Abstract
Lipid transfer proteins (LTPs), a class of small, ubiquitous proteins, play critical roles in various environmental stresses. However, their precise biological functions remain unknown. Here we isolated an extracellular matrix-localised LTP, NtLTP4, from Nicotiana tabacum. The overexpression of NtLTP4 in N. tabacum enhanced resistance to salt and drought stresses. Upon exposure to high salinity, NtLTP4-overexpressing lines (OE lines) accumulated low Na+ levels. Salt-responsive genes, including Na+/H+ exchangers (NHX1) and high-affinity K+ transporter1 (HKT1), were dramatically higher in OE lines than in wild-type lines. NtLTP4 might regulate transcription levels of NHX1 and HKT1 to alleviate the toxicity of Na+. Interestingly, OE lines enhanced the tolerance of N. tabacum to drought stress by reducing the transpiration rate. Moreover, NtLTP4 could increase reactive oxygen species (ROS)-scavenging enzyme activity and expression levels to scavenge excess ROS under drought and high salinity conditions. We used a two-hybrid yeast system and screened seven putative proteins that interact with NtLTP4 in tobacco. An MAPK member, wound-induced protein kinase, was confirmed to interact with NtLTP4 via co-immunoprecipitation and a firefly luciferase complementation imaging assay. Taken together, this is the first functional analysis of NtLTP4, and proves that NtLTP4 positively regulates salt and drought stresses in N. tabacum.
Similar content being viewed by others
Introduction
Plants frequently encounter environmental stresses, such as salinity, drought, heavy metal, low temperature and microbial infections1,2. Among these stresses, salinity and drought stresses are major constraints on economic development and world food supplies3. Approximately 7% of the land area worldwide is affected by salinity and 40% is affected by drought4,5. The detrimental effects of salt and drought on plants are consequences of osmotic stress, ion imbalance and oxidative toxicity6. During the millions of years of evolution, plants have evolved to have high plasticity and elaborate mechanisms to protect themselves from salt and drought stresses. Plants prevent water loss while maximising water uptake to mitigate osmotic stress7. In addition, many protective proteins, including osmolytes, antioxidants and late embryogenesis abundant proteins, are produced and are likely to enhance stress resistance8,9,10. Another strategy for attaining improved salt tolerance is helping plants re-establish ionic homeostasis by using terminal determinants, such as salt overly sensitive (SOS) pathway components, Na+/H+ antiporter (NHX1) and high-affinity K+ transporter (HKT1)11. The stress-related molecular regulatory network is sophisticated and mostly unexplored. Thus, studying the key and novel genes that regulate salt or drought stress to cultivate the new varieties has become increasingly important for modern agriculture.
Lipid transfer proteins (LTPs) are extensively responsive to abiotic and biotic stresses12. Plant LTPs are a conserved class of small (7–10 kDa), abundant, ubiquitous and mostly basic proteins (isoelectric point usually 8.5–12). All examples of LTPs contain eight cysteine residues (C…C…CC…CXC…C…C) that form four conserved disulfide bridges that stabilise a bundle of four alpha-helices13. This bundle forms a hydrophobic pocket with a unique plasticity to accommodate a fatty acid, phospholipids or acyl-CoA14,15. Thus, they can exchange lipids between membranes in vitro16.
LTPs are divided into two families based on polypeptide length of mature protein. The first family of LTPs, named type I-LTP, possesses molecular masses of approximately 10 kDa and contains a signal peptide that is generally between 21–27 amino acids at the N-terminus16. The second family (type II-LTP) consists of peptides that possess molecular masses of approximately 7 kDa and contains a signal peptide between 27–35 amino acids17. The importance of these small proteins in plant growth and development has become increasingly clear, such as in cutin synthesis16, post-meiotic anther development18, and pathogen defence19,20. Moreover, LTPs are involved in abiotic stresses, the transcriptions of CaLTP1 (Capsicum annuum L) and TdLTP4 (Triticum turgidum L) are induced by ABA, ethylene, methyl jasmonic acid (MeJA) and salicylic acid (SA)21,22; Moreover, ZmLTP3 and AtLTP3 are positively regulated in salt and drought stresses, respectively23,24. Interestingly, LTP in tree tobacco increases cuticular wax accumulation to enhance drought resistance in a non-specific mechanism25. However, the molecular mechanism of LTPs involved in abiotic stress remains unclear.
Given their important roles in abiotic stress responses, LTPs have been studied in many plant species, including Arabidopsis (Arabidopsis thaliana), pepper (Capsicum annuum L), wheat (Triticum turgidum L) and maize (Zea mays)21,22,23,24. In the economic crop tobacco (Nicotiana tabacum), only NtLTP1 and NtLTP2 have been functionally characterised, and these are involved in plant defence responses and cell wall loosening mediation, respectively19,26. Therefore, illuminating the specific functions of other LTPs in tobacco is important.
In this study, NtLTP4 was cloned from N. tabacum, which was dramatically induced by various stress treatments. Overexpression of NtLTP4 in tobacco plants significantly enhanced salt and drought tolerance, which were caused by dramatically improved activities of reactive oxygen species (ROS)-scavenging enzymes. The analysis of the Na+ content further clarified the role of NtLTP4 by partially regulating the transcription levels of NHX1 and HKT1 to alleviate the toxicity of Na+. Moreover, NtLTP4 interacted with the tobacco MAPK wound-induced protein kinase (WIPK), which was associated with abiotic and biotic stress responses in tobaccoo27,28. Together, these data show the novel mechanism by which NtLTP4 positively regulates salt and drought stresses.
Results
Characterisation of NtLTP4
The full-length cDNA of NtLTP4 was isolated from N. tabacum (AB625595.1). NtLTP4 contained a 354 bp open-reading frame (ORF) and encoded a 117-amino acid polypeptide. Blast analyses showed that NtLTP4 shared approximately 50% identity with LTP proteins of other plants (Fig. 1a). Similar to other LTPs, NtLTP4 had a highly conserved motif that contained pentapeptides (T/S-X-X-D-R/K) or (P-Y-X-I-S). Furthermore, NtLTP4 possessed a unique and conserved eight-cysteine motif (29C-39C-53C-54C-74C-76C-99C-113C) that appeared to be a structural scaffold of conserved helical regions connected by variable loops (Fig. 1b). Moreover, NtLTP4 was predicted to possess a signal peptide at the N-terminus, which might be indispensable for localisation and exact function (Fig. 1c). A phylogenetic tree was constructed based on the amino acid sequence alignment of N. tabacum LTP4 with other LTPs (Fig. 1d). The proposed 3D structural model of NtLTP4 indicated a characteristic globular shape of orthogonal four a-helices and a hydrophobic core as typical LTPs in plants (Fig. 1b).
Characterisation of NtLTP4. (a) Amino acid sequence alignment of N. tabacum NtLTP4 with other LTPs. The sequence of NtLTP4 was 42% identity with the LTP of Arabidopsis thaliana, 55% identity with T. durum, 47% identity with Zea mays and 52% identity with Hordeum vulgare. At, A. thaliana (At5g59310); Td, T. durum (ABO28527.1); Zm, Z. mays (NP_001105392.1); Hv, H. vulgare (CAA85484.1). The conserved cysteine residues were highlighted with asterisks. Conserved and semi-conserved amino acid residues were represented by different colours. The consensus pentapeptides were highlighted in the red box. (b) Amino acid sequence and ribbon model of NtLTP4 structure showing the location of four pairs of disulfide bonds indicated by connecting lines. 3-D structural model of NtLTP4 predicted by SWISS-MODLE. (c) Predicted signal peptide of NtLTP4 (http://www.cbs.dtu.dk/services/SignalP/). (d) Phylogenetic tree of N. tabacum NtLTP4, highlighted by red circle. Nt, N. tabacum (AB625593.1, AB518680.1, AB625594.1); Td, T. durum (JF799976.1); Ta, Triticum aestivum (AJ852557.1, AJ852549.1, AJ852552.1); Hv, H. vulgare (CAA85484, CAA42832); Os, Oryza sativa (AY466108.1, AY466109.1); Zm, Z. mays (ACG30536.1); Si, Setaria italica (LN810550.1); At, A. thaliana (At2g38540, At5g59320, At5g59310, etc.); Ca, C. annuum (AF118131.1); Ng, Nicotiana glauca (AF151214.1); Sb, Sorghum bicolor (X71667.1, X71668.1, XP_002458765.1).
Expression pattern analysis of NtLTP4
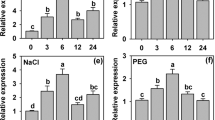
NtLTP4 was highly expressed in leaves and weakly expressed in roots, as observed by quantitative real-time polymerase chain reaction (qRT-PCR), which was similar to previous studies in 201219 (Fig. 2a). The transcription levels of NtLTP4 were measured after treatments representing drought (mannitol), salt (NaCl), wounding and Rastonia. solanacearum; and Murashige & Skoog (MS) treatment was used as control (Fig. 2b). A 12-fold induction of NtLTP4 was observed at 6 h post-treatment with NaCl, and a 7.2-fold induction was observed with mannitol at 8 h (Fig. 2c,d). Furthermore, wounding and infection with R. solanacearum strongly induced the transcription of NtLTP4 (Fig. 2f,i). In response to ABA, the NtLTP4 transcription levels were strongly increased to 25-fold (Fig. 2e). Moreover, the NtLTP4 transcription levels were enhanced by SA and JA by 5.7 and 4.3 times at 6 and 2 h, respectively (Fig. 2g,h). The rapid induction of NtLTP4 under various stress treatments reveals that NtLTP4 may be vital in responses to biotic or abiotic stresses in N. tabacum.
Expression pattern of NtLTP4. (a) Tissue-specific expressions of NtLTP4 in roots, leaves and stems were quantified by qRT-PCR. (b) Total RNA extracted from the whole plants of 14-day-old N. tabacum without treatment and used for qRT-PCR. (c–i) Total RNA extracted from the whole plants of 14-day-old N. tabacum exposed to 200 mM NaCl, 200 mM mannitol, 100 μM ABA, wounding, 5 μM SA, 500 μM MeJA and R. solanacearum were used for qRT-PCR, respectively. Lanes represented the various durations of treatments, the internal control gene was Actin. Experiment was repeated three times. Error bars indicated SEM. One-way ANOVA (Duncan’s multiple range test) was performed, and statistical significant differences were indicated by different lowercase letters (P < 0.01).
NtLTP4 was localised to extracellular matrix
To investigate the subcellular localisation and potential biological functions of NtLTP4 in plants, the full-length ORF was cloned into the binary vector pROKII-GFP and resulted in translational fusion with green fluorescent protein (GFP) (Fig. 3a). The transient expressions of GFP-fused NtLTP4 proteins were analysed in Agrobacterium-infiltrated Nicotiana benthamiana leaves. Confocal fluorescence imaging revealed that NtLTP4-GFP fusion was primarily observed at cellular boundary (Fig. 3b). Subsequently, to examine whether the fluorescence derived from NtLTP4-GFP remained in the plasma membrane or was secreted to the cell wall, Agrobacterium that contained 35S::NtLTP4-GFP or 35S::GFP plasmids were introduced into onion epidermal cells. The onion epidermal cells that carried the 35S::NtLTP4-GFP plasmid emitted fluorescence only in the peripheral cell layers upon plasmolysis (Fig. 3c,d). The location can be explained by the presence of a potential signal peptide in NtLTP4, as predicted in Fig. 1c, that drives LTPs into the extracellular matrix. This result may be linked to the cell-to-cell signal transduction function.
Subcellular localization analysis of the NtLTP4 in N. benthamiana leaves and onion epidermal cells. (a) Schematic illustration of the recombinant vector (35S::NtLTP4-GFP) and the control construct (35S::GFP). (b) Transient expression of NtLTP4-GFP and GFP in N. benthamiana leaves. Bars = 20 μm. (c) Transient expression of NtLTP4-GFP and GFP constructs in onion epidermal cells. GFP fluorescence visualised using fluorescence microscopy. Bar = 20 μm. (d) Images showing the magnified regions were indicated by red boxes in (c). Bar = 5 μm. Ten independent materials were analysed. The experiment was repeated with three biological replicates.
Overexpression of NtLTP4 enhanced salt tolerance in transgenic tobacco and yeast
NtLTP4 was highly induced by various abiotic stresses, thereby suggesting its role in multiple stress responses, especially to salt stress. To understand further the molecular mechanisms underlying the stress responses of NtLTP4, we produced NtLTP4-overexpressing transgenic tobacco plants. qRT-PCR analysis was conducted to detect the expression levels of NtLTP4 in different overexpressing lines (OE) (Supplementary Fig. S1). The transgenic lines with different expression levels were shown in Supplementary Fig. S1, and OE1 (high), OE2 (medium) and OE3 (low) were chosen for in-depth physiological analysis.
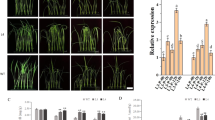
To investigate possible effects on salt stress, wild-type (WT) and OE lines on MS agar medium were germinated with or without NaCl. No obvious differences were observed between WT and OE lines under standard growth conditions, as illustrated in Fig. 4a,b. However, the germination of OE lines was more insensitive than that of WT upon exposure to high salinity. Only 50% of WT seeds were germinated; however, 55% of OE3, 65% of OE2 and 80% of OE1 seeds were germinated in 200 mM NaCl (Fig. 4b). Moreover, we tested whether NtLTP4 played a role in seedling establishment. N. tabacum seedlings were transferred to MS medium with or without NaCl. The WT showed significantly enhanced growth inhibition compared with those of OE lines (Fig. 4c). The primary root lengths of OE1, OE2 and OE3 lines were increased by 8, 7 and 4 mm, respectively, compared with those of WT in the presence of 200 mM NaCl (Fig. 4c,d). In addition, transgenic seedlings with NtLTP4 indicated up to 18–50% significantly higher fresh weight than the wild-type at 200 mM NaCl (Fig. 4d). To study further the function of NtLTP4 in the vegetative stage, 8-week-old WT and OE plants were irrigated with salt solution for 1 month. The WT plants indicated severe growth retardation, leaf curling and chlorosis compared with the OE lines (Fig. 4e). Furthermore, approximately 48–75% of OE plants survived under high salinity conditions, whereas only 40% of WT plants survived (Fig. 4e).
Overexpression of NtLTP4 in transgenic plants enhanced salt tolerance. (a) Seeds sown on MS medium containing various concentrations of NaCl. Bar = 1.5 cm. (b) Germination rates of WT and OE lines with or without 200 mM NaCl treatment. (c) Seedling phenotype of 14-day-old WT and OE lines with or without salt stress treatment. Bar = 1 cm. (d) Root length and fresh weight of seedlings shown in (c). (e) Phenotype and survival rates of 8-week-old WT and OE plants grew in soil irrigated with water and 250 mM NaCl solution for 1 month. Bar = 2 cm. (f) Representative phenotypes of leaf disks from detached leaves of WT and OE plants with MS supplied with or without 800 mM NaCl. Bar = 1 cm. (g) Quantification of chlorophyll contents in different plants. Chl.a = chlorophyll a. Chl.b = chlorophyll b. Data were mean values of three biological repeats. Error bars indicated SEM. One-way ANOVA (Duncan’s multiple range test) was performed, and statistical significant differences were indicated by different lowercase letters (P < 0.01).
As shown in Fig. 4f, no deformities were observed in all MS-treated plants for 3 d. The leaf disks of WT plants indicted severe signs of bleaching or chlorosis compared with those of OE lines after the leaf disks were floated in salt solution. This result was further confirmed by measuring the leaf disk chlorophyll contents with or without NaCl treatment (Fig. 4g).
A yeast system was used to further verify the function of NtLTP4 in response to salt stress, and pYES2-NtLTP4, pYES2-NtLTP427–118,aa (truncated without signal peptide) and pYES2 empty vectors were transformed into yeast (Gold Yeast, Clontech). The corresponding transformants were spotted on YPDA solid medium with or without NaCl. All the transformants indicated severe growth inhibition on the YPDA medium with NaCl (Supplementary Fig. 2a). The transgenic yeast that overexpressed NtLTP4 exhibited enhanced salt tolerance resistance; however, the pYES2-NtLTP427–118,aa transformants indicated same growth as the empty vector control (Supplementary Fig. 2a). Then, the transgenic strains were inoculated into the YPDA liquid medium that contained different NaCl concentrations, and absorbance of the stock cultures were determined 48 h later. The absorbance of the yeast strain that contained NtLTP4 was higher than those of the NtLTP427–118,aa and empty vector control (Supplementary Fig. 2b). In summary, NtLTP4 enhanced the salt stress tolerance in yeast, and the potential signal peptide might be indispensable to the involvement of NtLTP4 genes in abiotic stress responses.
These results indicate that NtLTP4 overexpression in tobacco and yeast increase salt stress tolerance.
NtLTP4 regulated Na+ homeostasis by HKT1 and NHX1
To understand the physiological role of NtLTP4 in salt stress response, CoroNa™ Green dye, a green fluorescent indicator of Na+, was used to specifically measure the Na+ contents in 14-day-old seedlings. Confocal microscopy revealed intense fluorescence signals in leaves and root tips of WT lines compared with those in transgenic plants after salt treatment, and the fluorescence in leaves of OE lines was barely detectable (Fig. 5a,b). To confirm the above results, Na+ levels in the shoots and roots were measured separately using atomic absorption spectrometry. OE lines contained lower concentrations of Na+ than WT plants treated with 250 mM NaCl, and statistically significant differences were observed mainly in the shoots (Fig. 5c). However, the K+ contents remained fairly similar among the Arabidopsis samples with or without salt treatment (Fig. 5c). Therefore, the salt-tolerant phenotype of OE lines may be caused by the reduced levels of Na+ contents, thereby suggesting that NtLTP4 may regulate Na+ homeostasis in plants.
NtLTP4 regulated Na+ homeostasis by controlling the expression levels of HKT1 and NHX1. (a) Na+ contents in 14-day-old seedling shoots and roots of WT and OE lines treated with MS or 200 mM NaCl for 48 h exhibited by CoroNa™ Green dye. Green-fluorescence detected by confocal microscopy. Ten independent plants used in detection. Bar = 40 μm. (b) Na+ content quantitatively analysed by ImageJ. (c) Contents of Na+ and K+ in shoots and roots of 8-week-old plants irrigated with water or 250 mM NaCl for one month and measured by atomic absorption spectrometry. (d) Relative expression analysis of NHX1 and HKT1 in 14-day-old WT and OE lines treated with MS or 200 mM NaCl for 48 h. Data as mean values of three biological repeats. Error bars indicated SEM. One-way ANOVA (Duncan’s multiple range test) was performed, and statistical significant differences were indicated by different lowercase letters (P < 0.01).
Na+ homeostasis is mainly mediated by Na+ transporters, including salt overly sensitive 1 (SOS1), SOS2, SOS3, Na+ transporter HKT1 and vacuolar membrane Na+/H+ exchangers NHX1. In addition, vacuolar membrane H+-pyrophosphatase (VP), vacuolar-type pyrophosphate-energised membrane proton pump (MVP) and high-affinity K+ transporters (HAK) are implicated in salt stress1. The sequences of salt stress-related Na+ transporters from N. tabacum genome website (https://solgenomics.net/organism/Nicotiana_tabacum/genome) were used to design primers for qRT-PCR assay. No obvious differences were observed in the expression levels of NtSOS1, NtSOS2, NtSOS3, NtVP1, NtHAK1 and NtHAK8 between OE and WT lines (Fig. 5d; Supplementary Fig. S3). Under saline conditions, the expression levels of NtHKT1 and NtNHX1 in OE lines were relatively higher compared with those in WT plants, and these transporters have been reported to be vital salt-tolerance determinants (Fig. 5d). Arabidopsis plants that overexpress HKT1 and NHX1 have been reported to enhance salt tolerance29,30. Therefore, NtLTP4 plays a role in tobacco at least partially by indirectly regulating the transcription of transporters, such as NtHKT1 and NtNHX1, to maintain Na+ homeostasis.
Overexpression of NtLTP4 enhanced drought tolerance in transgenic tobacco and yeast
To further investigate whether NtLTP4 functions in drought stress, seeds of WT and OE lines were evaluated by using MS medium supplemented with exogenous mannitol and PEG (Fig. 6a; Supplementary Fig. S4). The germination rate of WT plants was approximately 45% and those of OE lines were approximately 52–78% (OE1 78%, OE2 65%, OE3 52%) in the presence of 200 mM mannitol (Fig. 6b). In order to explore whether NtLTP4 functions at the seedling stage, root growth inhibition and fresh weight were measured on vertical growth plates with different concentrations of mannitol. Three OE lines indicated enhanced drought stress resistance compared with WT plants, regardless of root elongation or fresh weight (Fig. 6c,d). Moreover, similar phenotypes were observed on PEG treatment at germination and seedling stage (Supplementary Fig. S4).
Overexpression of NtLTP4 in transgenic plants enhanced drought tolerance. (a) Seed germination on MS medium containing various concentrations of mannitol. Bar = 1 cm. (b) Germination rates of WT and OE lines in MS medium with or without 200 mM mannitol. (c) Seedling phenotype of 14-day-old WT and OE lines with or without mannitol treatment. Bar = 1 cm. (d) Root length and fresh weight of seedlings shown in (c). (e) Phenotype and survival rate of 8-week-old WT and OE plants grew in soil with or without dehydration treatment for 15 days. Bar = 2 cm. (f) Photosynthetic performances of WT and OE plants with or without drought treatment. Pn: net photosynthetic rate (Pn, μmol CO2 m−2s−1) Gs: stomatal conductance (gs, mmol H2O m−2s−1) E: transpiration rate (E, mmol H2O m−2s−1). Data as mean values of three biological repeats. Error bars indicated SEM. One-way ANOVA (Duncan′s multiple range test) was performed, and statistical significant differences were indicated by different lowercase letters (P < 0.01).
To further assess the effect of drought stress on the growth of NtLTP4 transgenic lines during vegetative growth stage, 8-week-old WT and OE plants were grown with or without water for another 15 d. The WT plants showed worse leaf wilting and chlorosis than OE plants under drought treatment (Fig. 6e). Upon re-watering, the survival rate of transgenic plants reached approximately 55–80%, whereas WT plants showed a survival rate of 50% (Fig. 6e). Additionally, we evaluated the effects of drought stress on photosynthetic gas exchange parameters. As shown in Fig. 6f, no significant differences were observed in net photosynthesis rates (Pn) between transgenic and WT plants under standard conditions. Drought stress resulted in marked decrease in Pn in transgenic and WT plants, whereas this value was maintained at a higher level in OE lines than in WT under drought stress (Fig. 6f). The transpiration rates (Tr) of the transgenic and WT plants decreased after drought treatment, whereas the OE lines indicated lower transpiration rates than in WT plants (Fig. 6f). Similar results were observed for stomatal conductance (Fig. 6f). These results indicated that the NtLTP4-overexpressing lines were more tolerant to drought stress than WT plants secondary to regulation of transpiration rates and stomatal conductance.
The above results, in which overexpression of NtLTP4 enhanced drought tolerance, were confirmed in the yeast system treated with mannitol instead of NaCl, as described in result 4 (Supplementary Fig. S2). These data indicate that transgenic tobacco plants and yeast strains that overexpressed NtLTP4 demonstrate enhanced drought tolerance when subject to deficient water supply.
Overexpression of NtLTP4 enhanced tolerance to oxidative stress in transgenic plants
Abiotic stresses generally enhance the production of cellular ROS and cause oxidative damage31. To investigate whether NtLTP4 regulates ROS accumulation, 3,3′- diaminobenzidine (DAB) and nitro blue tetrazolium (NBT) staining were applied to evaluate the levels of two prominent ROS species, namely, H2O2 and O2−, in leaves that were detached from WT and transgenic plants after being subjected to salt and drought stresses. As shown in Fig. 7a,b, no difference was observed between OE and WT lines under normal conditions. However, DAB and NBT staining were stronger in WT than in OE lines after salt and drought treatments, and accumulation of H2O2 and O2− in WT plants were significantly elevated (Fig. 7a,b). Malondialdehyde (MDA) is the final product of lipid peroxidation, and the MDA contents in OE lines were lower than in WT plants, thereby indicating that the transgenic plants suffered slighter membrane damage (Fig. 7c).
Overexpression of NtLTP4 in transgenic plants decreased the ROS production and oxidative damage under salt and drought treatments. (a) Photographs of NBT staining and O2− concentrations measured by kit in WT and OE plants after drought and salt treatments. Bar = 2 cm. (b) Photographs of DAB staining and H2O2 concentrations measured by kit in WT and OE plants after drought and salt treatments. Bar = 2 cm. (c) MDA contents in WT and OE plants after salt and drought treatments. (d) SOD, POD and CAT activities measured by corresponding kit, respectively. (e) The expression levels of ROS-scavenging or ROS-producing genes APX, CAT, GST, SOD, RbohA and RbohB in WT and OE lines. Data as mean values of three biological repeats. Error bars indicated SEM. One-way ANOVA (Duncan′s multiple range test) was performed, and statistical significant differences were indicated by different lowercase letters (P < 0.01).
Therefore, plants evolve antioxidant systems for regulating intracellular ROS homeostasis31,32. The activities of three prominent antioxidant enzymes, namely, superoxide dismutase (SOD), peroxidase (POD) and catalase (CAT), were monitored before and after treatment, and no differences were found under normal growth conditions. After salt and drought treatments, SOD, CAT and POD activities were significantly higher in OE than in WT plants (Fig. 7d). Furthermore, the mRNA levels of several important ROS-scavenging enzymes encoding genes, such as SOD, APX, CAT and glutathione S-transferase (GST) and ROS-generating related gene, including respiratory burst oxidase homolog genes (RbohA and RbohB) were determined by qRT-PCR analysis. After salt and drought treatments, the expression levels of SOD, APX, CAT and GST dramatically increased in OE lines to levels higher than those in WT plants (Fig. 7e). By contrast, the expression levels of RbohA and RbohB were markedly decreased (Fig. 7e). These results suggest that NtLTP4 may improve ROS scavenging and suppress ROS production to relieve ROS toxicity caused by salt and drought stresses.
NtLTP4 interacted with WIPK
To further determine the mechanism by which NtLTP4 positively regulated salt and drought stresses in tobacco, we initially used yeast two-hybrid screening assay to investigate the candidate proteins that physically interact with NtLTP4 (Fig. 8a,b). Bait fusion vector (pGBKT7-NtLTP4) was constructed and transformed into the yeast strain Yeast Gold. Western Blot analysis with Myc monoclonal antibody was conducted to confirm the fusion protein of 14.6 kDa (11 kDa NtLTP4 + 3.6 kDa Myc fusion = 14.6 kDa) in yeast (Fig. 8a). We identified seven putative interactors, and stress-related WIPK was selected for further study (Supplementary Table S1).
Interaction of NtLTP4 with WIPK. (a) Detection of pGBKT7-NtLTP4-Myc bait fusion protein via Western Blot. Lane 1: pGBKT7 empty insert (negative control), lane 2: NtLTP4-pGBKT7-Myc. (b) Yeast two-hybrid screening assay was conducted in yeast. pGBKT7-p53 and pGADT7-T were used as positive control, and pGBKT7-pLam and pGADT7-T were used as negative control. (c) NtLTP4 interacted with WIPK by yeast-two-hybrid. Bait fusion vector and prey fusion vector pGBKT7-NtLTP4/pGADT7-WIPK and pGBKT7-WIPK/pGADT7-NtLTP4 were transformed into the yeast strain Gold Yeast. (d) NtLTP4 interacted with WIPK by LCI assay. Bar = 2 cm, (e) The interaction between NtLTP4 and WIPK was tested via co-immunoprecipitation (Co-IP) assay of transiently overexpressed WIPK-HA and NtLTP4-GFP in N. benthamiana leaves. N. benthamiana leaves that co-expressed GFP and WIPK-HA were used as negative controls.
The interaction between NtLTP4 and WIPK in yeast was confirmed by yeast two-hybrid assay (Fig. 8c). Then, the possible interaction between NtLTP4 and WIPK was further tested via a firefly luciferase complementation imaging (LCI) assay. As shown in Fig. 8d, coexpression of cLUC-WIPK and NtLTP4-nLUC or cLUC-NtLTP4 and WIPK-nLUC in N. benthamiana leaves strongly complemented the LUC activity compared to the negative control. Moreover, co-immunoprecipitation (Co-IP) assay of transiently overexpressed WIPK-HA and NtLTP4-GFP in N. benthamiana leaves was also performed. Total proteins were extracted and immunoprecipitated by using specific GFP-bead mixture, and then the proteins were analysed by Western Blot assay using an HA antibody. The results showed that WIPK was immunoprecipitated with NtLTP4 but not GFP (Fig. 8e). WIPK has been reported to be related to abiotic and biotic stress responses in previous studies27,33. Given that NtLTP4 is capable of interacting with WIPK, we conclude that NtLTP4 functions in salt and drought stress is associated with WIPK.
Discussion
LTPs are abundant, small and lipid-binding proteins that are involved in various physiological processes in plants, including cutin synthesis, somatic embryogenesis, post-meiotic anther development, pathogen defence and long-distance signalling of systemic acquired resistance (SAR)16,18,19,20,34,35. Moreover, LTPs can be implicated in abiotic stresses in various species, such as drought, cold and salt stresses25,36,37. In the present study, we identified a novel LTP protein from tobacco, NtLTP4, which was dramatically induced and upregulated rapidly by various abiotic stresses (Fig. 2). Furthermore, the NtLTP4-overexpressing tobacco plants enhanced tolerance to high salinity and drought stresses (Figs 4 and 6). The Na+ content analysis assay further clarified the role of NtLTP4 by partially regulating the transcription levels of NHX1 and HKT1 to alleviate the toxicity of Na+ (Fig. 5). In addition, NtLTP4 could reduce transpiration rate to reduce water loss rate (Fig. 6f). Interestingly, NtLTP4 was found to interact with stress-related MAPK, WIPK in plants (Fig. 8). Thus, the hyposensitivity phenotypes of NtLTP4-overexpressing tobacco plants might be associated with WIPK-mediated stress response.
Plants that expressed a mutant form of Defective in induced resistance 1 (DIR1), an apoplastic LTP, demonstrate normal local resistance to pathogens but fail to develop SAR or express pathogenesis-related proteins in their systemic leaves35. This observation suggests that DIR1 may function as a mobile SAR signal that transmits the information from inoculated leaves to distant leaves35. Moreover, induced SAR in grafted scion leaves reveals that CALTP also plays a role in long-distance systemic signalling in plants38. In our study, NtLTP4 was localised to the extracellular matrix (Fig. 3c), thereby suggesting that the stress-response LTP might also act as signal molecular involved in long-distance signalling transduction in tobacco. NtLTP4 was highly expressed in leaves and stems but weakly expressed in roots (Fig. 2a). However, Na+ was absorbed in large amounts by roots, and NtLTP4 might be transported from shoot to root via the phloem for roles in the latter. Future studies will test this hypothesis via grafted scion assay.
High salinity is a serious factor that seriously reduces crop yield, thereby inducing Na+/K+ imbalance and ion toxic effects. Plants have evolved high plasticity and elaborate mechanisms to adapt to ionic stress typically involve the removal of Na+ from root via the SOS1 Na+/H+ antiporter, the removal of Na+ from transpiration stream by HKT transporters and the vacuolar sequestration of Na+ via NHX1 tonoplast antiporter39. Na+ accumulation was lower in OE lines compared with those of WT lines after salt treatment (Fig. 5a–c), thereby suggesting that NtLTP4 could maintain Na+ homeostasis in plants. The expression levels of SOS1, SOS2, SOS3, HAK1, HAK8, MVP3 and VP1 were almost the same in OE lines and WT with or without salt treatment, whereas the OE lines obviously accumulated more transcripts of NtNHX1 and NtHKT1 (Fig. 5d; Supplementary Fig. S3). These results suggested that the salt tolerance phenotype of OE lines was not caused by the induction of Na+ efflux from root epidermis or regulation of K+ and H+ equilibrium but probably by compartmentalization of Na+ into vacuoles via NHX1 and removal of Na+ from transpiration stream by HKT1 in tobacco.
Salt and drought-induced secondary messengers activate diverse signalling proteins, including various members of MAPK family. MAPK3 is strongly associated with stress signalling, including salt and drought in Arabidopsis40. In Arabidopsis, MAPK3 seems to regulate the activity of heat shock factor HSFA4A or form complex with ZAT10, thereby conferring tolerance to salt stress41,42. Moreover, MAPK3 enhances tolerance to drought stress via drought-induced stomatal closure43. To date, LTPs have not been directly connected with MAPKs in tobacco. Previous study has implied that a LTP-related Hybrid proline-rich protein (HyPRP) acts as a direct component in the MAPK-mediated salt stress response in Arabidopsis44. In this study, NtLTP4 was detected as a direct interactor of WIPK for the first time (Fig. 8). WIPK, a homolog of MAPK3, was also reported to regulate the abiotic stresses in tobacco33. Thus, NtLTP4 that mediates salt and drought stress signalling in tobacco is at least linked with WIPK-related stress response. However, how NtLTP4-WIPK association is involved in abiotic stress remains obscure. Whether NtLTP4 can be phosphorylated by WIPK must also be tested via in vitro and in vivo kinase assays.
Tobacco LTP1-JA can bind to specific plasma membrane sites to activate plant defence, characterized as elicitin receptors45. Elicitins are small, cysteine-rich and lipid-binding proteins secreted by Phytophthora46, and they share some structural and functional properties with LTPs. The elicitin receptor has been assumed to be the allosteric calcium channel by isotopic tracing technique46,47. In addition, Ca2+ receptor, calmodulin (CaM)-binding site is found within the 69–80 amino acids of the C-terminal region of LTP1, thereby indicating that LTPs may be a new family of CaM-binding proteins that are involved in the CaM-mediated signalling pathway48,49. The Ca2+/CaM complex are evoked in response to different stimuli such as drought, salt stress, light and plant hormones, which represent intricate signalling networks involved in various abiotic stresses50. Ca2+/CaM complex have been reported to regulate the Na+ determiner NHX1, activate MAPK cascade pathway and induce stomatal closure in response to abiotic and biotic stresses51,52,53. All of the studies at least indirectly link NtLTP4 with Ca2+ signalling pathway and support the assumption that NtLTP4 functions in salt and drought stress in a Ca2+-dependent manner. Thus, we can hypothesise that salt and drought stresses elicit an increase in cytosolic Ca2+ and that CaM may detect the stress signal and then bind to NtLTP4 and evoke downstream responses in plants.
In summary, our findings provided the first evidence that NtLTP4 enhanced salt and drought tolerance in transgenic tobacco by alleviating Na+ and ROS toxic effects and regulating transpiration rate and stomatal conduction. Moreover, our study implied LTP as a direct interactor in the MAPK-mediated stress response in tobacco. Our results provided insight into the association of plant abiotic stress response and LTPs. To further understand the function of NtLTP4 in MAPK cascade pathway, we must clarify the physiological function of the WIPK-NtLTP4 complexes in tobacco.
Materials and Methods
Plant materials and growth conditions
N. tabacum seeds were germinated on MS medium and transferred to a growth chamber with a 16 h/8 h light/dark cycle at 28 °C. Plants were sprayed with 100 μM ABA, 500 μM MeJA or 5 μM SA after 14 days of growth. For the salt and drought treatments, the seedlings were transferred to MS medium with 200 mM NaCl or 200 mM mannitol. The seedlings were cut using scissors to mimic wounding and immersed into R. solanacearum suspension for biotic stress treatment. Plants grown under the normal condition were used as control.
RNA extraction and qRT-PCR assay
RNA was extracted from various issues or 14-day-old seedlings after different treatments using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions. Total RNA was digested to remove the genomic DNA and used for first-strand cDNA synthesis using reverse transcriptase (TIANGEN). qRT-PCR was performed using SYBR Premix Ex Taq (TaKaRa, Dalian, China) and CFX96TM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) to examine gene expression levels. The housekeeping gene Actin in N. tabacum was used as control. The expression levels were calculated using the 2−ΔΔCt comparative CT method. Three biological replicates were performed.
Sequence analysis
The sequence of NtLTP4 (AB625595.1) was obtained from National Center of Biotechnology Information (NCBI). The software ClustalW (1.82) from Europe Biotechnology Information was employed for the multiple sequence alignment of amino acid sequences of NtLTP4 with LTPs from various plants retrieved from NCBI. Subsequently, a phylogenetic tree was constructed by neighbour-joining (NJ) method using molecular evolutionary genetics analysis and sequence alignment tool MEGA 3.1. The reliability of the tree was measured using bootstrap analysis with 1500 trials. The theoretical calculation of molecular weight was analysed with ExPASy using Compute pI/Mw tool (http://www.expasy.ch/tools/pi_tool.html). For molecular modelling, SWISS-MODEL was used to predict the 3-D structure of NtLTP4.
Subcellular localisation of NtLTP4
To construct the 35S::NtLTP4-GFP expression plasmid, the CDS of NtLTP4 was inserted into the binary vector pROKII-GFP. Both 35S::GFP and the recombinant vectors were transferred into N. benthamiana and onion epidermal cells. The onion tissue was soaked in a 0.3% sucrose solution for 5 minutes. Fluorescence was observed using a fluorescence microscope (LSM 510 META, ZEISS, Germany). Ten independent plants were analysed. The experiment was repeated with three biological replicates.
Construction and genetic transformation
The full-length coding region of NtLTP4 was inserted into the binary vector pROKII. The vector was then electroporated into Agrobacterium tumefaciens (strain LBA4404) using a micropulser (BioRad, Hercules, CA, USA).
The A. tumefaciens-mediated leaf disc method was used to transform N. tabacum to achieve transgenic tobacco plants54. The infected leaf discs were placed in a selection medium composed of MS medium supplemented with 100 mg/L kanamycin, and transformations were confirmed using PCR. The seeds harvested from regenerated T0 plants were reproduced to produce the T1 and T2 generation plants for kanamycin resistance and confirmed as homozygotes using PCR.
Quantification of seed germination and root length
The N. tabacum seeds of each genotype were plated on MS medium containing 1% (w/v) sucrose with various concentrations of NaCl, mannitol or PEG. Seed-dotted plates were maintained in the dark at 4 °C for 3 days and transferred to a growth chamber with their germination percentages measured daily. Germination assays were done with three replicates of 120 seeds.
The 3-day-old seedlings that were sown on MS medium were transferred to MS medium with various concentrations of NaCl, mannitol or PEG. All seedlings were placed vertically in growth chamber. Root elongation and fresh weight were evaluated in the 14-day-old seedlings.
Salt stress treatment
The 8-week-old plants of WT and OE lines were irrigated with or without 250 mM NaCl solution for one month and survival rate was recorded. Roots and shoots were collected separately and enough materials were dried in an oven at 65 °C for 4 days, then were extracted with 16 M H2SO4 and heated to 300 °C for 8 hours. The Na+ and K+ contents of the supernatants were analysed by Atomic Abserption Spectrometer (Analytik Jena nevAA330) using a flame emission method.
For Na+ dying assay, the 14-day-old plants of WT and OE lines sown on MS medium were transferred to MS medium with or without 200 mM NaCl for 48 h. The CoroNa™ Green dye (Life, USA), which exhibited a fluorescence emission upon binding Na+, was used to specifically determine the contents of Na+. Seedlings were incubated in Na+ dye for 2 hours at 37 °C. Confocal laser scanning fluorescence microscopy (LSFM, Zeiss) was used to observe the fluorescence of root tips and leaves of seedlings. Ten independent plants were analysed. The experiment was repeated with three biological replicates.
Drought stress treatment
The 8-week-old seedlings of WT and OE lines were held without water for 15 days. The plants were then re-watered for 3 days and survival rate was recorded. In another experiment, 8-week-old WT and OE plants were held without water for 15 days, then the net photosynthetic rate (Pn), transpiration rate (E) and stomatal conductance (Gs) were measured by a portable photosynthetic system (CIRAS-3, PP Systems, Hitchin, UK) in the morning between 9:00–11:00. Photosynthesis was measured indoors. CO2 concentration was controlled at 380 ppm. Light intensity was controlled at 1000 μmolm−2·s−1 PPFD and relative humidity ranged from 60–70%. The temperature inside the leaf chamber was 25 °C.
Estimation of Chlorophyll Content
Leaf disc assay was performed. A hole-punch was used to punch similar-sized disks from 8-week-old control and transgenic tobacco plants. The disks were floated in MS supplemented with NaCl (0 and 800 mM) for 3 days. Furthermore, total chlorophylls were isolated from leaf discs and determined as the quantity per gram of fresh tissue weight. The experiment was repeated with three biological replicates.
Oxidative stress analyses
The 8-week-old plants of WT and OE lines were irrigated with 250 mM NaCl solution for one month, and other 8-week-old tobaccos were held without water for 15 days. Then, the leaves of WT and OE lines were detached for histochemical staining procedure. Staining was performed using DAB and NBT to detect the accumulation of H2O2 and O2−. A hydrogen peroxide test and a maleic dialdehyde assay kits (Nanjing Jiancheng Bioengineering Institute) were used to detect the H2O2 and O2− concentrations and MDA contents. The enzymes were extracted in 5 mL of extraction buffer and the protein concentrations were quantified by BCA Protein Assay Kit (Nanjing Jiancheng Bioengineering Institute). The activities of the antioxidant enzymes SOD, POD and CAT were measured by the kits from Nanjing Jiancheng Institute. Each assay was repeated three times.
Construction and generation of transgenic yeast
NtLTP4 and NtLTP427–118,aa were cloned into pYES2 vector. Afterwards, transformation into Gold Yeast cells was performed according to the manufacturer’s instruction (Clontech). The yeast transformed with empty pYES2 vector was used as control. Transgenic yeast cells with a series of dilution gradient were planted on YPDA solid medium with various concentrations of NaCl and mannitol. Photographs were taken at 30 °C for 48 h. The transgenic strains were inoculated to the liquid medium containing different concentrations of NaCl and mannitol, and the absorbance values of the stock cultures were determined 48 h later. The absorbance values were obtained and reported as the value relative to YPDA corresponded to different stock cultures in YPDA liquid medium. Each assay was repeated three times.
Yeast two-hybrid assay
Yeast transformation and screening were conducted following the manufacturer’s instructions (Clontech, USA). Yeast Gold cells were co-transformed with specific bait and prey constructs. All of the yeast transformants were grown on SD/-Leu/-Trp (DDO) and SD/-Leu/-Trp/-His/-Ade with X-a-gal (QDO) medium for selection or the interaction test.
Co-IP
For Co-IP assays, A. tumefaciens N. benthamiana leaves were harvested to extract proteins with IP buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% NP-40, 5 mM DTT, protease inhibitor cocktail) as previously described55. The leaf extracts were centrifuged to obtain a supernatant and incubated with GFP and Protein A beads (Invitrogen) for 6–8 h at 4 °C. The beads were recovered from the mixture by centrifugation and washing three times with cold washing buffer. Total protein was separated in 10% SDS-PAGE gel for Western Blot analysis by using an HA antibody (Sigma, Shanghai, China) and Pierce ECL western blot substrate (Thermo Fisher Scientific, Rockford, IL, USA).
LCI (firefly luciferase complementation imaging) assay
An LCI assay was performed following a previous study56. Different combinations of A. tumefaciens with related constructs were infiltrated into different positions in the same leaves of N. benthamiana for 60 h. Luciferin (Promega) was infiltrated into the leaves, and luciferase activity was measured with a low-light cooled CCD imaging apparatus (Lumina II, USA).
Statistical analysis
All of the experiments in this study were conducted three times. Error bars in each graph indicate the mean values ± standard error (SE) of replicates. Statistically significant differences between measurements were determined using one-way ANOVA in IBM Software version 24 (IBM, USA).
References
Zhang, S., Xu, R., Luo, X., Jiang, Z. & Shu, H. Genome-wide identification and expression analysis of MAPK and MAPKK gene family in Malus domestica. Gene 531, 377–387, https://doi.org/10.1016/j.gene.2013.07.107 (2013).
Li, P. et al. TheA UDP-glycosyltransferases UGT79B2 and 79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. The Plant journal: for cell and molecular biology, https://doi.org/10.1111/tpj.13324 (2016).
Zhou, Y. et al. Expression of Stipa purpurea SpCIPK26 in Arabidopsis thaliana Enhances Salt and Drought Tolerance and Regulates Abscisic Acid Signaling. International journal of molecular sciences 17, https://doi.org/10.3390/ijms17060966 (2016).
Rozema, J. & Flowers, T. Ecology. Crops for a salinized world. Science 322, 1478–1480, https://doi.org/10.1126/science.1168572 (2008).
Zhang, X. et al. Recent progress in drought and salt tolerance studies in Brassica crops. Breeding science 64, 60–73, https://doi.org/10.1270/jsbbs.64.60 (2014).
Belda-Palazon, B. et al. FYVE1/FREE1 Interacts with the PYL4 ABA Receptor and Mediates its Delivery to the Vacuolar Degradation Pathway. Plant Cell. https://doi.org/10.1105/tpc.16.00178 (2016).
Deinlein, U. et al. Plant salt-tolerance mechanisms. Trends in plant science 19, 371–379, https://doi.org/10.1016/j.tplants.2014.02.001 (2014).
Gnutt, D., Brylski, O., Edengeiser, E., Havenith, M. & Ebbinghaus, S. Imperfect crowding adaptation of mammalian cells towards osmotic stress and its modulation by osmolytes. Molecular bioSystems 13, 2218–2221, https://doi.org/10.1039/c7mb00432j (2017).
Jia, F. et al. Overexpression of Late Embryogenesis Abundant 14 enhances Arabidopsis salt stress tolerance. Biochemical and biophysical research communications 454, 505–511, https://doi.org/10.1016/j.bbrc.2014.10.136 (2014).
Wang, Y. et al. gamma-Aminobutyric Acid Imparts Partial Protection from Salt Stress Injury to Maize Seedlings by Improving Photosynthesis and Upregulating Osmoprotectants andAntioxidants. . Scientific reports 7, 43609, https://doi.org/10.1038/srep43609 (2017).
Zhu, J. K. Plant salt tolerance. Trends in plant science 6, 66–71 (2001).
Martin-Pedraza, L. et al. Two nonspecific lipid transfer proteins (nsLTPs) from tomato seeds are associated to severe symptoms of tomato-allergic patients. Molecular nutrition & food research 60, 1172–1182, https://doi.org/10.1002/mnfr.201500782 (2016).
Wang, N. J. et al. Construction and analysis of a plant non-specific lipid transfer protein database (nsLTPDB. BMC genomics 13(Suppl 1), S9, https://doi.org/10.1186/1471-2164-13-S1-S9 (2012).
Lee, J. Y. et al. Rice non-specific lipid transfer protein: the 1.6 A crystal structure in the unliganded state reveals a small hydrophobic cavity. Journal of molecular biology 276, 437–448, https://doi.org/10.1006/jmbi.1997.1550 (1998).
Liu, F. & Lu, C. M. [An overview of non-specific lipid transfer protein in plant]. Yi chuan = Hereditas 35, 307–314 (2013).
Han, G. W. et al. Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography. Journal of molecular biology 308, 263–278, https://doi.org/10.1006/jmbi.2001.4559 (2001).
Castro, M. S. et al. Purification and characterization of a small (7.3 kDa) putative lipid transfer protein from maize seeds. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences 794, 109–114 (2003).
Zhang, D. et al. OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant physiology 154, 149–162, https://doi.org/10.1104/pp.110.158865 (2010).
Choi, Y. E. et al. Tobacco NtLTP1, a glandular-specific lipid transfer protein, is required for lipid secretion from glandular trichomes. Plant J 70, 480–491, https://doi.org/10.1111/j.1365-313X.2011.04886.x (2012).
Kirubakaran, S. I., Begum, S. M., Ulaganathan, K. & Sakthivel, N. Characterization of a new antifungal lipid transfer protein from wheat. Plant physiology and biochemistry: PPB 46, 918–927, https://doi.org/10.1016/j.plaphy.2008.05.007 (2008).
Safi, H. et al. A wheat lipid transfer protein (TdLTP4) promotes tolerance to abiotic and biotic stress in Arabidopsis thaliana. Plant physiology and biochemistry: PPB 89, 64–75, https://doi.org/10.1016/j.plaphy.2015.02.008 (2015).
Park, C. J. et al. Induction of pepper cDNA encoding a lipid transfer protein during the resistance response to tobacco mosaic virus. Plant Mol Biol 48, 243–254 (2002).
Zou, H. W., Tian, X. H., Ma, G. H. & Li, Z. X. Isolation and Functional Analysis of ZmLTP3, a Homologue to Arabidopsis LTP3. International journal of molecular sciences 14, 5025–5035, https://doi.org/10.3390/ijms14035025 (2013).
Guo, L., Yang, H., Zhang, X. & Yang, S. Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J Exp Bot 64, 1755–1767, https://doi.org/10.1093/jxb/ert040 (2013).
Cameron, K. D., Teece, M. A. & Smart, L. B. Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant physiology 140, 176–183, https://doi.org/10.1104/pp.105.069724 (2006).
Nieuwland, J. et al. Lipid transfer proteins enhance cell wall extension in tobacco. The Plant cell 17, 2009–2019, https://doi.org/10.1105/tpc.105.032094 (2005).
Seo, S., Katou, S., Seto, H., Gomi, K. & Ohashi, Y. The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic and salicylic acids in wounded tobacco plants. The Plant journal: for cell and molecular biology 49, 899–909, https://doi.org/10.1111/j.1365-313X.2006.03003.x (2007).
Xu, J. et al. Reactive oxygen species in signalling the transcriptional activation of WIPK expression in tobacco. Plant, cell & environment 37, 1614–1625, https://doi.org/10.1111/pce.12271 (2014).
Apse, M. P., Aharon, G. S., Snedden, W. A. & Blumwald, E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258 (1999).
Davenport, R. J. et al. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ 30, 497–507, https://doi.org/10.1111/j.1365-3040.2007.01637.x (2007).
Choudhury, F. K., Rivero, R. M., Blumwald, E. & Mittler, R. Reactive oxygen species, abiotic stress and stress combination. The Plant journal: for cell and molecular biology. https://doi.org/10.1111/tpj.13299 (2016).
Mittler, R., Vanderauwera, S., Gollery, M. & Van Breusegem, F. Reactive oxygen gene network of plants. Trends in plant science 9, 490–498, https://doi.org/10.1016/j.tplants.2004.08.009 (2004).
Li, Y. et al. Rice MAPK phosphatase IBR5 negatively regulates drought stress tolerance in transgenic Nicotiana tabacum. Plant science: an international journal of experimental plant biology 188-189, 10–18, https://doi.org/10.1016/j.plantsci.2012.02.005 (2012).
Sterk, P., Booij, H., Schellekens, G. A., Van Kammen, A. & De Vries, S. C. Cell-specific expression of the carrot EP2 lipid transfer protein gene. The Plant cell 3, 907–921, https://doi.org/10.1105/tpc.3.9.907 (1991).
Maldonado, A. M., Doerner, P., Dixon, R. A., Lamb, C. J. & Cameron, R. K. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419, 399–403, https://doi.org/10.1038/nature00962 (2002).
Yubero-Serrano, E. M., Moyano, E., Medina-Escobar, N., Munoz-Blanco, J. & Caballero, J. L. Identification of a strawberry gene encoding a non-specific lipid transfer protein that responds to ABA, wounding and cold stress. Journal of experimental botany 54, 1865–1877, https://doi.org/10.1093/jxb/erg211 (2003).
Torres-Schumann, S., Godoy, J. A. & Pintor-Toro, J. A. A probable lipid transfer protein gene is induced by NaCl in stems of tomato plants. Plant molecular biology 18, 749–757 (1992).
Sarowar, S. et al. Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long-distance systemic signaling in tobacco. Plant cell reports 28, 419–427, https://doi.org/10.1007/s00299-008-0653-3 (2009).
Munns, R. & Tester, M. Mechanisms of salinity tolerance. Annual review of plant biology 59, 651–681, https://doi.org/10.1146/annurev.arplant.59.032607.092911 (2008).
Smekalova, V., Doskocilova, A., Komis, G. & Samaj, J. Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnology advances 32, 2–11, https://doi.org/10.1016/j.biotechadv.2013.07.009 (2014).
Shim, D. et al. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. The Plant cell 21, 4031–4043, https://doi.org/10.1105/tpc.109.066902 (2009).
Mittler, R. et al. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS letters 580, 6537–6542, https://doi.org/10.1016/j.febslet.2006.11.002 (2006).
Gudesblat, G. E., Iusem, N. D. & Morris, P. C. Guard cell-specific inhibition of Arabidopsis MPK3 expression causes abnormal stomatal responses to abscisic acid and hydrogen peroxide. The New phytologist 173, 713–721, https://doi.org/10.1111/j.1469-8137.2006.01953.x (2007).
Pitzschke, A., Datta, S. & Persak, H. Salt stress in Arabidopsis: lipid transfer protein AZI1 and its control by mitogen-activated protein kinase MPK3. Molecular plant 7, 722–738, https://doi.org/10.1093/mp/sst157 (2014).
Buhot, N. et al. Modulation of the biological activity of a tobacco LTP1 by lipid complexation. Molecular biology of the cell 15, 5047–5052, https://doi.org/10.1091/mbc.E04-07-0575 (2004).
Blein, J. P., Coutos-Thevenot, P., Marion, D. & Ponchet, M. From elicitins to lipid-transfer proteins: a new insight in cell signalling involved in plant defence mechanisms. Trends in plant science 7, 293–296 (2002).
Bourque, S. et al. Comparison of binding properties and early biological effects of elicitins in tobacco cells. Plant physiology 118, 1317–1326 (1998).
Wang, Z., Xie, W., Chi, F. & Li, C. Identification of non-specific lipid transfer protein-1 as a calmodulin-binding protein in Arabidopsis. FEBS letters 579, 1683–1687, https://doi.org/10.1016/j.febslet.2005.02.024 (2005).
Gao, G., Jin, L. P., Xie, K. Y. & Qu, D. Y. The potato StLTPa7 gene displays a complex Ca-associated pattern of expression during the early stage of potato-Ralstonia solanacearum interaction. Molecular plant pathology 10, 15–27, https://doi.org/10.1111/j.1364-3703.2008.00508.x (2009).
Batistic, O. & Kudla, J. Analysis of calcium signaling pathways in plants. Biochimica et biophysica acta 1820, 1283–1293, https://doi.org/10.1016/j.bbagen.2011.10.012 (2012).
Yamaguchi, T., Aharon, G. S., Sottosanto, J. B. & Blumwald, E. Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner. Proceedings of the National Academy of Sciences of the United States of America 102, 16107–16112, https://doi.org/10.1073/pnas.0504437102 (2005).
Yang, T. et al. Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant signaling & behavior 5, 991–994, https://doi.org/10.1074/jbc.M109.035659 (2010).
Xiao, Y., Chen, Y., Huang, R., Chen, J. & Wang, X. C. Depolymerization of actin cytoskeleton is involved in stomatal closure-induced by extracellular calmodulin in Arabidopsis. Science in China. Series C, Life sciences 47, 454–460 (2004).
A simple and general method for transferring genes into plants. Science 227, 1229-1231, https://doi.org/10.1126/science.227.4691.1229 (1985).
Jin, L. et al. Rice Dwarf Virus P2 Protein Hijacks Auxin Signaling by Directly Targeting the Rice OsIAA10 Protein, Enhancing Viral Infection and Disease Development. PLoS pathogens 12, e1005847, https://doi.org/10.1371/journal.ppat.1005847 (2016).
Xu, Y. et al. CYSTM, a Novel Non-secreted Cysteine-rich Peptide Family, Involved in Environmental Stresses in Arabidopsis thaliana. Plant & cell physiology, https://doi.org/10.1093/pcp/pcx202 (2017).
Acknowledgements
The present work was supported by the National Natural Science Foundation (Grant No. 31272113) and the China National Transgenic Plant Research and Commercialization Project (Grant No. 2016ZX08001–002) in China.
Author information
Authors and Affiliations
Contributions
C.-X.Z. and Y.X. conceived the original screening and research plans. Y.X., X.-X.Z., Y.-Z.S., L.-F.Z., Z.-P.Y. and L.-M.G. performed experiments. Y.X. and C.-X.Z. conceived the project and wrote the article with contributions of all the authors. S.-M.Z., H.-M.L and F.-J.W. provided suggestions and proofed the article. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, Y., Zheng, X., Song, Y. et al. NtLTP4, a lipid transfer protein that enhances salt and drought stresses tolerance in Nicotiana tabacum. Sci Rep 8, 8873 (2018). https://doi.org/10.1038/s41598-018-27274-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-27274-8
This article is cited by
-
Functional annotation of proteins in Catharanthus roseus shoot cultures under biogenic zinc nanotreatment
Plant Molecular Biology (2024)
-
Emission of floral volatiles is facilitated by cell-wall non-specific lipid transfer proteins
Nature Communications (2023)
-
Application of seaweed extracts to mitigate biotic and abiotic stresses in plants
Physiology and Molecular Biology of Plants (2023)
-
Lipid transfer proteins: structure, classification and prospects of genetic engineering for improved disease resistance in plants
Plant Cell, Tissue and Organ Culture (PCTOC) (2023)
-
Potato (Solanum tuberosum L.) non-specific lipid transfer protein StLTP6 promotes viral infection by inhibiting virus-induced RNA silencing
Planta (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.