Abstract
PRC2 is a major regulator of gene expression in eukaryotes. It catalyzes the repressive chromatin mark H3K27me3, which leads to very low expression of target genes. NRT2.1, which encodes a key root nitrate transporter in Arabidopsis, is targeted by H3K27me3, but the function of PRC2 on NRT2.1 remains unclear. Here, we demonstrate that PRC2 directly targets and down-regulates NRT2.1, but in a context of very high transcription, in nutritional conditions where this gene is one of the most highly expressed genes in the transcriptome. Indeed, the mutation of CLF, which encodes a PRC2 subunit, leads to a loss of H3K27me3 at NRT2.1 and results, exclusively under permissive conditions for NRT2.1, in a further increase in NRT2.1 expression, and specifically in tissues where NRT2.1 is normally expressed. Therefore, our data indicates that PRC2 tempers the hyperactivity of NRT2.1 in a context of very strong transcription. This reveals an original function of PRC2 in the control of the expression of a highly expressed gene in Arabidopsis.
Similar content being viewed by others
Introduction
Polycomb Repressive Complex 2 (PRC2) is a major and conserved regulatory complex of gene expression in eukaryotes. PRC2 is essential for growth and development and in both plants and animals loss-of-function of PRC2 subunits leads to serious phenotypic defects1,2. PRC2-mediated regulation of gene expression relies on the modification of chromatin state, by catalyzing the tri-methylation of Lys 27 of histone H3 (H3K27me3). Point mutation in H3K27 leads to similar phenotypes to those of PRC2 mutants, demonstrating that H3K27me3 is the main effector of PRC2-mediated regulation3. In Arabidopsis, CURLY LEAF (CLF) and SWINGER (SWN) are two different PRC2 enzymatic subunits that tri-methylate H3K27 in vegetative tissues2. CLF and SWN are thought to have overlapping functions, but the predominant contribution of CLF to H3K27me3 enrichment, as well as the more severe phenotype of clf mutant plants compared to that of swn mutant plants, suggest that CLF is the major H3K27 tri-methyltransferase during Arabidopsis vegetative development4,5. Although the molecular mechanisms by which PRC2 and H3K27me3 mediate transcriptional regulation are not fully understood, a large number of epigenomic analyses have demonstrated that H3K27me3 and PRC2 members are associated with strong repression of gene expression5,6,7,8,9. In the Arabidopsis genome, 20–25% of genes are marked by H3K27me3 and globally display low or very low expression6,7, and mutations in CLF lead to up-regulation of several hundred H3K27me3-associated genes5,10.
Many genes controlled by PRC2-mediated H3K27me3 levels in Arabidopsis correspond to genes involved in the regulation of development, and in particular transcription factors5,6,8,9,11. One of the best described examples corresponds to the repression of the FLOWERING LOCUS C (FLC) gene. FLC repression depends on H3K27me3 enrichment, and further experiments have demonstrated that FLC exists in bistable on/off expression states whether it is marked or not by H3K27me3, suggesting that H3K27me3 is a major molecular determinant of strong gene repression12,13. On the other hand, H3K27me3 levels have been also proposed to quantitatively regulate gene expression. This has been notably illustrated by the effect of mutations for PRC2 subunits in the control of the rate of induction of the VERNALIZATION INSENSITIVE 3 (VIN3) gene in response to cold treatment14.
PRC2 target loci also often correspond to genes showing tissue-specific expression. Such genes are heavily marked with H3K27me3 in the tissues where they are silent, and at the reverse largely depleted in H3K27me3 enrichment in tissues where they are normally expressed11,15. Accordingly, numerous studies have described in Arabidopsis that loss of PRC2-mediated regulation leads to an aberrant expansion of the expression territory of tissue-specific genes10,16,17,18,19. Altogether, the observations listed above led to the conclusion that PRC2 and associated H3K27me3 enrichment are strong negative transcriptional regulators ensuring the correct spatio-temporal pattern of expression of developmental genes. Nevertheless, decrease in PRC2-mediated H3K27me3 levels on target genes is not systematically associated with increase in gene expression or modifications of tissue-specific expression pattern20,21,22.
In Arabidopsis, NITRATE TRANSPORTER 2.1 (NRT2.1) encodes a key high-affinity root nitrate (NO3−) transporter, crucial for root uptake of NO3− and thus for nitrogen (N) nutrition of the plant23,24. Accordingly, nrt2.1 mutants show a dramatic reduction of growth under low and limiting NO3− availability23,25,26. In agreement with its major physiological role, the NRT2.1 gene is strongly regulated at the transcriptional level by environmental factors affecting root NO3− uptake27. In particular, NRT2.1 is very differentially expressed depending on the level of N supply, with very low expression under N-rich media, and exceptionally high expression under low and limiting NO3− availability28. In addition, NRT2.1 displays a very strict tissue-specific transcriptional profile, with expression confined to the outer layers of the root tissues29,30. It has recently been observed that NRT2.1 is marked by H3K27me36,9, indicating that PRC2 activity could be a potential determinant of the repression of NRT2.1 gene expression under N-rich condition31. In contrast to the regulation of genes involved in cell differentiation and plant development, the role of PRC2 in the regulation of environmentally-responsive and nutrition-related genes like NRT2.1 remains to be fully investigated.
To address this question, we investigated in detail the role of H3K27me3 and PRC2 in the regulation of NRT2.1 expression under both strongly repressive (high N supply) or highly inductive (low NO3− availability) conditions. We unexpectedly found that PRC2 downregulates NRT2.1 expression only in a context of very strong transcription, and specifically in tissues where NRT2.1 is highly expressed. We observed that a loss of H3K27me3 under conditions of very high expression results in a further increase in NRT2.1 promoter activity. We thus reveal here an original role for PRC2 in modulating the transcriptional level of NRT2.1 specifically under conditions where it is one of the most highly expressed genes in Arabidopsis roots.
Results
PRC2 directly regulates NRT2.1 in the context of very strong expression
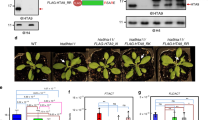
NRT2.1 is differentially expressed depending on the level of N supply, with very low expression under N-rich media, and very high expression under low and limiting NO3− availability28. To investigate the role of PRC2 in NRT2.1 regulation, we measured H3K27me3 enrichment at the NRT2.1 locus in WT and mutant lines for CLF and SWN, under highly contrasted conditions for expression, and compared it with an actively transcribed gene (ACTIN2, ACT2) or a known PRC2 target gene (LEAFY COTYLEDON 2, LEC2). Under N-rich repressive conditions, H3K27me3 enrichment at the NRT2.1 locus was indeed elevated in the roots of a WT line, and significantly reduced in clf-29 mutant but not in swn-3 mutant (Figs 1A and S1 for information about primers position). Under a NO3− limiting environment, which corresponds to the most favorable condition for NRT2.1 expression29, we surprisingly also observed a strong H3K27me3 enrichment at the NRT2.1 locus, similar to those observed for typical PRC2-controlled genes such as LEC2 (Fig. 1B). This was completely unexpected as, strikingly, NRT2.1 is ranked among the 3 most highly expressed genes in the whole Arabidopsis root transcriptome obtained under exactly the same NO3− limiting condition (Table S1). Under NO3− limitation, H3K27me3 levels at the NRT2.1 locus were also significantly diminished in clf-29, and not in swn-3 (Fig. 1B), revealing that CLF is the main methyltransferase operating at the NRT2.1 locus. In order to have a more complete view of the effect of clf mutation at the NRT2.1 locus, we screened the whole locus for H3K27me3 enrichment in WT and clf-29 lines. In agreement with published epigenomic dataset, a high H3K27me3 enrichment was observed in the NRT2.1 promoter and in the 5’ part of the gene, and was maintained throughout the whole NRT2.1 gene body (Figs 1C and S2). Reduction of H3K27me3 enrichment at the NRT2.1 locus in clf-29 was found throughout all the locus, but the extent of the reduction was maximal at the promoter region (Fig. 1C). When we measured NRT2.1 transcript levels in WT, clf-29 and swn-3 lines, we observed that decrease in H3K27me3 levels under N-rich repressive conditions did not lead to induction of NRT2.1 expression, which is, under this condition, still close to zero in mutant lines for PRC2 components (Fig. 2A). Surprisingly, and unlike under repressive N-rich condition, we observed that the reduction of H3K27me3 enrichment in clf-29 mutant under NO3− limitation led to significantly higher NRT2.1 transcripts level than in the WT line (Fig. 2A). This unexpectedly suggests that PRC2, and in particular CLF, regulates NRT2.1 in a context of very strong expression. Since we observed that the effect of clf mutation on H3K27me3 enrichment at the NRT2.1 locus was maximal at the promoter region, we crossed the clf-29 mutant line with the reporter construct ProNRT2.1:GUS29. ProNRT2.1:GUS reporter gene has been previously characterized, and faithfully transposes the transcriptional regulations targeted to NRT2.1, including N-responsiveness and tissue-specificity expression29. We compared, specifically under NO3− limitation, changes in transcript levels and H3K27me3 enrichment at the ProNRT2.1:GUS locus in WT and clf-29 plants. Under NO3− limitation, we observed in the clf-29 mutant a strong increase in GUS transcripts level (Fig. 2B). Strikingly, the induction of GUS expression in clf-29 was higher than the one of NRT2.1, again suggesting that the regulation mediated by CLF is mainly directed to the promoter activity. In agreement with these observations, we actually found a strong H3K27me3 enrichment at the GUS locus in a WT line, and a reduction of this enrichment in the clf-29 mutant (Fig. 1D). This means that the NRT2.1 promoter is able to instruct H3K27me3 enrichment to downstream sequences, and that the ProNRT2.1:GUS follows the same behavior as NRT2.1 in response to clf mutation. In order to further confirm our observations, we crossed an independent ProNRT2.1:LUC transcriptional reporter line28 with the clf-29 mutant. We observed an increase in LUC transcripts level in the clf-29 mutant as compared to the WT background, confirming our observations made on NRT2.1 and ProNRT2.1:GUS (Fig. S3).
CLF controls H3K27me3 enrichment at the NRT2.1 locus under both repressive and active conditions for expression. ChIP analysis of H3K27me3 in WT, clf-29, and swn-3 roots of 7 days-old plants grown under (A) high nitrogen (10 mM NH4NO3) or (B) low nitrate (0.3 mM NO3−) conditions. LEC2 and ACT2 served as positive or negative control for H3K27me3, respectively. Positions of primers used in qRT-PCR are available in Fig. S1. (C) ChIP analysis of H3K27me3 in WT and clf-29 covering the NRT2.1 locus. (D) ChIP analysis of H3K27me3 at the ProNRT2.1:GUS locus in WT and clf-29 roots of 7 days-old plants grown under low nitrate (0.3 mM NO3−) condition. Quantification by qRT-PCR is shown as the percentage of H3. Error bars represent standard errors of the mean based on 3 biological replicates. Statistical significance was computed using a two-tailed Student’s t-test. Significance cutoff: *p < 0.05, **p < 0.01, ***p < 0.001.
Reduction of H3K27me3 in clf-29 increases the expression of NRT2.1 in a context of very high expression. (A) Relative expression of NRT2.1 by qRT-PCR in roots of 7-days old of WT, clf-29, and swn-3 plants grown under high nitrogen (10 mM NH4NO3) or low nitrate (0.3 mM NO3−) conditions. Quantification by qRT-PCR is shown as the percentage of ACT2 transcript levels. (B) Relative GUS expression by qRT-PCR in roots of 7-days old ProNRT2.1:GUS WT and clf-29 plants grown under low nitrate (0.3 mM NO3−) condition. Quantification by qRT-PCR is shown as the percentage of ACT2 transcript levels. Error bars represent standard errors of the mean based on 3 biological replicates. Statistical significance was computed using a two-tailed Student’s t-test. Significance cutoff: *p < 0.05, **p < 0.01, ***p < 0.001.
To support the observation that CLF regulates NRT2.1 in a context of very high expression, we checked the presence of CLF at the NRT2.1 locus in this condition. We therefore performed ChIP using a ProCLF:CFP:CLF;clf-29 line17 to test whether NRT2.1 is bound by CLF. In comparison to negative and positive controls, we found that CLF indeed associates with the NRT2.1 locus (Fig. S4). As mutations in CLF lead to up-regulation of several hundred genes, we also checked that the expression of transcriptional regulators of NRT2.1 under NO3− limitation was not perturbed in clf-29. We therefore measured, in WT and clf-29 lines, transcript levels for the main transcriptional regulators of NRT2.1 that have been previously identified32. None of the NRT2.1 transcriptional regulator that we tested shows a significant de-regulation in clf-29 (Fig. S5), strongly reinforcing the idea of a direct action of CLF-PRC2 in the regulation of NRT2.1 under highly permissive condition for expression.
To further analyze the chromatin-based regulation of NRT2.1 by CLF under inductive conditions, we analyzed specifically under NO3− limitation the pattern of chromatin marks associated with transcriptional activation. In the WT, NRT2.1 was surprisingly weakly enriched in H3K4me3, H3K36me3 and H3K9ac (Fig. 3A,B and C), in spite of very high expression levels. In the clf-29 mutant, reduction of H3K27me3 level and higher transcripts level were not associated with an increase in any of the chromatin marks associated with transcriptional activation (Fig. 3A,B and C). We observed for active chromatin marks at the ProNRT2.1:GUS the same profile as the one observed at the NRT2.1 locus, except a slight increase in H3K9ac enrichment in clf-29, in agreement with a higher induction of expression for ProNRT2.1:GUS than for NRT2.1 (Fig. 3C). This suggests that reduction of H3K27me3 could be by itself the cause of the overexpression of NRT2.1. All together, these results led us to conclude that the absence of functional PRC2, and subsequent reduction in H3K27me3 levels, consequently increase NRT2.1 expression, exclusively under highly permissive NO3− limiting condition.
Reduction of H3K27me3 in clf-29 in the context of active transcription does not lead to an increase in H3K4me3, H3K36me3 or H3K9ac at the NRT2.1 locus. ChIP analysis of (A) H3K4me3, (B) H3K36me3, (C) H3K9ac in WT and clf-29 roots of 7 days-old plants grown under low nitrate (0.3 mM NO3−) condition. Quantification by qRT-PCR is shown as the percentage of H3. ACT7 served as positive for H3K4me3 and H3K9ac, ACT2 served as positive for H3K36me3, LEC2 served as negative control for H3K4me3, H3K36me3 and H3K9ac. Error bars represent standard errors of the mean based on at least 3 biological replicates. Statistical significance was computed using a two-tailed Student’s t-test. Significance cutoff: *p < 0.05, **p < 0.01, ***p < 0.001.
PRC2 modulates the expression of NRT2.1 specifically in NRT2.1-expressing tissues
PRC2 and associated H3K27me3 enrichment are strong negative transcriptional regulators, which also ensure the correct spatio-temporal expression pattern of target genes. We therefore addressed the question whether down-regulation of NRT2.1 by CLF under highly inductive conditions corresponds to transcriptional repression in tissues where NRT2.1 is not expressed, or to modulation of expression in tissues where NRT2.1 is strongly expressed. Since ProNRT2.1:GUS faithfully transposes chromatin-based regulation of NRT2.1 by CLF, we first performed transversal root sections using the WT or clf-29 lines containing the ProNRT2.1:GUS reporter grown under highly inductive NO3− limiting condition. In the WT, as previously described, we observed that NRT2.1 expression is confined, in a very strict manner, to the outer tissues of the root (cortex and epidermis) (Fig. 4A). Strikingly, NRT2.1 expression in clf-29 was similarly confined in cortex and epidermis, showing that tissue-specific expression of NRT2.1 is maintained in spite of a decrease in H3K27me3 enrichment. On the other hand, we observed in clf-29 specifically and homogeneously in every cortex or epidermis cell a strong increase in GUS staining, reflecting the overexpression ProNRT2.1 activity specifically in these tissues (Fig. 4B). Altogether, our results demonstrate that the level of H3K27me3, which has been fully characterized as a repressive chromatin mark associated with strongly repressed genes, directly modulates the expression of NRT2.1, one of the most highly expressed genes in the transcriptome under limiting NO3− availability.
Genes with very strong expression targeted by H3K27me3 are principally involved in response to stimulus, metabolism, and nutrition
Our results demonstrate an original role for PRC2 and H3K27me3 in the modulation of gene with very high expression. In order to explore the extent of this original function for PRC2, we compared the profile of highly expressed genes in the Arabidopsis root transcriptome31 with the genome-wide distribution of H3K27me3 in the roots6. We used in this case the transcriptome of plants grown under N-rich condition in order to be consistent with the conditions used to perform the epigenomic map. Among the most highly expressed genes in the transcriptome, we observed that 139 genes are targeted by H3K27me3 (Fig. 5, Table S2). This amount is obviously much lower than for poorly expressed genes, but it suggests that the regulation identified on NRT2.1 may affect a substantial number of genes. We also analyzed whether highly expressed genes marked by H3K27me3 could be down-regulated by CLF, as we observed for NRT2.1. We therefore compared the list of highly expressed genes marked by H3K27me3 with the genes regulated by CLF in Arabidopsis roots10. 9 of the 139 highly expressed genes marked by H3K27me3 were found to be regulated by CLF (Fig. 5, Table S2), which is a similar proportion to that of poorly expressed genes (61 genes regulated by CLF on 803 genes with very low expression in the roots and marked by H3K27me3). Most compelling is the finding that the functional categories of hyperactive genes targeted by H3K27me3 in Arabidopsis roots are different from those of low expression genes marked by H3K27me3 (Table S3). Indeed, the set of low expression genes targeted by H3K27me3 is principally enriched in genes involved in the regulation of development, transcription and gene expression, as previously described5,6,11. In contrast, the list of very highly expressed genes targeted by H3K27me3 shows a significant enrichment in genes involved in metabolic processes and response to diverse stimuli (Table S3). In particular, it included gene categories related to nitrate transport and assimilation, as well as several other processes linked to mineral nutrition and secondary metabolism (Table S3). Such observation lends support to the conclusion that this original regulation by PRC2 in plants could mostly affect genes that are relevant to plant physiology and to response to the environment, including those linked to an essential function like mineral nutrition.
Comparison of genes with very low or high expression showing H3K27me3 enrichment and regulation by CLF. Venn diagram representing a comparison of the proportion of genes marked by H3K27me3 and regulated by CLF among the very low expressed or very highly expressed genes. Very low expressed genes correspond to the 10 percent of genes the most poorly expressed in the transcriptome; very highly expressed genes correspond to the 10 percent of genes the most highly expressed in the transcriptome. Data of H3K27me3-marked genes in Arabidopsis roots are from6, expression data are from31, CLF-regulated genes are from10.
Discussion
NRT2.1 is a major root NO3− transporter and is essential for plant growth under limiting NO3− availability23,24,25. The molecular mechanisms that control the level of NRT2.1 expression are therefore crucial for plant growth and development. We show here that NRT2.1 expression, which is one of the highest in the transcriptome, is modulated by PRC2, a chromatin-based regulator of gene expression known to be associated with strong gene repression. Examination of NRT2.1 chromatin state reveals that this locus is targeted by H3K27me3 under both repressive and inductive conditions for gene expression, mainly directed by CLF. Decrease in H3K27me3 levels did not lead to induction of NRT2.1 expression under repressive conditions. These observations lend support to the conclusion that H3K27me3 enrichment is not the main determinant of NRT2.1 repression under N-rich condition, and are in agreement with the global view that loss of H3K27me3 alone is generally not sufficient to lead to a gain of expression21. Indeed, it is possible that induction factors specific to low N conditions are also required to activate NRT2.1 in the absence of repressive chromatin marks. Unexpectedly, in the context of very high expression, loss of H3K27me3 leads to further increase in NRT2.1 expression. This reveals a greatly unusual targeting by H3K27me3 to such a very highly expressed gene. Further increase in NRT2.1 expression is observed in NRT2.1-expressing tissues, and coincides with an absence of misregulation of known regulators of NRT2.1 expression. However, it should be noted that we cannot entirely rule out indirect effect of PRC2 loss-of-function. Notably, cell type-specific chromatin analysis in cortex and/or epidermis cells will be required to further explore the role of PRC2 in the regulation of NRT2.1 high expression. We also observed that chromatin marks associated with active transcription, which would have been expected to be strongly present at NRT2.1, were surprisingly low at this locus. A strong enrichment of H3K27me3 at NRT2.1 could explain such observation, at least for H3K36me3, which has been shown to be mutually exclusive with H3K27me333. However, a dilution of NRT2.1-expressing cells in chromatin analysis performed with whole roots may also explain the lower than expected enrichment in chromatin marks associated with active transcription at the NRT2.1 locus. Nevertheless, our results reveal an unusual chromatin state with high level of H3K27me3 and H3K4me3, and an original function for PRC2 in the regulation of the target gene.
Our results reveal that NRT2.1 promoter under highly active conditions for expression is sufficient to drive H3K27me3 targeting to downstream sequences, and show that the effect of clf mutation occurs mainly at the level of promoter activity. Moreover, the induction of expression following loss of H3K27me3 varies substantially between NRT2.1 and reporters of NRT2.1 promoter activity. Indeed, induction of ProNRT2.1:GUS expression was clearly higher than the one observed for NRT2.1 itself. This might translate additional mechanisms of transcriptional or post-transcriptional modulation targeted specifically to NRT2.1 gene body or to NRT2.1 mRNA. In addition, although regulation by CLF seems clearly directed to the NRT2.1 promoter region, a larger genomic context at the NRT2.1 locus is certainly also important for the regulation of its expression, and for the regulation by chromatin complexes. It is for instance interesting to observe that, although under repressive conditions, local chromatin interactions have been identified at the NRT2.1 locus34, suggesting that chromatin conformation may have an influence on the expression of NRT2.1. The presence of multiple mechanisms to regulate NRT2.1 is consistent, as this gene is essential for plants to survive in the majority of soil environments27.
Our analysis of previously published datasets reveals that a considerable number of genes showing very high expression also display H3K27me3 targeting. Although we cannot rule out that this overlap could be due to a combination of different cell types in which genes are either highly expressed or marked by H3K27me3, it supports the possibility that our observations made on the regulation of NRT2.1 apply to many other genes. Interestingly, genes that would be targeted by this regulation fall into specific functional categories. Most of them are directly involved in metabolic processes, including mineral nutrition. These genes may be representative of fundamental mechanisms, for which a balance between high expression and gene integrity would be essential for plant physiology. In conclusion, our work provides the first example of a totally unexpected function of PRC2 in Arabidopsis in the modulation of one of the most highly expressed gene in the transcriptome, in a context of very strong transcription. This study opens a new route for further investigation concerning the role of PRC2 in the control of the expression of highly transcribed genes.
Material and Methods
Plant material and growth conditions
The Arabidopsis thaliana accession used in this study was Col-0. Mutant alleles and transgenic plants used in this study are clf-2935, swn-34, ProNRT2.1:GUS29, ProNRT2.1:LUC28, ProCLF:CFP:CLF;clf-2917. Most of experiments were performed using roots from 7 days-old seedlings grown under a long-day photoperiod (16 h light and 8 h dark) on vertical MS/2 plates without nitrogen (PlantMedia) supplied with 0.8% agar, 0.1% of sucrose, 0.5 g/L MES and the appropriate concentration of nitrogen as described in figure legends.
RNA extraction and expression analysis
Root samples were frozen in liquid nitrogen and total RNA was extracted using TRI REAGENT (MRC), DNase treated (RQ1 Promega), and reverse transcription was achieved with M-MLV reverse transcriptase (RNase H minus, Point Mutant, Promega) using an anchored oligo(dT)20 primer. Accumulation of transcripts was measured by qRT-PCR (LightCycler 480, Roche Diagnostics) using the SYBRR Premix Ex TaqTM (TaKaRa). Gene expression was normalized using ACT2 as an internal standard. Sequences of primers used in qPCR for gene expression analysis are listed in Supplementary File 1.
ChIP experiments
ChIP experiments were performed as previously described36 with minor modifications. Nuclei were isolated using Nuclei Isolation Buffer (20 mM PIPES-KOH pH 7.6, 1 M hexylene glycol, 10 mM MgCl2, 0.1 mM EGTA, 15 mM NaCl, 60 mM KCl, 0.5% Triton X100, 5 mM beta-mercaptoethanol, protease inhibitor cocktail (complete tablets EASYpack, Roche)) and then resuspended in Nuclei Lysis Buffer. Chromatin was precipitated with 2.5 μg of antibodies against H3 (Abcam 1791), H3K27me3 (Millipore 07-449), H3K4me3 (Diagenode C15410030), H3K36me3 (Abcam 9050), H3K9ac (Agrisera AS163198). Immunoprecipitation of CFP::CLF was performed using GFP-Trap MA (Chromotek). Immunoprecipitated DNA was purified with IPURE Kit (Diagenode) and resulting DNA was analyzed by qPCR analysis. ChIP experiments were normalized using H3 level as an internal standard. Data are presented as the percentage of H3K27me3 enrichment over H3 enrichment, using the following formula: 2−(Cp IP H3K27me3 – Cp IP H3) × 100. For CFP::CLF immunoprecipitation, experiments were normalized using an INPUT (10% of sample adjusted to 100%). Data are presented as the percentage of CFP:CLF enrichment over input, using the following formula: (2−(Cp IP H3K27me3 – Cp Input) × 100)/10. Sequences of primers used in qPCR for ChIP experiments are listed in Supplementary File 1.
GUS histochemical staining and Arabidopsis root cross-section
Plants were harvested and prefixed 45 minutes at room temperature in 50 mM NaPO4 pH 7, 1.5% formaldehyde, 0.05% Triton X100. Plants were washed 3 times in 50 mM NaPO4 pH 7 before staining in 50 mM NaPO4 pH 7, 0.5 mM ferricyanide, 0.5 mM ferrocyanide, 0.05% Triton X100, 1 mM X-Gluc 30 minutes under vacuum following by 2 h incubation at 37 °C. Three other washes in 50 mM NaPO4 pH 7 are performed before another fixation under vacuum for 15 minutes in 2% paraformaldehyde, 0.5% glutaraldehyde, 100 mM NaPO4 pH 7 following by 24 h incubation at 4 °C. Samples were cut into 1 cm fragments and mature parts of roots were subjected to gradual dehydration to overnight incubation in 100% EtOH. Inclusions were performed using Technovit 7100 cold-curing resin (Heraeus Kulzer performed according manufacturer’s recommendations). Transversal sections of 5 µm were realized using a microtome (Leica RM2165) and observed in water under BH2 microscope with color view soft imaging system (camera) and Cell^A software.
Gene Ontology analysis
Gene ontology has been analyzed using BINGO under Cytoscape environment, using Biological Process file, and a significance level of 0.05.
Data analysis and presentation
Mean ± SE is shown for all numerical values, and based on at least 3 biological replicates. Statistical significance was computed using a two-tailed Student’s t-test. Significance cutoff: *p < 0.05, **p < 0.01, ***p < 0.001.
References
Margueron, R. & Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349, https://doi.org/10.1038/nature09784 (2011).
Mozgova, I. & Hennig, L. The polycomb group protein regulatory network. Annu Rev Plant Biol 66, 269–296, https://doi.org/10.1146/annurev-arplant-043014-115627 (2015).
Pengelly, A. R., Copur, O., Jackle, H., Herzig, A. & Muller, J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339, 698–699, https://doi.org/10.1126/science.1231382 (2013).
Chanvivattana, Y. et al. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131, 5263–5276, https://doi.org/10.1242/dev.01400 (2004).
Wang, H. et al. Arabidopsis Flower and Embryo Developmental Genes are Repressed in Seedlings by Different Combinations of Polycomb Group Proteins in Association with Distinct Sets of Cis-regulatory Elements. PLoS Genet 12, e1005771, https://doi.org/10.1371/journal.pgen.1005771 (2016).
Roudier, F. et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 30, 1928–1938, https://doi.org/10.1038/emboj.2011.103 (2011).
Sequeira-Mendes, J. et al. The Functional Topography of the Arabidopsis Genome Is Organized in a Reduced Number of Linear Motifs of Chromatin States. Plant Cell, https://doi.org/10.1105/tpc.114.124578 (2014).
Turck, F. et al. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3, https://doi.org/10.1371/journal.pgen.0030086 (2007).
Zhang, X. et al. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nature structural & molecular biology 14, 869–871, https://doi.org/10.1038/nsmb1283 (2007).
Liu, J. et al. CURLY LEAF Regulates Gene Sets Coordinating Seed Size and Lipid Biosynthesis. Plant Physiol 171, 424–436, https://doi.org/10.1104/pp.15.01335 (2016).
Lafos, M. et al. Dynamic Regulation of H3K27 Trimethylation during Arabidopsis Differentiation. PLoS Genet 7, https://doi.org/10.1371/journal.pgen.1002040 (2011).
Angel, A., Song, J., Dean, C. & Howard, M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476, 105–108, https://doi.org/10.1038/nature10241 (2011).
Angel, A. et al. Vernalizing cold is registered digitally at FLC. Proc Natl Acad Sci USA 112, 4146–4151, https://doi.org/10.1073/pnas.1503100112 (2015).
Jean Finnegan, E. et al. Polycomb proteins regulate the quantitative induction of VERNALIZATION INSENSITIVE 3 in response to low temperatures. Plant J 65, 382–391, https://doi.org/10.1111/j.1365-313X.2010.04428.x (2011).
Deal, R. B. & Henikoff, S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell 18, 1030–1040, https://doi.org/10.1016/j.devcel.2010.05.013 (2010).
Berger, N., Dubreucq, B., Roudier, F., Dubos, C. & Lepiniec, L. Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell 23, 4065–4078, tpc.111.087866 (2011).
de Lucas, M. et al. Transcriptional Regulation of Arabidopsis Polycomb Repressive Complex 2 Coordinates Cell-Type Proliferation and Differentiation. Plant Cell 28, 2616–2631, https://doi.org/10.1105/tpc.15.00744 (2016).
Goodrich, J. et al. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51, https://doi.org/10.1038/386044a0 (1997).
Makarevich, G. et al. Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep 7, 947–952, https://doi.org/10.1038/sj.embor.7400760 (2006).
Aichinger, E., Villar, C. B. R., Di Mambro, R., Sabatini, S. & Köhler, C. The CHD3 Chromatin Remodeler PICKLE and Polycomb Group Proteins Antagonistically Regulate Meristem Activity in the Arabidopsis Root. Plant Cell 23, 1047–1060, https://doi.org/10.1105/tpc.111.083352 (2011).
Bouyer, D. et al. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet 7, e1002014, https://doi.org/10.1371/journal.pgen.1002014 (2011).
Farrona, S. et al. Tissue-specific expression of FLOWERING LOCUS T in Arabidopsis is maintained independently of polycomb group protein repression. Plant Cell 23, 3204–3214, https://doi.org/10.1105/tpc.111.087809 (2011).
Cerezo, M. et al. Major alterations of the regulation of root NO(3)(-) uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol 127, 262–271 (2001).
Filleur, S. et al. An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Letters 489, 220–224, https://doi.org/10.1016/s0014-5793(01)02096-8 (2001).
Lezhneva, L. et al. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J 80, 230–241, https://doi.org/10.1111/tpj.12626 (2014).
Orsel, M., Eulenburg, K., Krapp, A. & Daniel-Vedele, F. Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 219, 714–721, https://doi.org/10.1007/s00425-004-1266-x (2004).
Nacry, P., Bouguyon, E. & Gojon, A. Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant and Soil 370, 1–29, https://doi.org/10.1007/s11104-013-1645-9 (2013).
Girin, T. et al. Identification of Arabidopsis Mutants Impaired in the Systemic Regulation of Root Nitrate Uptake by the Nitrogen Status of the Plant. Plant Physiology 153, 1250–1260, https://doi.org/10.1104/pp.110.157354 (2010).
Girin, T. et al. Identification of a 150 bp cis-acting element of the AtNRT2.1 promoter involved in the regulation of gene expression by the N and C status of the plant. Plant, Cell & Environment 30, 1366–1380, https://doi.org/10.1111/j.1365-3040.2007.01712.x (2007).
Nazoa, P. et al. Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: responses to nitrate, amino acids and developmental stage. Plant Molecular Biology 52, 689–703, https://doi.org/10.1023/A:1024899808018 (2003).
Widiez, T. et al. High Nitrogen Insensitive 9 (HNI9)-mediated systemic repression of root NO3- uptake is associated with changes in histone methylation. Proceedings of the National Academy of Sciences 108, 13329–13334, https://doi.org/10.1073/pnas.1017863108 (2011).
Bellegarde, F., Gojon, A. & Martin, A. Signals and players in the transcriptional regulation of root responses by local and systemic N signaling in Arabidopsis thaliana. J Exp Bot, https://doi.org/10.1093/jxb/erx062 (2017).
Yang, H., Howard, M. & Dean, C. Antagonistic Roles for H3K36me3 and H3K27me3 in the Cold-Induced Epigenetic Switch at Arabidopsis FLC. Curr Biol 24, 1793–1797, https://doi.org/10.1016/j.cub.2014.06.047 (2014).
Veluchamy, A. et al. LHP1 Regulates H3K27me3 Spreading and Shapes the Three-Dimensional Conformation of the Arabidopsis Genome. PLoS One 11, e0158936, https://doi.org/10.1371/journal.pone.0158936 (2016).
Xu, L. & Shen, W. H. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18, 1966–1971, https://doi.org/10.1016/j.cub.2008.11.019 (2008).
Gendrel, A. V., Lippman, Z., Yordan, C., Colot, V. & Martienssen, R. A. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297, 1871–1873, https://doi.org/10.1126/science.1074950 (2002).
Acknowledgements
We thank every members of A.G. lab for discussion. This work was supported by a grant from the National Agency for Research (ANR) (ANR14-CE19-0008 IMANA to A.G., A.M., C.F., E.C. and F.R.) and from INRA BAP department to A.M., A.G. and C.F. F.B. was the recipient of a PhD fellowship from INRA BAP department. We thank M. De Lucas and S. Brady for ProCLF:CFP:CLF;clf-29 line. We thank C. Alcon, the Montpellier Rio Imaging platform, and the Plant Histocytology and cell imaging (PHIV) platform for microscopy observations and histocytology.
Author information
Authors and Affiliations
Contributions
Conceptualization, A.M., A.G., F.R. and F.B.; Investigation, F.B., L.H., D.S., J.B., E.C., C.F. and A.M.; Writing, A.M., A.G., and F.B; Funding Acquisition, A.M., A.G. and F.R.; Supervision, A.M. and A.G.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bellegarde, F., Herbert, L., Séré, D. et al. Polycomb Repressive Complex 2 attenuates the very high expression of the Arabidopsis gene NRT2.1. Sci Rep 8, 7905 (2018). https://doi.org/10.1038/s41598-018-26349-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-26349-w
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.