Abstract
The number of service visits of Alzheimer’s disease (AD) patients is different from each other and their visit time intervals are non-uniform. Although the literature has revealed many approaches in disease progression modeling, they fail to leverage these time-relevant part of patients’ medical records in predicting disease’s future status. This paper investigates how to predict the AD progression for a patient’s next medical visit through leveraging heterogeneous medical data. Data provided by the National Alzheimer’s Coordinating Center includes 5432 patients with probable AD from August 31, 2005 to May 25, 2017. Long short-term memory recurrent neural networks (RNN) are adopted. The approach relies on an enhanced “many-to-one” RNN architecture to support the shift of time steps. Hence, the approach can deal with patients’ various numbers of visits and uneven time intervals. The results show that the proposed approach can be utilized to predict patients’ AD progressions on their next visits with over 99% accuracy, significantly outperforming classic baseline methods. This study confirms that RNN can effectively solve the AD progression prediction problem by fully leveraging the inherent temporal and medical patterns derived from patients’ historical visits. More promisingly, the approach can be customarily applied to other chronic disease progression problems.
Similar content being viewed by others
Introduction
Significance of predicting dementia progression
As of 2017, approximately 5.4 million Americans in the US live with Alzheimer’s disease (AD), which is the most common form of dementia. According to the US National Alzheimer’s Coordinating Center (NACC), AD is one of leading causes of death in the US. Moreover, for a patient with AD, his or her AD condition will chronically and progressively deteriorate over a long period of time. However, as of April of 2018 there exists no effective cure for AD. In other words, AD cannot be reversed or cured with today’s medicines and treatments. Unless a method of prevention or treatment will be discovered, the estimated total cost of care of people with Alzheimer’s and other dementias in the US will grow to about $1 trillion in 2050 from an estimated $226 billion in 20151. It is known that the social and psychological burden on individuals and families will be even more daunting than the costs of care.
While waiting for significant progress of developing AD cure medicines, many researchers have been looking for alternative, viable, and cost-effective solutions that help fill in the gap of the needed care and treatment for AD patients2,3,4,5,6,7,8. A very promising approach has been widely explored, focusing on early prediction and positive intervention at the personalized and comfortable level, which inherently and truly varies with patients and keeps changing over time. An appropriate and positive intervention includes ways of facilitating AD patients with right and effective levels of lifestyle changes and brain training. Therefore, understanding and predicting how AD develops on an individual patient basis over time is the key to the success of enabling early intervention of AD and accordingly providing personalized healthcare services in an effective manner1,2.
Studies relevant to modeling disease progression
Traditional time series methods and machine learning algorithms have been widely applied to AD progression modeling and severity classification problems. Sukkar et al.3 applied hidden Markov chains to model AD progression; Zhou et al.4 proposed a convex fused sparse group Lasso formulation to predict AD patients’ cognitive scores at different time points; Zhou et al.5 then used a multi-task regression model to predict Mini Mental State Examination score and AD Assessment Scale Cognitive subscale score; Liu et al.6 identified the transitional patterns of AD using a series of joint random-effects transition models; Huang et al.7 proposed a nonlinear regression-based random forest model to predict longitudinal AD clinical scores; Gavidia-Bovadilla et al.8 proposed aging-based null models for early diagnosis of AD; Hou et al.9 proposed a model to predict AD longitudinal scores by estimating clinical scores from MRI data at multiple time points; O’Kelly10 used supervised machine learning algorithms to classify the statuses of AD patients; Qiu et al.2 applied decision tree algorithms to classify the progression statuses of AD patients. Note that many researchers have also focused on other diseases progression modeling11,12,13,14,15,16,17,18,19.
Although the above-mentioned researchers have made promising progress in studies related to diseases progression modeling2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19, many modeling challenging issues still remain20. For example, it is difficult for traditional time series methods to handle high-dimensional longitudinal data. Rather than predicting disease’s future status, the literature largely focuses on disease progression modeling using hidden Markov models and multi-task regression models, which predict the progression statuses of diseases at known time points based on the collected information relevant to those time points, or explores classifying progression stages only within a narrow observation window2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19.
Unlike the problem of disease progression modeling covered by the existing literature, we aim to use AD patients’ medical information at historical time points to predict their disease progression stages at a future time point. In general, it is extremely challenging to capture and derive temporal patterns to help predict accurately the future progressions of AD patients due to the fact that AD patients’ data are highly heterogeneous2,21. Indeed, regarding longitudinal electronic health records (EHR) describing the length of progression staging and the progressive rate at different AD stages over the years, each patient’ EHR is unique22. Moreover, time intervals between two consecutive visits are often irregular or uneven. However, for the future progression prediction problem under study, traditional machine learning algorithms predict diseases’ staging simply by aggregating longitudinal features rather than leveraging their longitudinal temporal patterns. As a result, modeling accuracies suffer to some extent. Therefore, it is necessary to explore a new approach for the future disease progression predictive problems.
Predictive modeling with RNN in healthcare
RNN is naturally good at capturing longitudinal temporal patterns23,24. Recently, RNN models have shown great potential in healthcare applications: predicting the diagnosis and medications of the subsequent visit for a patient with gated recurrent units (GRU) RNN25, predicting kidney transplantation endpoints within the future six or twelve months using different RNN variants26, early detecting heart failure onsets using GRU models27, predicting the onsets of multiple conditions using Long Short-Term Memory (LSTM) models28, classifying diagnoses for pediatric intensive care unit (PICU) patients using LSTM networks29, predicting PICU’s mortality with LSTM networks30, predicting risk of mortality, physiologic decline, length of stay, and phenotype simultaneously with a Multitask LSTM model31, learning patients’ similarities for patients with Parkinson’s disease using a 2-D GRU model32. These studies applied various RNN models for medical prediction tasks through effectively leveraging the temporal relations among longitudinal patient data. As of April of 2018, however, RNN has not been well adopted in the AD future progression predictive modeling.
Objective
To address this above-mentioned interesting, challenging, while long-overdue problem, we investigate whether RNN can predict the future progression of AD through fully leveraging inherent longitudinal temporal information of patients’ historical visits. Particularly, we aim to predict the AD progression stage of the next hospital visit in the future for a patient only based on the information of his/her historical visits. The Global Staging Clinical Dementia Rating (CDR) score is a clinical comprehensive metric to assess the dementia levels, which has been well adopted to define Dementia (including AD) progression stages7,33. Note that the Global CDR score is derived from the rating scores of six cognitive elements defined as the standard CDR scale according to clinical scoring rules. The standard CDR scale was primarily developed by Washington University School of Medicine for staging the dementia severity of AD34. A patient who is clinically diagnosed with dementia can be at five different stages with respect to the Global CDR score: 0 (no impairment), 0.5 (questionable impairment), 1 (mild impairment), 2 (moderate impairment), and 3 (severe impairment)34. Hence, the Global CDR score (i.e., CDRGLOB define in the NACC database) is utilized to define the AD progression stages in this study. Note that the proposed model is neutral to etiologic diagnoses, which implies that other metrics defining AD progression stages can be used to replace the Global CDR score if necessary.
Materials and Methods
Data description, analysis and preprocessing
Data description and analysis
The patient dataset used in this study was provided by NACC. The data includes patients’ demographics, health history, physical information, elements of the CDR scale34,35, Geriatric Depression Scale (GDS)36,37,38,39,40, and Functional Activities Questionnaire (FAQ)41,42,43. The detailed information about these feature categories is provided in the Supplementary Table S1.
To focus on the future progression predictive modeling of AD, patients who were marked with probable AD and had more than three visits were chosen from the original dataset. As a result, 5432 patients with probable AD between August 31, 2005 and May 25, 2017 as a subset of the NACC dataset are included in this study. The number of visits still varies with patients in the subset, with an average of 4.98 visits for a patient while the maximal number reaches as high as 12. In addition, the time intervals between patients’ two consecutive visits are quite different too. As illustrated in Fig. 1, although about 46% of time intervals are about 12 months, other time intervals span from 5 months to as far as 5 years.
Table 1 shows the likelihood that a patient changing from one Global CDR score to another between two consecutive visits in the dataset under study. \({{y}}_{{{t}}_{{n}}}\) denotes the global CDR score of a patient in its \({n}^{th}\) visit and \({y}_{{t}_{n+1}}\) denotes the global CDR score of a patient in its \({(n+1)}^{th}\) visit.
With about 60% of visits, the global CDR score of a patient had no change at a given stage, except for the fifth stage (\({{y}}_{{{\boldsymbol{t}}}_{{\boldsymbol{n}}}}=3\)). With about 40% of visits, a patient got worsen by one stage, and with about 4% of visits, a patient got worsen by two stages. By contrast, with about 4% of visits, a patient got better by one stage with respect to the global CDR score. In short, Table 1 clearly shows that AD progresses slowly over time. Therefore, to help an AD patient with effective healthcare guidance, it is critical to identify if the patient will have pejorative progression in his/her next visit.
Figure 2 shows the distribution of demented patients at different progression stages with respect to their first visits and their last visits in the selected subset. 47.7% and 34.9% patients as the majority are at the second and third stages respectively on their first visits, which indicates that most patients began to visit AD related healthcare service centers when they were at either questionable impairment or mild impairment stages. However, 30.8% and 29.3% patients had progressed to or remained at the fourth and fifth stages respectively on their last visits. In fact, when we simply look at Fig. 2 and compare bars of their first and last visits, those AD patients under study had become more severe as time passed. Therefore, it is important and promising to explore how and predict the disease of an AD patient progresses over time.
In total, there are 78 features/predictors (including time interval) and a label (Global CDR score) in the created data subset. The variable time interval is derived from two successive visit dates, which is measured in months. Detailed descriptions of other variables are listed in the Supplementary Table S1. There are different data types in this data subset, such as continuous variables, ordinal variables, and nominal variables (including dummy variables). Most variables have different percentages of missing data, and the distributions of those missing data are given in Table 2. Thus, it needs to perform data imputations to obtain quality data for predictive modeling.
Data preprocessing
Two steps of data preprocessing were adopted: (a) we first take advantage of some simple and fast imputation techniques to impute the missing data to obtain high quality data, (b) we then apply normalization or encoding on the imputed data to get appropriate inputs for the adopted deep learning model.
In general, we can apply mean imputation to continuous variables, median imputation to ordinal variables, and mode imputation to nominal variables. Nevertheless, in this study we also applied customized imputation schemes to certain variables by taking their feature characteristics into consideration as shown in Table 3.
Then normalization or encoding was performed on the imputed data. For the continuous variables, we normalized the variables with their corresponding mean and variance. For the categorical variables (including ordinal and nominal variables), one-hot encoding was performed on each of those variables. For example, if a categorical variable had five classes, we used five-dimension one-hot vectors to represent the variable. After the completion of one-hot encoding for the categorical variables, the dimension of the input features for the proposed model was increased to 234.
RNN model for AD progression stage prediction
Long short-term memory (LSTM) RNN model
We applied RNN to the AD progression modeling in this study. Different from traditional neural networks, RNN models allow temporal information to be passed from one time step to the next time step in the network23. The proposed RNN model is structured with one input layer, two hidden layers (Fig. 3), and one output layer. The input layer can accommodate well the needed information of all historical visits and irregular visit time intervals for patients.
Formally, given that there are N visits (time points or steps) for a patient, the input at time \({t}_{n}\), \(n=1,\,2,\cdots ,\,N\), includes patient’s features \({{\boldsymbol{x}}}_{{t}_{n}}\) and time interval \({\rm{\Delta }}{t}_{n}={t}_{n+1}-{t}_{n}\). More specifically, \({{\boldsymbol{x}}}_{{t}_{n}}\) is a row vector with \(Q\,\)dimensions where Q indicates the number of input features. Note that a feature can be continuous, ordinal or nominal. If the Nth visit represents the current visit of a patient, then the \((N+1){}^{{\rm{th}}}\) visit represents the next visit of the patient. The prediction model proposed in our study can be written as:
where the output \({\hat{{\boldsymbol{y}}}}_{{t}_{N+1}}\) is the AD stage of the next visit of the patient (Fig. 3) and the function f(·) represents the proposed model. When the prediction model is applied in the real world, \({\rm{\Delta }}{t}_{N}\) can be set according to a user’s need, which means the future prediction time point can be customized as needed. As mentioned earlier, demented patients will experience five stages with reference to CDRGLOB. Therefore, \({\hat{{\boldsymbol{y}}}}_{{t}_{N+1}}\) is a one-hot vector with five dimensions, and each dimension indicates a corresponding AD stage. Since the proposed model has a time window shift between the input and the output, a pad with zero vectors for the (N + 1)th visit was performed on the input to create a “many-to-one” model24.
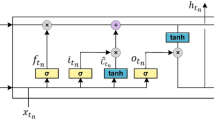
Each module in the proposed RNN architecture is a very special kind of RNN cell, called LSTM (Fig. 4)44,45. \({{\boldsymbol{C}}}_{{t}_{n-1}}\) is the cell state of the last time step of the same hidden layer, and \({{\boldsymbol{h}}}_{{t}_{n-1}}\) is the output of the last cell state of the same hidden layer. \({{\boldsymbol{C}}}_{{t}_{n}}\) is the cell state of the current time step, and will connect to the next time step of the same hidden layer. \({{\boldsymbol{h}}}_{{t}_{n}}\) is the output part of the current time step, which will pass information to both the next time step of the same hidden layer and the same time step of the next hidden layer, as shown in Fig. 3.
The equations of Fig. 4 are defined as follows23,24,45:
There are three gates in an LSTM cell (Fig. 4): \({{\boldsymbol{f}}}_{{t}_{n}}\), \({{\boldsymbol{i}}}_{{t}_{n}}\) and \({{\boldsymbol{o}}}_{{t}_{n}}\) represents a forget gate, an input gate and an output gate, respectively. \({\tilde{{\boldsymbol{C}}}}_{{t}_{n}}\) represents the candidate value of a new cell. The new cell state, \({{\boldsymbol{C}}}_{{t}_{n}}\), is derived from the cell state of the last time step through the forget gate while the candidate value derived from the input gate. The output part of the current cell state, i.e., \({{\boldsymbol{h}}}_{{t}_{n}}\), is obtained from a tanh (·) activation function.
The proposed RNN model has a chain of the same LSTM module in each hidden layer. Hence, to predict the AD stage of the \((N+1){}^{{\rm{th}}}\) visit for a patient, we used a softmax function:
where \({{\boldsymbol{h}}}_{{t}_{N}}^{(2)}\) represents the output of the Nth step of the second hidden layer.
Loss function with regularization
To avoid overfitting, regularization was applied. Formally, the loss function of the model becomes \({\rm{J}}({\boldsymbol{\theta }})+{\rm{\lambda }}\cdot {\rm{\Omega }}(\,\cdot \,)\), where \({\boldsymbol{\theta }}\) indicates the vector of parameters of the model (including weights and biases) and \({\rm{J}}({\boldsymbol{\theta }})\) is the original loss function (before regularization). The regularization item, Ω(·), represents the model complexity while λ is the ratio of the cost of the model complexity and the total cost. In general, the complexity of a model depends on weights. So the regularization item can be simply computed based on weights, i.e., \({\rm{\Omega }}({\bf{W}})\). There are two common kinds of regularization: L1 regularization, and L2 regularization46.
Both L1 and L2 regularization techniques focus on limiting the weight values to avoid overfitting. L1 regularization will make some weights zeroes in the training process, which is equivalently considered as conducting feature selections. While L2 regularization will not make some weights zeroes in the training process. When some weights have small values, their corresponding square values will become smaller, which is small enough to ignore. In this study, L2 regularization is used so that all features can be well considered. The cross-entropy function with regularization for M patients thus is defined as follows:
where M is the total number of patients, \({{\boldsymbol{y}}}_{{t}_{N+1}}^{(m)}\) indicates the true AD stage of the mth patient, \({\hat{{\bf{y}}}}_{{t}_{N+1}}^{(m)}\) is the value predicted by the model, λ is a hyperparameter that controls the L2 regularization, and \({w}_{i}\) denotes a weight in the built network.
Model evaluation
Our goal is to predict the AD progress of a patient at the time he/she visits in the future. Hence, accuracy that defines the percentage of correct predictions can be used as the basic metric to evaluate the performance of a multi-class prediction model. To a patient, he/she might be more concerned with the correct prediction of his/her AD progression with respect to CDRGLOB. In practice, an accurate prediction of disease progression provides a significant value to the patient, doctors, and caregivers.
Based on the definition of CDRGLOB, if the value of a patient’s CDRGLOB from the Nth visit to the (N + 1)th visit increases by 0.5, 1, 2 or 3, we consider that the patient’s AD has developed or made pejorative progress. Otherwise, we simply consider that the patient’s condition has been stable or not deteriorated. In theory, if the value of CDRGLOB of the patient changes by any negative number, the patient would have got better. We know this wouldn’t happen until an effective drug or treatment is discovered. Thus, the pejorative progression identification accuracy (PPIA) is a meaningful and important metric for now.
In addition, patients can be divided into two different groups with respect to CDRGLOB. One group consists of the patients whose CDRGLOB values are 0, 0.5, or 1 on their last visits; the other group includes the patients whose CDRGLOB values are 2 or 3 on their last visits. Obviously, the second group includes severe AD patients, while the first group is considered as a non-severe one. Thus, the prediction accuracy of the second group can be denoted as severe patient identification accuracy (SPIA), which is another performance metric for the proposed model that users may also have interest.
Model training and comparisons
Baseline models for performance comparisons
For the progression prediction problem under study, one may wonder if classical time series methods might be applied to solve the problem. That is, one might use the information of the target variable (i.e., AD stage) of historical visits to predict its future values, i.e., \({\hat{{\boldsymbol{y}}}}_{{t}_{N+1}}=\psi ({{\boldsymbol{y}}}_{{t}_{1}},{{\boldsymbol{y}}}_{{t}_{2}},\,\ldots ,{{\boldsymbol{y}}}_{{t}_{N}})\), where \(\psi (\,\cdot \,)\) represents a classical time series method. Unfortunately, as mentioned earlier, the number of visits has an average of 4.98 visits for a patient and the target variable is an ordinal variable. As a result, traditional time series methods fail to capture the AD progression patterns. In other words, traditional time series methods can’t be well applied to the problem under study. Therefore, four machine-learning algorithms were chosen as baselines in this study. These four baseline models include logistic regression (LR), support vector machine (SVM), decision tree (DT), and random forest (RF), which will be used to be compared with the proposed RNN model in terms of their performances. Since these baseline models cannot handle various lengths of longitudinal temporal data, three training approaches are adopted for each of the baseline models to predict AD patients’ future progression stages.
Approach 1: we can train the baseline models with aggregated features of all patients’ historical visits, which is a common method for longitudinal data in predictive modeling27. The aggregated features’ values from all the historical visits can be defined as:
where \({\bar{{\boldsymbol{x}}}}_{{t}_{1},\cdots ,{t}_{N}}\,\)is a vector with Q dimensions, and the aggregated value of the qth dimension (\(q=1,\,2,\,\cdots ,Q\)) is defined as follows:
-
If the qth feature is a continuous type variable, then the aggregated value \({\bar{{\boldsymbol{x}}}}_{q}\) is the mean of the feature values of all the historical visits.
-
If the qth feature is an ordinal type variable, then the aggregated value \({\bar{{\boldsymbol{x}}}}_{q}\) is the median of the feature values of all the historical visits.
-
If the qth feature is a nominal type variable, then the aggregated value \({\bar{{\boldsymbol{x}}}}_{q}\) is the mode value of the feature values of all the historical visits.
Based on the aggregated features’ values, the baseline models used to predict the future progression stage can be defined as follows:
where \({f}_{1}(\,\cdot \,)\) represents a baseline model.
Approach 2: similar to time series methods, the baseline models can also be implemented with the information of the most recent r visits among historical visits. We merge the feature values of most recent r visits into one feature vector, i.e.,
where \({\tilde{{\boldsymbol{x}}}}_{{t}_{N-r+1},\cdots ,{t}_{N}}\) is a merged vector with r*Q dimensions, note that \({{\boldsymbol{x}}}_{{t}_{N-r+1}}\), \({{\boldsymbol{x}}}_{{t}_{N-r+2}},\,\,\ldots ,\,{{\boldsymbol{x}}}_{{t}_{N}}\) are feature vectors with Q dimensions at the \((N-r+1){}^{{\rm{th}}}\), the \((N-r+2){}^{{\rm{th}}}\), \(\ldots \), and the Nth visit, respectively.
Based on the merged feature vector, the baseline models used to predict the future progression stage can be then defined as follows:
where \({f}_{2}(\,\cdot \,)\) represents a baseline model and r should be smaller than the minimum number of visits of patients. Note that the minimal number of visits among patients is 3 in the dataset under study, and in this study r is therefore set to 2 in order to evaluate the prediction performance of built models. That is, one visit was left for each patient that was used to evaluate the prediction performance of the models.
Approach 3: the baseline models can be easily implemented with the feature information at the Nth visit, i.e.,
where \({f}_{3}(\,\cdot \,)\) represents a baseline model.
Regarding the above training approaches, Approach 3 is essentially a special case of Approach 2. As mentioned above, r is set to 2 for Approach 2 in this study, which makes Approach 2 and Approach 3 very similar. It is worth mentioning that when the minimum number of visits of patients increases, r can be set to other higher values.
Different from the baseline models, we can train our proposed model with or without time intervals (TI) that are derived from two consecutive visits. Essentially, we aim to investigate whether the proposed RNN model can fully leverage irregular time intervals information to improve AD progression modeling performances.
Impact analysis of various feature categories
To get a better understanding of factors or variables impacting the cognitive declines of patients, we perform feature analysis based on different subsets of variables. More specifically, more experiments are conducted to investigate how each category of features would impact the prediction performance. In this study, the full dataset based model (or simply called the full model) contains 6 feature categories: Subject Demographics, Subject Health History, Physical information, CDR, GDS, and FAQ. Since the visit time intervals, demographical, health historical physical information and MMSE score are basic information for patients, we conduct various sub-models using different combinations of CDR, GDS and/or FAQ to analyze their impacts on corresponding modeling performances.
Data availability
The data that support the findings of this study are available from NACC but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of NACC.
Results
In this study, we implemented our RNN model of 2 hidden layers with 100 hidden units at each layer. The learning rate decay and moving average decay mechanism were applied in the training process. L2 regularization was added to the loss function, and Adam Optimizer was used in the loss function optimization47. We used 10-fold cross validation in our experiments. Thus the results illustrated later are the average values of the 10-fold cross validation runs. The detailed implementation information for the proposed model is provided in the Supplementary File.
First, we conducted different experiments to train the proposed model with or without TIs. Then we compared the performances of the proposed models with the baseline models. The results are shown in Table 4.
As shown in Table 4, LSTM with TI performs better than LSTM w/o TI by about 0.6%, 1.0% and 0.6% regarding the metrics - Accuracy, PPIA, and SPIA, respectively. In other words, a model of AD progression with irregular TI is better than one only with logical temporal information. This confirms that LSTM based RNN models can truly account for the longitudinal temporal patterns in the problem under study.
Note that the baseline models that are trained with the aggregated information of all the historical visits outperform the corresponding models based on these methods trained with the information of two most recent historical visits or the most recent historical visit. The proposed models based on LSTM (both LSTM with TI and LSTM w/o TI) are far superior to the baseline models as shown in Table 4. Specially, the accuracy of LSTM with TI is about 19.5%, 24.6%, 28.7% and 29.9% better than LR, SVM, DT and RF models trained with the aggregated information of all the historical visits, respectively. Moreover, the proposed models also perform far better than the baseline models with respect to other two metrics PPIA and SPIA.
In addition, to analyze the impact of various feature categories on the model performance, we conducted experiments of LSTM with TI using different combinations of feature categories (Table 5).
As illustrated in Table 5, a model without GDS shows a slight negative impact on the model’s capability to predict, while a model without CDR or FAQ performs much worse than the full model, with accuracy being decreased by about 3.5% or 2.9% respectively. Note that the performance is considerably decreased when CDR and FAQ are not considered by the model at the same time. Therefore, when the model is applied in practice, it is very important to take into account CDR data and/or FAQ data, and if available, both CDR and FAQ should be included for better results.
To further show the effect of each specific feature in the CDR/FAQ category on the model performance, experiments with variable control approaches have been conducted. A model only with the basic information (i.e., a model without CDR, FAQ, GDS) is defined as our basic model. There are 9 features in the CDR category and 10 features in the FAQ category. Each time one specific feature of the CDR/FAQ category was incorporated into the basic model to investigate the impact of the specific feature on the model performance. Table 6 shows the model improvement by incorporating a specific feature of the CDR/FAQ category when compared with the basic model.
As shown in Table 6, the values of all the three metrics that had more than 10% improvements are marked in bold. It is clear that HOMEHOBB, COMMUN, ORIENT, JUDGMENT, and CDRSUM in the CDR category significantly contribute to the model performance improvement. Note that the feature CDRSUM is actually the sum of the scores of six features including MEMORY, ORIENT, JUDGMENT, COMMUN, HOMEHOBB, PERSCARE. Therefore, HOMEHOBB, COMMUN, ORIENT, and JUDGMENT as independent features are ones that substantially impact the model performance. By contrast, top contributed features in the FAQ category help improve the model performance by about 7% respectively, which are less significant when compared to top contributed CDR features.
Discussion
As indicated in Table 4 the proposed LSTM RNN model when it is trained with irregular TIs performs better than one without accounting for TIs. Generally, the proposed model performs far better than models based on traditional machine learning methods such as LR, SVM, DT and RF. This mainly attributes to the fact that the proposed model can fully capture and leverage patients’ temporal information patterns along with their historical visits.
Interestingly, the results of the feature analysis show that CDR and FAQ play an important role in the AD progression prediction, while GDS contains less effective predictors when compared to CDR and FAQ. In fact, these findings are in line with the medical implications of these data categories:
-
CDR includes six cognitive elements in the standard CDR scale version and two additional variables defining the language and the behavior-comportment and personality of a patient. Together they can comprehensively describe the clinical dementia status of patients35.
-
FAQ is used to access a patient’s instrumental activities of daily living (IADLs), which is effective when used to monitor the functional change of the patient over time41,42,43. For an individual with AD, his/her functional ability usually declines over time. Hence, FAQ has been well considered as risk prognosis for cognitive impairment48,49,50.
-
GDS is a self-administered instrument, so when a person with mild dementia of the Alzheimer’s type, GDS is not an effective tool to identify depression37. Specially, when a patient is not able to complete a GDS test according to clinician’s best judgment, the corresponding GDS items will be marked as “did not answer”, which essentially become missing data in the archived database. Hence, GDS variables are not considered as effective predictors in the AD progression modeling.
The feature analysis mentioned earlier in fact provides sufficient evidences that are aligned well with the intentions or implications of these data categories.
To the best of our knowledge, we are the first group to apply RNN to the AD progression predictive modeling. The contributions of this study can be summarized as follows:
-
First, unlike classification modeling, without a patient’s next visit feature information the proposed LSTM RNN model can predict his/her next visit’s AD progression stage. This is attributed to the adopted RNN with an enhanced structure. Because we have successfully incorporated the shift of time steps into the proposed model, the longitudinal medical patterns of his/her previous visits on record can be well captured and leveraged.
-
Secondly, because the “many-to-one” RNN structure is applied, the prediction can also be executed without target variables (i.e., AD stages in this study) in the time steps that are marked as preceding visits from the very last one. In other words, only the values of the target variable in the last time step are needed in a developed model based on the adopted RNN structure. Therefore, the adopted RNN structure for this kind of study works well for the target variables that are partially or totally missing for the preceding time steps except the very last ones of patients in the dataset.
-
Thirdly, the proposed progression prediction model can be very adaptive over time since it allows irregular visit TIs and various numbers of visits. Note that TIs are always irregular or uneven, which frequently depends on the patient’s preference or need. It is worth mentioning that the prediction time interval between the current visit and the next visit is part of the input information of a model, which can be chosen by users according to their prediction requirements.
-
Finally, other metrics defining AD progression stages can be used to replace the Global CDR score if necessary. The severity stages of AD progressions can be adjusted for a variety of metrics with different granularity levels. Furthermore, the proposed model can be applied to other chronic disease progression modeling.
In the near future, we will develop models that can account for deeper and more granular details in patients’ information, for example different deteriorating rates among patients. If possible, more accurate classification schemes (e.g., biomarker indicators) will be taken into account. Furthermore, we will extend the proposed approach to broader studies with a focus on early disease stage predictions, such as exploring how patients progress from non-demented to demented stages.
Conclusion
This paper proposed a disease progression model based on a deep RNN with LSTM cells. A “many-to-one” architecture with enhancements in support of time step shifts was fully leveraged in this study. As a result, the proposed model can accurately predict the AD stage of the next visit of a patient when the information of the patient’s previous visits becomes available. By relying on the real-world dataset, i.e., the NACC dataset for patients with AD, we tested and validated the developed model to confirm its applicability in practice, and also compared its performance with four classic approaches. We also explored several sub-models using different combinations of feature categories to analyze their impacts on the model’s performances. In overall, the results of this project show that the proposed LSTM RNN model can effectively predict the future statuses of AD patients. Most promisingly, we can easily apply the proposed model to other chronic disease progression predictive modeling.
References
Borson, S. et al. Report on milestones for care and support under the US National Plan to Address Alzheimer’s Disease. Alzheimers Dement. 12(3), 334–369 (2016).
Qiu, R. G., Qiu, J. L. & Badr, Y. Predictive modeling of the severity/progression of Alzheimer’s diseases. 2017 International Conference on Grey Systems and Intelligent Services (GSIS). 400–403 (2017).
Sukkar, R., Katz, E., Zhang, Y., Raunig, D. & Wyman, B. T. Disease progression modeling using hidden Markov models. Conf Proc IEEE Eng Med Biol Soc. 2012, 2845–2848 (2012).
Zhou, J., Liu, J., Narayan, V. A. & Ye, J. Modeling disease progression via fused sparse group lasso. KDD. 2012, 1095–1103 (2012).
Zhou, J. et al. Modeling disease progression via multi-task learning. Neuroimage. 78, 233–248 (2013).
Liu, W., Zhang, B., Zhang, Z. & Zhou, X. H. Joint Modeling of Transitional Patterns of Alzheimer’s Disease. PLoS One. 8(9), e75487 (2013).
Huang, L., Jin, Y., Gao, Y., Thung, K. H. & Shen, D. Longitudinal clinical score prediction in Alzheimer’s disease with soft-split sparse regression based random forest. Neurobiol Aging. 46, 180–191 (2016).
Gavidia-Bovadilla, G. et al. Early Prediction of Alzheimer’s Disease Using Null Longitudinal Model-Based Classifiers. PLoS One. 12(1), e0168011 (2017).
Hou, W., Lei, B., Zou, W., Li, X. & Zhang, C. Ensemble prediction of longitudinal scores of Alzheimer’s disease based on ℓ2, 1-norm regularized correntropy with spatial-temporal constraint. 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017). 891–894 (2017).
O’Kelly, N. Use of Machine Learning Technology in the Diagnosis of Alzheimer’s Disease (Dublin City University 2016).
Jackson, C. H., Sharples, L. D., Thompson, S. G., Duffy, S. W. & Couto, E. Multistate Markov models for disease progression with classification error. Biometrics. 66(3), 742–752 (2010).
Tangri, N. et al. A predictive model for progression of chronic kidney disease to kidney failure. JAMA. 305(15), 1553–1559 (2011).
Liu, Y. Y. et al. Longitudinal modeling of glaucoma progression using 2-dimensional continuous-time hidden markov model. Med Image Comput Comput Assist Interv. 16, 444–451 (2013).
Schulam, P. & Saria, S. A framework for individualizing predictions of disease trajectories by exploiting multi-resolution structure. Advances in Neural Information Processing Systems. 748–756 (2015).
Wang, X., Sontag, D., Wang, F. Unsupervised learning of disease progression models. In: Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining. 85–94 (2014).
Burckhardt, P., Nagin, D. S. & Padman, R. Multi-Trajectory Models of Chronic Kidney Disease Progression. AMIA Annu Symp Proc. 2016, 1737–1746 (2017).
Futoma, J., Sendak, M., Cameron, C. B. & Heller, K. Scalable Modeling of Multivariate Longitudinal Data for Prediction of Chronic Kidney Disease Progression. Preprint at https://arxiv.org/abs/1608.04615 (2016).
Kannan, V. et al. Conditional disease development extracted from longitudinal health care cohort data using layered network construction. Sci Rep. 6, 26170 (2016).
Miotto, R., Li, L., Kidd, B. A. & Dudley, J. T. Deep patient: An unsupervised representation to predict the future of patients from the electronic health records. Sci Rep. 6, 26094 (2016).
Goldstein, B. A., Navar, A. M., Pencina, M. J. & Ioannidis, J. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J Am Med Inform Assoc. 24(1), 198–208 (2017).
Lemos, R. et al. The free and cued selective reminding test for predicting progression to Alzheimer’s disease in patients with mild cognitive impairment: a prospective longitudinal study. J Neuropsychol. 11(1), 40–55 (2017).
Yang, J., McAuley, J., Leskovec, J., LePendu, P. & Shah, N. Finding progression stages in time-evolving event sequences. In: Proceedings of the 23rd international conference on World wide web. 783–794 (2014).
Graves, A. Supervised Sequence Labelling with Recurrent Neural Networks (Springer 2012).
Lipton, Z. C., Berkowitz, J. & Elkan, C. A critical review of recurrent neural networks for sequence learning. Preprint at https://arxiv.org/abs/1506.00019 (2015).
Choi, E., Bahadori, M. T., Schuetz, A., Stewart, W. F. & Sun, J. Doctor ai: Predicting clinical events via recurrent neural networks. JMLR Workshop Conf Proc. 56, 301–318 (2016).
Esteban, C., Staeck, O., Baier, S., Yang, Y. & Tresp, V. Predicting clinical events by combining static and dynamic information using recurrent neural networks. 2016 IEEE International Conference on Healthcare Informatics (ICHI). 93–101 (2016).
Choi, E., Schuetz, A., Stewart, W. F. & Sun, J. Using recurrent neural network models for early detection of heart failure onset. J Am Med Inform Assoc. 24(2), 361–370 (2017).
Razavian, N., Marcus, J. & Sontag, D. Multi-task prediction of disease onsets from longitudinal laboratory tests. Machine Learning for Healthcare Conference. 73–100 (2016).
Lipton, Z. C., Kale, D. C., Elkan, C. & Wetzel, R. Learning to diagnose with LSTM recurrent neural networks. Preprint at https://arxiv.org/abs/1511.03677 (2015).
Aczon, M. et al. Dynamic Mortality Risk Predictions in Pediatric Critical Care Using Recurrent Neural Networks. Preprint at https://arxiv.org/abs/1701.06675 (2017).
Harutyunyan, H., Khachatrian, H., Kale, D. C. & Galstyan, A. Multitask Learning and Benchmarking with Clinical Time Series Data. Preprint at https://arxiv.org/abs/1703.07771 (2017).
Che, C. et al. An RNN Architecture with Dynamic Temporal Matching for Personalized Predictions of Parkinson’s Disease. In: Proceedings of the 2017 SIAM International Conference on Data Mining. Society for Industrial and Applied Mathematics. 198–206 (2017).
Williams, M. M., Storandt, M., Roe, C. M. & Morris, J. C. Progression of Alzheimer’s disease as measured by Clinical Dementia Rating Sum of Boxes scores. Alzheimers Dement. 9(1), S39–S44 (2013).
Morris, J. C. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 43(11), 2412–2414 (1993).
Knopman, D. S., Weintraub, S. & Pankratz, V. S. Language and behavior domains enhance the value of the clinical dementia rating scale. Alzheimers Dement. 7(3), 293–299 (2011).
Yesavage, J. A. et al. Development and validation of a geriatric depression screening scale: a preliminary report. J Psychiatr Res. 17(1), 37–49 (1983).
Burke, W. J., Houston, M. J., Boust, S. J. & Roccaforte, W. H. Use of the Geriatric Depression Scale in dementia of the Alzheimer type. J Am Geriatr Soc. 37(9), 856–860 (1989).
Almeida, O. P. & Almeida, S. A. Short versions of the geriatric depression scale: a study of their validity for the diagnosis of a major depressive episode according to ICD‐10 and DSM‐IV. Int J Geriatr Psychiatry. 14(10), 858–865 (1999).
Van Marwijk, H. W. et al. Evaluation of the feasibility, reliability and diagnostic value of shortened versions of the geriatric depression scale. Br J Gen Pract. 45(393), 195–199 (1995).
de Craen, A. J., Heeren, T. J. & Gussekloo, J. Accuracy of the 15‐item geriatric depression scale (GDS‐15) in a community sample of the oldest old. Int J Geriatr Psychiatry. 18(1), 63–66 (2003).
Pfeffer, R. I., Kurosaki, T. T., Harrah, C. H. Jr., Chance, J. M. & Filos, S. Measurement of functional activities in older adults in the community. J Gerontol. 37(3), 323–329 (1982).
Marshall, G. A. et al. Executive function and instrumental activities of daily living in mild cognitive impairment and Alzheimer’s disease. Alzheimers Dement. 7(3), 300–308 (2011).
Hall, J. R., Vo, H. T., Johnson, L. A., Barber, R. C. & O’Bryant, S. E. The link between cognitive measures and ADLs and IADL functioning in mild Alzheimer’s: what has gender got to do with it? Int J Alzheimers Dis. 2011, 1–6 (2011).
Hochreiter, S. & Schmidhuber, J. LSTM can solve hard long time lag problems. Advances in neural information processing systems. 473–479 (1997).
Greff, K., Srivastava, R. K., Koutník, J., Steunebrink, B. R. & Schmidhuber, J. LSTM: A search space odyssey. IEEE Trans Neural Netw Learn Syst. 28(10), 2222–2232 (2017).
Merity, S., Keskar, N. S. & Socher, R. Regularizing and optimizing LSTM language models. Preprint at https://arxiv.org/abs/1708.02182 (2017).
Kingma, D. & Ba, J. Adam: A method for stochastic optimization. Preprint at https://arxiv.org/abs/1412.6980 (2014).
Péres, K. et al. Natural history of decline in instrumental activities of daily living performance over the 10 years preceding the clinical diagnosis of dementia: a prospective population‐based study. J Am Geriatr Soc. 56(1), 37–44 (2008).
Gamble, K. J., Boyle, P. A., Yu, L. & Bennett, D. A. Aging and financial decision making. Manage Sci. 61(11), 2603–2610 (2015).
Tabert, M. H. et al. Functional deficits in patients with mild cognitive impairment Prediction of AD. Neurology. 58(5), 758–764 (2002).
Acknowledgements
This work was supported by National Alzheimer’s Coordinating Center (NACC Proposal ID #776), USA and partially supported by IBM Faculty Awards (RDP-Qiu2016 and RDP-Qiu2017). NACC is funded by the National Institute on Aging (U01 AG016976). The NACC database is funded by NIA/NIH Grant U01 AG016976. NACC data are contributed by the NIAfunded ADCs: P30 AG019610 (PI Eric Reiman, MD), P30 AG013846 (PI Neil Kowall, MD), P50 AG008702 (PI Scott Small, MD), P50 AG025688 (PI Allan Levey, MD, PhD), P50 AG047266 (PI Todd Golde, MD, PhD), P30 AG010133 (PI Andrew Saykin, PsyD), P50 AG005146 (PI Marilyn Albert, PhD), P50 AG005134 (PI Bradley Hyman, MD, PhD), P50 AG016574 (PI Ronald Petersen, MD, PhD), P50 AG005138 (PI Mary Sano, PhD), P30 AG008051 (PI Steven Ferris, PhD), P30 AG013854 (PI M. Marsel Mesulam, MD), P30 AG008017 (PI Jeffrey Kaye, MD), P30 AG010161 (PI David Bennett, MD), P50 AG047366 (PI Victor Henderson, MD, MS), P30 AG010129 (PI Charles DeCarli, MD), P50 AG016573 (PI Frank LaFerla, PhD), P50 AG016570 (PI MarieFrancoise Chesselet, MD, PhD), P50 AG005131 (PI Douglas Galasko, MD), P50 AG023501 (PI Bruce Miller, MD), P30 AG035982 (PI Russell Swerdlow, MD), P30 AG028383 (PI Linda Van Eldik, PhD), P30 AG010124 (PI John Trojanowski, MD, PhD), P50 AG005133 (PI Oscar Lopez, MD), P50 AG005142 (PI Helena Chui, MD), P30 AG012300 (PI Roger Rosenberg, MD), P50 AG005136 (PI Thomas Montine, MD, PhD), P50 AG033514 (PI Sanjay Asthana, MD, FRCP), P50 AG005681 (PI John Morris, MD), and P50 AG047270 (PI Stephen Strittmatter, MD, PhD).
Author information
Authors and Affiliations
Contributions
R.G.Q. and T.Y.W. conceived of the study and formulated the research problem. R.G.Q. prepared the medical data. T.Y.W. designed the method and conducted the experiments. T.Y.W. and R.G.Q. analyzed model performances. T.Y.W. drafted and revised the manuscript. R.G.Q. and M.Y. revised the manuscript and provided advice. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, T., Qiu, R.G. & Yu, M. Predictive Modeling of the Progression of Alzheimer’s Disease with Recurrent Neural Networks. Sci Rep 8, 9161 (2018). https://doi.org/10.1038/s41598-018-27337-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-27337-w
This article is cited by
-
Visual acuity prediction on real-life patient data using a machine learning based multistage system
Scientific Reports (2024)
-
Cortical thickness modeling and variability in Alzheimer’s disease and frontotemporal dementia
Journal of Neurology (2024)
-
Genetic algorithm-based hybrid deep learning model for explainable Alzheimer’s disease prediction using temporal multimodal cognitive data
International Journal of Data Science and Analytics (2024)
-
Prediction of chronic kidney disease progression using recurrent neural network and electronic health records
Scientific Reports (2023)
-
Computer aided progression detection model based on optimized deep LSTM ensemble model and the fusion of multivariate time series data
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.