Abstract
Increasing evidence shows that microbes are closely related to various human diseases. Obtaining a comprehensive and detailed understanding of the relationships between microbes and diseases would not only be beneficial to disease prevention, diagnosis and prognosis, but also would lead to the discovery of new drugs. However, because of a lack of data, little effort has been made to predict novel microbe-disease associations. To date, few methods have been proposed to solve the problem. In this study, we developed a new computational model based on network consistency projection to infer novel human microbe-disease associations (NCPHMDA) by integrating Gaussian interaction profile kernel similarity of microbes and diseases, and symptom-based disease similarity. NCPHMDA is a non-parametric and global network based model that combines microbe space projection and disease space projection to achieve the final prediction. Experimental results demonstrated that the integrated space projection of microbes and diseases, and symptom-based disease similarity played roles in the model performance. Cross validation frameworks and case studies further illustrated the superior predictive performance over other methods.
Similar content being viewed by others
Introduction
Joshua Lederberg systematically explained the concept of the microbiome for the first time as “Microbiome signifies the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space”1. Many microorganisms inhabit the human body, comprising mainly bacteria, archaea, viruses, fungi and protozoa2. The number of bacteria inhabiting the human body is at least 10 times more than the number of human cells3. Thus, nearly 90% of the cells in the human body are microbial cells. These microbes exist in different organs of the human body, such as the gastrointestinal tract, respiratory tract, mouth, and skin4. Over the past few decades, there has been increasing interest in microbes that inhabit the human body5. 16S rRNA gene sequencing is generally used to study these microbes6,7,8,9. Moreover, the Human Microbiome Project (HMP) has successfully described the microbes in terms of their structure, function and diversity10; its goal is to generate a comprehensive catalogue of human associated microbes11.
The relationship between the microbiome and the host is complex. The microbiome inhabiting the human body can do both good and harm to the host. On the one hand, the microbiome is conducive to developing the immune system12,13, maintaining homeostasis14, protecting against pathogens, and drug metabolism15. On the other hand, there is strong evidence that microbes are associated with various diseases, such as obesity16, diabetes17,18, asthma19, and cancer20. Therefore, a comprehensive and detailed understanding of the relationships between microbes and diseases would be not only beneficial for disease prevention, diagnosis and prognosis, but would also promote the discovery of new drugs.
Currently, certain computational methods have been proposed to study microbes and human diseases21,22. These studies aimed to predict the impact of microbes on biological events and to identify functional subnetworks in microbiome-related diseases. However, because of a lack of data, little effort has been made to study the relationships between microbes and diseases. In 2016, Ma et al. established the first Human Microbe-Disease Association Database (HMDAD) using large-scale text mining, which provides experimental data for the study of microbe-disease associations. On the basis of the database, Chen et al. developed a method called KATZ measure for Human Microbe-Disease Association prediction (KATZHMDA)23, while Huang et al. proposed a method called Path-Based Human Microbe-Disease Association prediction (PBHMDA)24. Both methods achieve satisfactory predictive results. However, the microbes in HMDAD belong to different taxonomic levels, such as phylum, class, genus and species. An entire phylum or class contains thousands of individual species; therefore using an entire phylum or class to predict novel associations is inaccurate, which would lead to optimistic claims of the model performance. Hence, to improve the reliability and accuracy of experimental data, we reprocessed the data in HMDAD and kept the known microbe-disease associations at defined taxonomic levels: genus level and species level, respectively.
In this study, we developed a computational model based on network consistency projection to infer novel human microbe-disease associations (NCPHMDA) by integrating Gaussian interaction profile kernel similarity of microbes and diseases, and symptom-based disease similarity. The most significant difference from previous models is that our model is more simple and effective without any parameters. Additionally, symptom-based disease similarity is introduced to predict human microbe-disease association for the first time. To evaluate the prediction performance of NCPHMDA, cross validation frameworks (leave-one-out and 5-fold cross validation) were implemented on two datasets: genus level dataset and species level dataset, respectively. The experimental results illustrated that symptom-based disease similarity and integrated space projection have effects on the prediction performance of the model. Moreover, the results also demonstrated that our model has a favourable advantage over the other two state-of-the-art models. Furthermore, case studies of asthma and type 2 diabetes were implemented to evaluate the predictive performance of our model. Seven and eight of the top 10 predictions for these two diseases have been confirmed by recent research, respectively. Both cross validation frameworks and case studies fully demonstrated the powerful ability of NCPHMDA to predict novel microbe-disease associations.
Results
Construction and analysis of the microbe-disease association networks
The genus level dataset includes 155 known microbe-disease associations between 94 microbes and 20 diseases, while the species level dataset contains 180 known microbe-disease associations between 147 microbes and 30 diseases. Based on these two datasets, we constructed two different microbe-disease association networks. The association network of genus level can be seen in Fig. 1, and the association network of species level is shown in Supplementary Information S1. In each heterogeneous network, the nodes denote either microbes or diseases, and the edges correspond to the associations between microbes and diseases25.
To obtain a comprehensive view of these two microbe-disease association networks, we further analysed some of their statistical characteristics26 (Table 1). The degree distributions of microbes and diseases in the microbe-disease association networks of genus level and species level are illustrated in Fig. 2 and Supplementary Information S2, respectively. The degree of a microbe node is the number of diseases associated with the given microbe. The degree of a disease node is defined similarly. On average, each microbe is associated with 1.65 and 1.22 diseases, while each disease is associated with 7.75 and 6 microbes in the microbe-disease association networks of genus level and species level, respectively.
Performance evaluation
In this study, leave-one-out cross validation (LOOCV) and 5-fold cross validation (5-fold CV) were implemented on the known microbe-disease associations to evaluate the predictive performance of NCPHMDA. In each trail of LOOCV, every known microbe-disease association was left out as test samples, while the remaining associations were taken as training samples. It should be noted that the microbe similarity and disease similarity should be recalculated in each trail. For each disease, the microbes that do not have known associations with the given disease were considered as candidate microbes. The scores of all microbe-disease pairs could be obtained by implementing NCPHMDA. We then acquired the rank of all candidate microbes. The test samples that received higher ranks than a given threshold could be regarded as correct predictions. In 5-fold CV, the known microbe-disease associations were divided randomly and equally into five subsets. For each trial, one subset was processed as test samples and the other four subsets were processed as training samples. Moreover, receiver-operating characteristic (ROC) curves were implemented by plotting the true positive rate (TPR, sensitivity) against the false positive rate (FPR, 1-specificity) at different thresholds to check the performance of the model. Sensitivity refers to the percentage of test samples that ranked higher than the given threshold, while specificity means the percentage of test samples that ranked lower than the given threshold. The area under the ROC curve (AUC) can thus be calculated to reflect the predictive performance, where an AUC value of 1 indicates perfect performance, and 0.5 indicates random performance. As a result, when using genus level dataset, our method achieved AUC values of 0.9129 based on LOOCV and 0.9108 based on 5-fold CV. When using species level dataset, our method achieved AUC values of 0.9748 based on LOOCV and 0.9782 based on 5-fold CV. These results indicated a reliable and effective predictive performance.
To evaluate the effectiveness of NCPHMDA, we tested its performance in different situations based on LOOCV. The results can be seen in Fig. 3 and Supplementary Information S3. Taking the experiment results of genus level for instance (Fig. 3), when using only Gaussian interaction profile kernel similarity, the AUC value of NCPHMDA decreased to 0.9039. This showed that integrating symptom-based disease similarity is conducive to improving the predictive performance of NCPHMDA. In addition, NCPHMDA achieved AUC values of 0.7916 and 0.7672 in disease space projection and microbe space projection, respectively. It demonstrated that integrated space projection could contribute to improve predictive performance.
Comparison with other methods
As far as we know, KATZHMDA and PBHMDA are the state-of-the-art computational models for microbe-disease association prediction. KATZHMDA calculates correlations between nodes in the heterogeneous network to predict links, which was initially proposed to solve the friend prediction problem in a social network. PBHMDA is a path-based method that applies a special depth-first search algorithm to traverse all paths between microbes and diseases. The similarities of these two methods are: they are both achieved based on a heterogeneous network, which is constructed by connecting a microbe similarity network and a disease similarity network via the known microbe-disease associations; moreover, Gaussian interaction profile kernel similarity is applied to measure microbe similarity and disease similarity in these two methods. Importantly, in NCPHMDA, besides Gaussian interaction profile kernel similarity, symptom-based disease similarity was also introduced to measure disease similarity, which could improve the predictive performance. In addition, NCPHMDA does not need any parameters, which simplifies the model and improves the computational efficiency. Moreover, NCPHMDA is still applicable in situations where there are very few known microbe-disease associations.
To further evaluate the predictive performance of NCPHMDA, using the same parameters and datasets, we compare KATZHMDA and PBHMDA with NCPHMDA based on LOOCV and 5-fold CV. The results can be seen in Fig. 4 and Supplementary Information S4. Here, we also take the results of genus level for instance (Fig. 4): in LOOCV, NCPHMDA achieved a superior performance among all the methods, with an AUC value of 0.9129, while KATZHMDA and PBHMDA yielded AUC values of 0.8584 and 0.8232, respectively; in 5-fold CV, NCPHMDA still performed well, with an AUC value of 0.9108, which was significantly better than the performance of KATZHMDA and PBHMDA, with the AUC values of 0.8394 and 0.8693, respectively.
CaseStudies
To illustrate the application of NCPHMDA to infer novel microbe-disease associations, we performed case studies of asthma and type 2 diabetes. The microbe-disease pairs that were not known to be associated in the dataset were the candidate microbe-disease associations. For each disease, the candidate microbes were ranked according to the prediction scores calculated by NCPHMDA. We used the top 10 that have been confirmed to date as the indicator to measure the predictive performance.
Asthma is a common long-term inflammatory disease of the airways of the lungs, which over the past few decades has increased noticeably in prevalence. Increasing studies showed that microbes might play important roles in the causation and exacerbation of asthma, hence in the co-morbidities due to asthma27. In the predicted list of asthma, seven microbes (genus level) ranking in the top 10 have been validated (Table 2). It has been found that Bacteriodes could be used as an early indicator of asthma later in life24. Veillonella is less represented in asthmatic patients24. A study demonstrated that the decrease of Lactobacillus could do help to prevent asthma24. Orally administered probiotic strain Bifidobacterium has a positive effect on atopic asthma28. Compared to healthy controls, there is a high level of Fusobacterium in asthmatic patients24. Research found that severe asthmatics are enriched in several taxa, with the largest fold-difference seen in a Klebsiella29. Streptococcus was found to be associated with pediatric asthma and allergic asthma30. To date, no relevant study has found that Enterococcus, Desulfovibrio and Burkholderia are related to asthma, but they could be considered as potential asthma-causing microbes.
Type 2 diabetes is a long-term metabolic disorder that is characterized by high blood sugar, insulin resistance, and relative lack of insulin. Until 2015, there were nearly 392 million people diagnosed with type 2 diabetes. In the predicted list of type 2 diabetes, eight microbes (species level) ranking in the top 10 have been validated (Table 3). It has been found that Helicobacter pylori infection is higher in diabetic obese patients than non-diabetic subjects31. Clostridum difficile infection is increasingly seen among hospitalised patients with type 2 diabetes32. A study showed that Staphylococcus aureus plays a role in the development of type 2 diabetes33. The level of Collinsella aerofaciens in type 2 diabetes group is significantly lower than those in normal glucose tolerance group34. It has been confirmed that Bacteroides vulgatus and Prevotella copri species are associated with the development of type 2 diabetes35. Glycemic level in diabetes is affected by the persistence of Porphyromonas gingivalis36. High prevalence of Escherichia coli in diabetes patients would result in high mortality37. Although Tropheryma whipplei and Bacteroides uniformis have not been proved to be associated with type 2 diabetes, their presence in the top 10 predicted list could provide direction for future research.
Discussion
Microbes play an important part in human health and disease; therefore, it is imperative that we obtain a comprehensive and detailed understanding of microbe-disease associations, and then use this knowledge to promote disease prevention, diagnosis and prognosis.
In this study, to improve the reliability and accuracy of the experimental data, we first reprocessed the data in HMDAD by keeping the known microbe-disease associations at defined taxonomic levels, then we obtained two independent datasets named genus level dataset and species level dataset. Next, we developed a computational model based on network consistency projection to infer novel microbe-disease associations. NCPHMDA is a global based method that combines microbe space projection and disease space projection to obtain the final prediction results. The important differences from previous methods are as follows: symptom-based disease similarity is introduced to integrate with Gaussian interaction profile kernel similarity to construct disease similarity; in addition, our method does not acquire any parameters, which simplifies the model and reduces the computation time. Moreover, the method is still applicable in situations where there are very few verified microbe-disease associations. NCPHMDA and cross validation frameworks (LOOCV and 5-fold CV) were implemented on the above-mentioned two datasets, respectively. The experiment results demonstrated that integrated network consistency projection and symptom-based disease similarity played roles in the predictive performance. Additionally, our method was demonstrated to be superior compared with two other state-of-the-art methods. Case studies of asthma and type 2 diabetes were implemented to illustrate the favourable performance of NCPHMDA. Taken together, the results demonstrated that NCPHMDA could be utilized as an efficient and effective model to reveal novel microbe-disease associations.
Despite the favourable results, some limitations still exist in this model. Firstly, the number of known microbe-disease associations is relatively few, which would have a negative effect on the prediction results. Studies should aim to discover more verified microbe-disease associations to expand the size of the database. Secondly, Gaussian interaction profile kernel similarities of microbes and diseases are calculated based on the known microbe-disease associations, which may cause bias towards diseases with more associated microbes and microbes with more associated diseases. Different datasets of microbes and diseases need to be integrated to reduce this bias. Thirdly, as far as we know, there is no specific standard dataset of microbe similarity; therefore, the microbe similarity network was constructed only based on known microbe-disease associations. A reasonable method needs to be developed to measure microbe similarity, which could then be applied to microbe-disease association inference in a future study.
A similar method38 was published while our manuscript was under consideration for publication. It happened that we and Bao et al.38 adopted the same method network consistency projection to predict novel human microbe-disease associations. Nevertheless, there are several important differences between these two papers. Firstly, to improve the reliability and accuracy of experimental data, we reprocessed the data in HMDAD and kept the known microbe-disease associations at defined taxonomic levels: genus level and species level, respectively. Secondly, it was the first time in this paper that symptom-based disease similarity has been introduced to integrate with Gaussian interaction profile kernel similarity of diseases to obtain the final disease similarity. Thirdly, based on two different datasets, we constructed two different microbe-disease association networks and analysed some of their statistical characteristics. As a result, experiments on these two datasets showed that our method in this paper performed better than Bao et al.’s in both LOOCV and 5-fold CV.
Methods
Dataset
The human microbe-disease association data can be retrieved from the Human Microbe-Disease Association Database (HMDAD, http://www.cuilab.cn/hmdad), which has recorded 483 verified microbe-disease associations between 39 human diseases and 292 microbes. After removing repeated microbe-disease entries and keeping the microorganisms at defined taxonomic levels, we finally acquired 155 microbe-disease associations between 94 microbes and 20 diseases at the genus level and 180 microbe-disease associations between 147 microbes and 30 diseases at the species level. In this study, they were called genus level dataset and species level dataset, respectively.
Symptom-based disease similarity data were also downloaded from HMDAD, which are calculated based on the term co-occurrence of diseases and symptoms. After converting the symptom disease into the corresponding microbe disease, we finally obtained 141 symptom similarity scores between 25 human diseases. Accordingly, there were 44 symptom similarity scores between 13 diseases in genus level dataset and 101 symptom similarity scores between 21 diseases in species level dataset.
Microbe similarity
In this study, based on the assumption that microbes that are associated with highly similar diseases tend to be more similar, Gaussian interaction profile kernel similarity was applied to measure similarities between microbes23. Firstly, we construct the adjacency matrix A of the microbe-disease association network. A(i, j) is 1, if a known association exists between disease i and microbe j; otherwise it is 0. We then defined the microbe interaction profile m(j), a binary vector denoting the presence or absence between microbe j with every disease. Actually, it is the jth column of the adjacency matrix A. As a result, Gaussian interaction profile kernel similarity between microbe j and microbe k can be calculated from their interaction profiles:
where γ m is the kernel bandwidth, which can be calculated from a new bandwidth γ′ m by the average number of associations with diseases per microbe; and nm is the number of all microbes. Here, γ′ m is simply set to 1.
Disease similarity
Symptom-based disease similarity
Symptom-based disease similarity was measured by the symptoms shown by one specific disease. The association between diseases and symptoms were quantified by term co-occurrence39,40. For each disease i and each symptom m, the quantitative strength of their association could be measured as:
where wi,m is defined as the term frequency-inverse document frequency. Wi,m denotes the co-occurrence (number of disease i and symptom m appear together). N is the number of all diseases and n m is the number of diseases appearing together with symptom m. Log(N/n m ) decreases the weights of symptoms that are generally related to many diseases and increases the weights of symptoms that are specifically related to some diseases. Then, each disease i can be represented as a vector:
where M is the number of all symptoms. Here, cosine similarity is applied to measure the symptom-based disease similarity between disease i and disease n:
The cosine similarity ranges from 0 to 1, where 0 denotes no shared symptoms between two diseases and 1 denotes these two diseases have identical symptoms.
Gaussian interaction profile kernel similarity for diseases
Similarly, based on the assumption that diseases that are associated with highly similar microbes tend to be more similar, the Gaussian interaction profile kernel similarity for diseases could be calculated. We still use the adjacency matrix A constructed above. Here, we defined the disease interaction profile d(i), a binary vector denoting the presence or absence between disease i with every microbe. Actually, it is the ith row of the adjacency matrix A. As a result, the Gaussian interaction profile kernel similarity between disease i and disease n could be calculated from their interaction profiles:
where γ d is the kernel bandwidth, which can be calculated based on a new bandwidth γ′ d by the average number of associations with microbes per disease; and nd is the number of all diseases. Similarly, γ′ d is also set to 1.
Integrated disease similarity
Based on the symptom-based disease similarity and Gaussian interaction profile kernel similarity for diseases mentioned above, the integrated disease similarity could be constructed as follows:
where DS(i, n) is the integrated similarity between disease i and disease n; SS(i, n) is the symptom-based similarity between disease i and disease n; GS(i, n) is the Gaussian interaction profile kernel similarity between disease i and disease n.
NCPHMDA
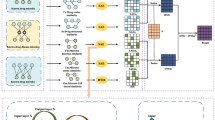
In 2016, Gu et al. proposed a method called Network Consistency Projection for miRNA-Disease Associations (NCPMDA) to reveal the potential associations between miRNAs and diseases41. Inspired by its superior performance, in this study, we developed NCPHMDA to infer novel microbe-disease associations. The flowchart of NCPHMDA is shown in Fig. 5.
Network consistency means that the spatial similarity between microbe j associated microbes in the microbe similarity network and disease i associated microbes in the microbe-disease association network (or the spatial similarity between disease i associated diseases in the disease similarity network and microbe j associated diseases in the microbe-disease association network) is positively related to the association between disease i and microbe j. We projected the microbe similarity network and disease similarity network on the microbe-disease association network, respectively, and then combined these two space projections to obtain the final network consistency projection score. Vector space projection is applied to represent this process. Microbe space projection is defined as:
where msp(i, j) is the network consistency projection of MS j on AS i . AS i is the ith row of the microbe-disease association network; actually, it is the vector encoding the associations between disease i and all microbes. MS j is the jth column of the microbe similarity network; actually, it is the vector denoting the similarities between microbe j and all microbes. |AS i | is the length of vector ASi. To avoid the denominator being 0, we use a small value δ instead of 0 in the adjacency matrix of the microbe-disease association network. Here, δ was set to 10−30.
Similarly, disease space projection can be defined as:
where dsp(i, j) is the network consistency projection of DS i on AS j . DS i is the ith row of the disease similarity network; meanwhile, it is the vector denoting the similarities between disease i and all diseases. AS j is the jth column of the microbe-disease association network; actually, it is the vector encoding the associations between microbe j and all diseases. |AS j | is the length of vector AS j .
Based on the microbe space projection and disease space projection calculated above, the final solution can thus be integrated as follows:
where ncp(i, j) is the final score of network consistency projection of disease i and microbe j. The code is available in Supplementary Information S5.
Data availability
The dataset analyzed in the study is available in the Human Microbe-Disease Association Database (HMDAD), http://www.cuilab.cn/hmdad.
References
Lederberg, J. & Mccray, A. T. ‘Ome Sweet’ Omics-A Genealogical Treasury of Words. Scientist 15, 22–27 (2001).
Sommer, F. & Bäckhed, F. The gut microbiota–masters of host development and physiology. Nature Reviews Microbiology 11, 227–238 (2013).
Savage, D. C. Microbial ecology of the gastrointestinal tract. Annual Review of Microbiology 31, 107 (1977).
Pflughoeft, K. J. & Versalovic, J. Human microbiome in health and disease. Annual Review of Pathology 7, 99–122 (2012).
Peterson, J. et al. The NIH Human Microbiome Project. Genome Research 19, 2317–2323 (2009).
Eckburg, P. B. et al. Diversity of the Human Intestinal Microbial Flora. Science 308, 1635–1638 (2005).
Palmer, C., Bik, E. M., Digiulio, D. B., Relman, D. A. & Brown, P. O. Development of the Human Infant Intestinal Microbiota. Plos Biology 5, e177 (2007).
Faveri, M. et al. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol Immunol 23, 112–118 (2008).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Human gut microbes associated with obesity. Nature 444, 1022–1023 (2011).
The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Fodor, A. A. et al. The “Most Wanted” Taxa from the Human Microbiome for Whole Genome Sequencing. Plos One 7, e41294 (2012).
Round, J. L. & Mazmanian, S. K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America 107, 12204–12209 (2010).
Gollwitzer, E. S. et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nature Medicine 20, 642–647 (2014).
Bouskra, D. et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510 (2008).
Kreth, J., Zhang, Y. & Herzberg, M. C. Streptococcal Antagonism in Oral Biofilms: Streptococcus sanguinis and Streptococcus gordonii Interference with Streptococcus mutans. Journal of Bacteriology 190, 4632–4640 (2008).
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006).
Giongo, A. et al. Toward defining the autoimmune microbiome for type 1diabetes. Isme Journal Multidisciplinary Journal of Microbial Ecology 5, 82–91 (2011).
Cai, L., Wu, H., Li, D., Zhou, K. & Zou, F. Type 2 Diabetes Biomarkers of Human Gut Microbiota Selected via Iterative Sure Independent Screening Method. Plos One 10, e0140827 (2015).
Dang, H. T. et al. Analysis of Oropharyngeal Microbiota between the Patients with Bronchial Asthma and the Non-Asthmatic Persons. Journal of Bacteriology & Virology 43, 270–278 (2013).
Schwabe, R. F. & Jobin, C. The microbiome and cancer. Nature Reviews Cancer 13, 800–812 (2013).
Coelho, E. D., Santiago, A. M., Arrais, J. P. & Oliveira, J. L. Computational methodology for predicting the landscape of the human-microbial interactome region level influence. Journal of Bioinformatics & Computational Biology 13, 1550023 (2015).
May, A. et al. metaModules identifies key functional subnetworks in microbiome-related disease. Bioinformatics 32, 1678–1685 (2016).
Chen, X., Huang, Y. A., You, Z. H., Yan, G. Y. & Wang, X. S. A novel approach based on KATZ measure to predict associations of human microbiota with non-infectious diseases. Bioinformatics 33, 733–739 (2016).
Huang, Z. A. et al. PBHMDA: Path-Based Human Microbe-Disease Association Prediction. Frontiers in Microbiology 8, 233 (2017).
Zou, Q. et al. Prediction of MicroRNA-Disease Associations Based on Social Network Analysis Methods. Biomed Research International 2015, 810514 (2015).
Sun, J. et al. Inferring novel lncRNA–disease associations based on a random walk model of a lncRNA functional similarity network. Molecular Biosystems 10, 2074–2081 (2014).
Brar, T., Nagaraj, S. & Mohapatra, S. Microbes and Asthma in the Elderly: The Missing Cellular and Molecular Links. Current Opinion in Pulmonary Medicine 18, 14–22 (2012).
Gutkowski, P. et al. Effect of orally administered probiotic strains Lactobacillus and Bifidobacterium in children with atopic asthma. Central-European Journal of Immunology 35, 233–238 (2010).
Huang, Y. J. et al. The airway microbiome in patients with severe asthma: Associations with disease features and severity. Journal of Allergy & Clinical Immunology 136, 874–884 (2015).
Shen, X. et al. Prioritizing disease-causing microbes based on random walking on the heterogeneous network. Methods 124, 120–125 (2017).
Bener, A. et al. Association between type 2 diabetes mellitus and Helicobacter pylori infection. Turkish Journal of Gastroenterology the Official Journal of Turkish Society of Gastroenterology 18, 225–229 (2007).
Hassan, S. A., Rahman, R. A., Huda, N., Wan, B. W. & Lee, Y. Y. Hospital-acquired Clostridium difficile infection among patients with type 2 diabetes mellitus in acute medical wards. Journal of the Royal College of Physicians of Edinburgh 43, 103–107 (2013).
Schlievert, P. M., Salgadopabón, W. & Klingelhutz, A. J. Does Staphylococcus aureus have a role in the development of Type 2 diabetes mellitus? Future Microbiology 10, 1549–1552 (2015).
Xiong, J. H. et al. The relationship of Atopobium cluster and Collinsella aerofaciens levels in gut microbiota with type 2 diabetes mellitus in Uygurs and Kazaks of Xinjiang. Chinese Journal of Microecology 27, 633–641 (2015).
Leite, A. Z. et al. Detection of Increased Plasma Interleukin-6 Levels and Prevalence of Prevotella copri and Bacteroides vulgatus in the Feces of Type 2 Diabetes Patients. Front Immunol 8, 1107 (2017).
Makiura, N. et al. Relationship of Porphyromonas gingivalis with glycemic level in patients with type 2 diabetes following periodontal treatment. Oral Microbiology & Immunology 23, 348–351 (2010).
Deraje, A., Shenoy, S., Dhanshree, B. & Adhikari, P. Asymptomatic bacteriuria with Escherichia coli in type 2 diabetic patients: an unresolved riddle. British Journal of Medicine & Medical Research 11, 1–9 (2016).
Bao, W., Jiang, Z. & Huang, D. S. Novel human microbe-disease association prediction using network consistency projection. Bmc Bioinformatics 18, 543 (2017).
Zhou, X., Menche, J., Barabási, A. L. & Sharma, A. Human symptoms-disease network. Nature Communications 5, 4212 (2014).
Ma, W. et al. An analysis of human microbe-disease associations. Briefings in Bioinformatics 18, 85–97 (2017).
Gu, C., Liao, B., Li, X. & Li, K. Network Consistency Projection for Human miRNA-Disease Associations Inference. Scientific Reports 6, 36054 (2016).
Vael, C., Nelen, V., Verhulst, S. L., Goossens, H. & Desager, K. N. Early intestinal Bacteroides fragilis colonisation and development of asthma. Bmc Pulmonary Medicine 8, 19 (2008).
Ege, M. J. The Hygiene Hypothesis in the Age of the Microbiome. Annals of the American Thoracic Society 14, S348–S353 (2017).
Park, H., Shin, J. W., Park, S. G. & Kim, W. Microbial communities in the upper respiratory tract of patients with asthma and chronic obstructive pulmonary disease. Plos One 9, e109710 (2014).
Arrieta, M. C. et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Science Translational Medicine 7, 307ra152 (2015).
Hevia, A. et al. Allergic Patients with Long-Term Asthma Display Low Levels of Bifidobacterium adolescentis. Plos One 11, e0147809 (2016).
Durack, J. et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. Journal of Allergy & Clinical Immunology 140, 63–75 (2016).
Hollams, E. M. et al. Vitamin D over the first decade and susceptibility to childhood allergy and asthma. Journal of Allergy & Clinical Immunology 139, 472–481 (2016).
Li, J. Z. et al. Helicobacter pylori Infection Is Associated with Type 2 Diabetes, Not Type 1 Diabetes: An Updated Meta-Analysis. Gastroenterology Research and Practice 2017, 5715403 (2017).
Devrajani, B. R., Shah, S. Z. A., Soomro, A. A. & Devrajani, T. Type 2 diabetes mellitus: a risk factor for Helicobacter pylori infection: a hospital based case-control study. International Journal of Diabetes in Developing Countries 30, 22–26 (2010).
Olanipekun, T. O., Salemi, J. L., Mc, M. D. G., Gonzalez, S. J. & Zoorob, R. J. Clostridium difficile infection in patients hospitalized with type 2 diabetes mellitus and its impact on morbidity, mortality, and the costs of inpatient care. Diabetes Research & Clinical Practice 116, 68–79 (2016).
Shakov, R., Salazar, R. S., Kagunye, S. K., Baddoura, W. J. & Debari, V. A. Diabetes mellitus as a risk factor for recurrence of Clostridium difficile infection in the acute care hospital setting. American Journal of Infection Control 39, 194–198 (2011).
Gottschalk, F. et al. Staphylococcus aureus Infections in German Patients with Type 2 Diabetes Mellitus after Orthopedic Surgery: Incidence, Risk Factors, and Clinical and Health-Economic Outcomes. Surgical Infections 18, 915–923 (2017).
Tamer, A., Karabay, O. & Ekerbicer, H. Staphylococcus aureus nasal carriage and associated factors in type 2 diabetic patients. Japanese Journal of Infectious Diseases 59, 10–14 (2006).
Yang, B. T., Xu, J. L., He, L., Meng, H. X. & Xu, L. Porphyromonas gingivalis FimA genotype distribution among periodontitis patients with type 2diabetes. Chinese journal of stomatology 51, 20–24 (2016).
Quintero, A. J. et al. Presencia de Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola y Aggregatibacter actinomycetemcomitans en el biofilm subgingival de pacientes diabéticos tipo 2: Estudio transversal. Revista Clínica De Periodoncia Implantología Y Rehabilitación Oral 4, 54–58 (2011).
Wang, F. et al. Drug resistant analysis of 87 type 2 diabetes patients complicated with urinary tract infection caused by Escherichia coli. China Tropical Medicine 15, 1228–1240 (2015).
Ye, A. L. & Li, W. Clinical features of type 2 diabetes patients complicated with urinary tract infections. Chinese Journal of Nosocomiology 23, 5943–5952 (2013).
Acknowledgements
This study is supported by the National Natural Science Foundation of China (Grant No. 61379109, M1321007) and Science and Technology Plan of Hunan Province (Grant No. 2014GK2018, 2016JC2011) and Fundamental Research Funds for the Central Universities of Central South University (Grant No. 2018zzts064).
Author information
Authors and Affiliations
Contributions
S.Z. designed and implemented the experiments, analyzed the result, and wrote the paper. J.Z. analyzed the result and wrote the paper. Z.Z. analyzed the result and wrote the paper. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zou, S., Zhang, J. & Zhang, Z. Novel human microbe-disease associations inference based on network consistency projection. Sci Rep 8, 8034 (2018). https://doi.org/10.1038/s41598-018-26448-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-26448-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.