suggesting there may have been a significant change in relationships between dogs and people in the Pacific at this time. We have previously suggested that this may signal changes in technology and animal husbandry that facilitated the successful movement of dogs further into the Pacific than ever before, possibly also linked with the impetus for the initial colonisation of East Polynesia46. Other possibilities involve a movement of new people and their dogs into Polynesia50 or the incorporation of dogs into existing cultures via exchange networks. The presence of this A2b2* lineage in Near Oceania may also be associated with a westwards (as well as eastwards) expansion of Remote Oceanic groups resulting in the establishment of the Polynesian Outliers in the western Pacific.
The ancient sequences from the major and widespread A2b2* sub-clade are most similar to partial sequences we obtained from two dog specimens from Bronze Age archaeological sites in Thailand (MS10331 and MS10332). Although we were unable to obtain complete mitogenomes from the archaeological Thai specimens, we were able to examine enough nucleotide positions with adequate coverage (covered by at least three reads) to indicate that they are consistent with the A2 sub-clade19. Compared to the wider dataset of modern dog mitogenomes, the ancient Pacific dog mitogenomes are part of the A2b2* sub-clade19, also shared with modern dogs sampled in Thailand5, Indonesia31 and Fiji19. This indicates that dogs from Hg A2b2* in the Pacific may have ultimately been derived from MSEA dogs, rather than being incorporated into Austronesian expansions southward from Taiwan. High frequencies of two short control region haplotypes, found in ancient Polynesian dogs, have also been observed in modern Indonesian dogs13. A similar southwestern route through ISEA has been suggested for the dispersal of pigs51. It is possible that some modern village dogs in ISEA carry ancient lineages associated with the dispersal of dogs into the Pacific, but as yet there is very little comparative data for dog mitogenomes in that region. Shannon and colleagues52 surveyed autosomal, mitochondrial and Y-chromosome diversity in modern purebred and village dogs. They found that while some populations of village dogs are almost entirely descended from European dogs, especially in the neotropics and Remote Oceania, others show little evidence of admixture. Notably, village dog populations sampled from Papua New Guinea and the Solomon Islands comprise a mix of indigenous and European ancestry, while those from Borneo have no observable European ancestry and are similar to Vietnamese dogs. The Vietnamese dogs sampled also showed little evidence of European admixture. These results suggest that studies of modern village dogs in MSEA and ISEA may be especially valuable in advancing our understanding of dog dispersals through Southeast Asia into Oceania, particularly where aDNA data are unavailable.
Conclusion
The dispersal of dogs across the Pacific is inseparably linked to the relationships between dogs and people. Unlike movement across continental landmasses, Pacific dogs must have been transported by people across the waters that separate islands. The ancient mitogenomes sequenced from archaeological dog specimens presented here offer a novel series of individual insights into the history of dog translocation from Southeast Asia as it occurred prior to the influence of modern European dog breeds. We generated seven mitogenomes and five partial sequences from ancient MSEA, ISEA and Pacific dogs, and four modern dingoes. Despite the small sample size, our results reveal levels of complexity and discontinuity in the introduction and movement of dogs, which are mirrored in the archaeological and linguistic evidence, suggesting at least three introductions of dogs to the wider Pacific region, in addition to the earlier appearance of the dingo in Australia. Further mtDNA studies of ancient dogs and modern village populations throughout the region may contribute additional data that can be used to evaluate these hypothesised dispersals. Autosomal and Y-chromosome analyses also have the potential to generate additional information about dog dispersal, which could reveal different dispersal signatures based on sex, or phenotypic characteristics, though the environmental conditions in the region are not particularly conducive to aDNA preservation.
Our molecular genetic analyses reveal one of the earliest dogs present in ISEA around 3,000 years ago from Timor-Leste possesses a mtDNA lineage not found elsewhere in the region. We also found similarities between mtDNA of modern dingoes and NGSDs and an ancient Taiwanese sequence, which supports previous observations about possible links between Y-chromosome markers of modern dingoes and a modern Taiwanese sample32. More work is required to address whether these connections reflect the genetic diversity of a shared ancestral population in mainland China, or attest to a currently unknown dispersal event linking the two populations. Archaeological evidence for the introduction of dogs to Oceania as part of the LCC is extremely limited. Nonetheless, we demonstrate that mitogenomes from dogs in terminal Lapita and post-Lapita levels of archaeological sites along the south coast of mainland New Guinea also show affinities with an Iron Age dog specimen from Taiwan, raising the possibility of at least one introduction of dogs during Austronesian expansions ultimately from the north. Finally, we have identified a major late introduction of dogs across the islands of Oceania beginning around 2,000 years ago, which appears to have originated in MSEA, not Taiwan, and culminated in the establishment of dog populations in initial colonisation-era sites throughout East Polynesia.
Materials and Methods
Ancient dna extraction, library preparation, sequencing and data processing
Twelve dog specimens comprising bone and teeth from thirteen archaeological sites were used for this study (Supplementary Table S1), in conjunction with three ancient specimens previously published (Supplementary Table S2)17,26. The mitogenomes reported here were generated using hybridisation capture and were sequenced on the Illumina MiSeq platform as previously described17, in accordance with protocols to ensure the authenticity of aDNA sequences. Throughout DNA extraction and library preparation, blank extractions were processed alongside samples to provide negative controls.
We processed the raw sequence reads using a computational protocol previously described17. A notable feature of this pipeline is the use of a composite mitochondrial reference genome (made up of likely contaminant genomes such as pig, cow, chicken and human) to check for contamination (Supplementary Table S4). Coverage and read depth plots were created for all samples (Supplementary Figs S4 and S5). All reads displayed the expected characteristics of aDNA, including fragment length and deamination patterns (Supplementary Fig. S6). To maximize sequence usability in downstream analyses, imputation was performed and sites with greater than 80% confidence were retained. Consensus sequences containing insertions and/or deletions (indels) were produced for each sample that passed quality control, and these were deposited in Genbank (accession numbers KT168373, KY98508–KY98516). In these Genbank sequences non-variant sites that were supported by fewer than three reads were changed to ‘Ns’.
Modern dingo mitogenomes
Blood samples from four dingoes from Wellington Zoo were collected by the Veterinary Science Manager, Dr Lisa Argilla and sent to the University of Otago. DNA was extracted using the MagJET magnetic bead (ThermoFisher) protocol (as per manufacturer’s instructions). Four long-range primer pairs were used to amplify the complete mitogenome (Supplementary Text). Blunt end repair, ligation of sequencing adaptors and barcoding were carried out following53, with modifications for Illumina sequencing adaptors. Sequencing was carried out on the Illumina MiSeq platform. Reads were processed using a modified version of the ancient DNA pipeline previously described17.
Phylogenetic reconstructions
The phylogenetic structure of the archaeological dog samples, modern dingoes and published modern dog sequences used in this study (Supplementary Tables S1 and S2) was characterised using network analysis Popart v1.7.154. Imputed VCF files were converted to FASTA and then to NEXUS format, using a custom python script and Biopython55 respectively. This dataset was then used to create a median-joining network using Popart with default settings (Fig. 1). We used jmodeltest2 (v2.1.7)56 to assess different models of evolution for the ancient and modern dog sequences. The best model was then selected based on the Bayesian information criterion (BIC). According to this criterion a HKY + I model was most appropriate. Using this model, a maximum likelihood (ML) tree was generated using MEGA728 with 10,000 bootstrap replicates (Supplementary Fig. S2). Bayesian trees were created with BEAST (v1.8.2)57 using the Birth-Death tree model and 10,000,000 generations, and sampling every 1000th generation (Supplementary Fig. S1). The maximum credibility tree was determined using TreeAnnotator with 25% of the trees being discarded as burn-in. Convergence diagnostics were investigated using Tracer (v1.4.0) and the phylogenetic tree was visualised in FigTree (v1.4.0).
To compare the newly sequenced Pacific dog sequences with published data, the 582 bp control region fragment was aligned with control region fragments available in GenBank, using mafft v758. This dataset comprised a total of 408 sequences and was used to create a median-joining network using Popart v1.7.154 (Supplementary Fig. S3).
Data availability statement
Consensus sequences are available from the GenBank database (accession numbers KY798508-KY798516, MH035673-MH035676).
References
Anderson, A. The rat and the octopus: initial human colonization and the prehistoric introduction of domestic animals to Remote Oceania. Biological Invasions 11, 1503–1519 (2009).
Matisoo-Smith, E. In Genes, language and culture history in the Southwest Pacific (ed J S Friedlaender) 157–170 (Oxford University Press, 2007).
Bellwood, P. Holocene Population History in the Pacific Region as a Model for Worldwide Food Producer Dispersals. Current Anthropology 52, S363–S378 (2011).
Kirch, P. V. On the road of the winds: an archaeological history of the Pacific Islands before European contact. (Univ of California Press, 2017).
Pang, J. F. et al. mtDNA Data Indicate a Single Origin for Dogs South of Yangtze River, Less Than 16,300 Years Ago, from Numerous Wolves. Molecular Biology and Evolution 26, 2849–2864, https://doi.org/10.1093/molbev/msp195 (2009).
Savolainen, P., Zhang, Y.-P., Luo, J., Lundeberg, J. & Leitner, T. Genetic Evidence for an East Asian Origin of Domestic Dogs. Science 298, 1610–1613, https://doi.org/10.1126/science.1073906 (2002).
Wang, G. D. et al. Out of southern East Asia: the natural history of domestic dogs across the world. Cell Res 26, 21–33, https://doi.org/10.1038/cr.2015.147 (2016).
Savolainen, P., Leitner, T., Wilton, A. N., Matisoo-Smith, E. & Lundeberg, J. A detailed picture of the origin of the Australian dingo, obtained from the study of mitochondrial DNA. Proceedings of the National Academy of Sciences of the United States of America 101, 12387–12390, https://doi.org/10.1073/pnas.0401814101 (2004).
Milham, P. & Thompson, P. Relative antiquity of human occupation and extinct fauna at Madura Cave, southeastern Western Australia. Mankind 10, 175–180 (1976).
Reponen, S. E. M., Brown, S. K., Barnett, B. D. & Sacks, B. N. Genetic and morphometric evidence on a Galápagos Island exposes founder effects and diversification in the first-known (truly) feral western dog population. Molecular Ecology 23, 269–283, https://doi.org/10.1111/mec.12595 (2013).
Smith, B. P. & Litchfield, C. A. A Review of the Relationship between Indigenous Australians, Dingoes (Canis dingo) and Domestic Dogs (Canis familiaris). Anthrozoos: A Multidisciplinary Journal of The Interactions of People & Animals 22, 111–128, https://doi.org/10.2752/175303709X434149 (2009).
Koler-Matznick, J., Yates, B. C., Bulmer, S. & Brisbin, I. L. J. The New Guinea singing dog: its status and scientific importance. Australian Mammalogy 29, 47–56, https://doi.org/10.1071/AM07005 (2007).
Oskarsson, M. C. R. et al. Mitochondrial DNA data indicate an introduction through Mainland Southeast Asia for Australian dingoes and Polynesian domestic dogs. Proceedings of the Royal Society B: Biological Sciences 279, 967–974, https://doi.org/10.1098/rspb.2011.1395 (2012).
Paijmans, J. L. A., Gilbert, M. T. P. & Hofreiter, M. Mitogenomic analyses from ancient DNA. Molecular Phylogenetics and Evolution 69, 404–416, https://doi.org/10.1016/j.ympev.2012.06.002 (2013).
Horsburgh, K. A. et al. The Genetic Diversity of the Nguni Breed of African Cattle (Bos spp.): Complete Mitochondrial Genomes of Haplogroup T1. PLoS One 8, e71956, https://doi.org/10.1371/journal.pone.0071956 (2013).
Lancioni, H. et al. Phylogenetic Relationships of Three Italian Merino-Derived Sheep Breeds Evaluated through a Complete Mitogenome Analysis. PLoS One 8, e73712, https://doi.org/10.1371/journal.pone.0073712.s006 (2013).
Greig, K. et al. Complete Mitochondrial Genomes of New Zealand’s First Dogs. PLoS One 10, e0138536, https://doi.org/10.1371/journal.pone.0138536 (2015).
Cairns, K. M. & Wilton, A. N. New insights on the history of canids in Oceania based on mitochondrial and nuclear data. Genetica 144, 553–565, https://doi.org/10.1007/s10709-016-9924-z (2016).
Duleba, A., Skonieczna, K., Bogdanowicz, W., Malyarchuk, B. & Grzybowski, T. Complete mitochondrial genome database and standardized classification system for Canis lupus familiaris. Forensic Science International: Genetics 19, 123–129, https://doi.org/10.1016/j.fsigen.2015.06.014 (2015).
Fregel, R. et al. Mitochondrial DNA haplogroup phylogeny of the dog: Proposal for a cladistic nomenclature. Mitochondrion. https://doi.org/10.1016/j.mito.2015.04.001 (2015).
Bulmer, S. Settlement and economy in prehistoric Papua New Guinea: a review of the archeological evidence. Journal de la Société des Océanistes 31, 7–75 (1975).
McNiven, I. J. et al. Midden formation and marine specialisation at Goemu village, Mabuyag, Torres Strait, before and after European contact. Memoirs of the Queensland Museum, Culture 8, 377 (2015).
Gibbs, M., Duncan, B. & Kiko, L. Spanish Maritime Exploration in the South‐west Pacific: the search for Mendaña’s lost almiranta, Santa Isabel, 1595. International Journal of Nautical Archaeology 44, 430–438 (2015).
Addison, D. J. et al. Archaeology of Atafu, Tokelau: Some initial results from 2008. Rapa Nui Journal 23, 5–9 (2009).
Graves, M., Field, J. & McElroy, W. An overview of site 50-30-01-196, Nu ‘alolo Kai, Kaua ‘i: Features, excavation, stratigraphy, and chronology of historic and prehistoric occupation. Na Mea Kahiko o Kaua ‘i: Archaeological studies in Kaua ‘i. Special Publication, 149–187 (2005).
Greig, K., Boocock, J., Allen, M. S., Walter, R. & Matisoo-Smith, E. Ancient DNA evidence for the introduction and dispersal of dogs in New Zealand. Journal of Pacific Archaeology 9, 1–10 (2018).
Tsang, C. Recent advances in the Iron Age archaeology of Taiwan. Bulletin of the Indo-Pacific Prehistory Association 20, 153–158 (2000).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, 1870–1874 (2016).
Gonzalez, A., Clark, G., O’Connor, S. & Matisoo-Smith, L. A 3000 year old dog burial in Timor-Leste. Australian Archaeology 76, 13–20 (2013).
Cairns, K. M., Brown, S. K., Sacks, B. N. & Ballard, J. W. O. Conservation implications for dingoes from the maternal and paternal genome: Multiple populations, dog introgression, and demography. Ecology and Evolution 7, 9787–9807, https://doi.org/10.1002/ece3.3487 (2017).
Cairns, K. Population differentiation in the dingo: biogeography and molecular ecology of the Australian Native Dog using maternal, paternal and autosomal genetic markers PhD thesis, University of New South Wales, (2014).
Sacks, B. N. et al. Y Chromosome Analysis of Dingoes and Southeast Asian Village Dogs Suggests a Neolithic Continental Expansion from Southeast Asia Followed by Multiple Austronesian Dispersals. Molecular Biology and Evolution 30, 1103–1118, https://doi.org/10.1093/molbev/mst027 (2013).
Ardalan, A. et al. Narrow genetic basis for the Australian dingo confirmed through analysis of paternal ancestry. Genetica 140, 65–73, https://doi.org/10.1007/s10709-012-9658-5 (2012).
Koler-Matznick, J., Brisbin, I. L., Feinstein, M. & Bulmer, S. An updated description of the New Guinea singing dog (Canis hallstromi, Troughton 1957). Journal of Zoology 261, 109–118, https://doi.org/10.1017/S0952836903004060 (2003).
Dwyer, P. D. & Minnegal, M. Wild dogs and village dogs in New Guinea: were they different? Australian Mammalogy 38, 1–11 (2016).
J. M. I. et al. Mask Cave: Red‐slipped pottery and the Australian‐Papuan settlement of Zenadh Kes (Torres Strait). Archaeology in Oceania 41, 49–81, https://doi.org/10.1002/j.1834-4453.2006.tb00610.x (2006).
Balme, J. & O’Connor, S. Dingoes and Aboriginal social organization in Holocene Australia. Journal of Archaeological Science: Reports 7, 775–781, https://doi.org/10.1016/j.jasrep.2015.08.015 (2016).
Spriggs, M. Archaeology and the Austronesian expansion: where are we now? Antiquity 85, 510–528 (2011).
Piper, P. J. et al. Early Evidence for Pig and Dog Husbandry from the Neolithic Site of An Son, Southern Vietnam. International Journal of Osteoarchaeology 24, 68–78, https://doi.org/10.1002/oa.2226 (2014).
Grant, A. & Higham, C. In The Excavation of Khok Phanom Di. Vol II The Biological Remains (Part I) Vol. 2 Research Report of the Society of Antiquaries of London (eds C. F. W. Higham & R. Bannanurag) 147–191 (1991).
Specht, J., Denham, T., Goff, J. & Terrell, J. E. Deconstructing the Lapita Cultural Complex in the Bismarck Archipelago. Journal of Archaeological Research 22, 89–140, https://doi.org/10.1007/s10814-013-9070-4 (2014).
McNiven, I. J. et al. In Peopled Landscapes: Archaeological and Biogeographic Approaches to Landscapes (eds S. G. Haberle and B. David) 121–156 (Terra Australis, 2012).
Petchey, F. et al. 14C marine reservoir variability in herbivores and deposit-feeding gastropods from an open coastline, Papua New Guinea. Radiocarbon 54, 967–978 (2012).
David, B. et al. A new ceramic assemblage from Caution Bay, south coast of mainland PNG: the linear shell edge-impressed tradition from Bogi 1. Journal of Pacific Archaeology 3, 73–89 (2012).
Kahn, J. G. & Sinoto, Y. Refining the Society Islands cultural sequence: Colonisation phase and developmental phase coastal occupation on Mo’orea Island. Journal of the Polynesian Society, 126, 33 (2017).
Greig, K., Walter, R. & Matisoo-Smith, E. In The Routledge Handbook of Bioarchaeology in Southeast Asia and the Pacific Islands (eds M. Oxenham & H. Buckley) Ch. 21, 462–482 (Routledge, 2016).
Osmond, M. & Pawley, A. In The lexicon of Proto Oceanic: the culture and environment of ancestral Oceanic society, Volume 4: Animals (eds Malcolm Ross, Andrew Pawley, & Meredith Osmond) 217–269 (Pacific Linguistics, 2011).
Sutton, N., Summerhayes, G. & Ford, A. Regional Interaction Networks in Southern Papua New Guinea During the Late Holocene: Evidence from the Chemical Characterisation of Chert Artefacts. Proceedings of the Prehistoric Society, 1–17, https://doi.org/10.1017/ppr.2015.14 (2015).
Allen, J., Summerhayes, G., Mandui, H. & Leavesley, M. New Data from Oposisi: Implications for the Early Papuan Pottery Phase. Journal of Pacific Archaeology 2, 69–81 (2011).
Addison, D. J. & Matisoo-Smith, E. Rethinking Polynesians origins: A West-Polynesia Triple-I Model. Archaeology in Oceania 45, 1–12, https://doi.org/10.1002/j.1834-4453.2010.tb00072.x (2010).
Larson, G. et al. Phylogeny and ancient DNA of Sus provides insights into neolithic expansion in Island Southeast Asia and Oceania. Proceedings of the National Academy of Sciences of the United States of America 104, 4834–4839, https://doi.org/10.1073/pnas.0607753104 (2007).
Shannon, L. M. et al. Genetic structure in village dogs reveals a Central Asian domestication origin. Proceedings of the National Academy of Sciences 112, 13639–13644 (2015).
Kircher, M. Analysis of high-throughput ancient DNA sequencing data. Methods in molecular biology (Clifton, NJ) 840, 197–228 (2012).
Leigh, J. W. & Bryant, D. popart: full-feature software for haplotype network construction. Methods in Ecology and Evolution 6, 1110–1116, https://doi.org/10.1111/2041-210X.12410 (2015).
Cock, P. J. A. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423, https://doi.org/10.1093/Bioinformatics/Btp163 (2009).
Posada, D. & Crandall, K. A. MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818 (1998).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29, 1969–1973, https://doi.org/10.1093/molbev/mss075 (2012).
Katoh, K. & Standley, D. M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 30, 772–780, https://doi.org/10.1093/molbev/mst010 (2013).
Acknowledgements
We gratefully acknowledge Dr. Argilla and the Wellington Zoo for providing samples from their dingoes, and the Bishop Museum for specimens from the Nu’alolo Kai archaeological site, on Kauai, Hawai’i. The late Dr. Susan Bulmer provided the Taurama dog bone and was involved in many valuable discussions about dog introductions to the region. We would like to thank Nick Sutton for discussions about the dating of the Taurama archaeological site. Les O’Neill, from the Department of Anthropology at the University of Otago, produced the map showing sample distribution.
Author information
Authors and Affiliations
Contributions
K.G. and E.M.-S. conceived of and designed the experiments, and wrote the manuscript text; K.G., A.G., and K.M. performed the experiments; K.G., J.B. and C.C. analysed the data; A.G., C.C., J.B., K.M., D.A., M.A., B.D., M.G., C.H., F.L., I.M., S.O., C.T. and R.W. reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Greig, K., Gosling, A., Collins, C.J. et al. Complex history of dog (Canis familiaris) origins and translocations in the Pacific revealed by ancient mitogenomes. Sci Rep 8, 9130 (2018). https://doi.org/10.1038/s41598-018-27363-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-27363-8
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.