Abstract
In tumor microenvironment, interactions among multiple cell types are critical for cancer progression. To understand the molecular mechanisms of these complex interplays, the secreted protein analysis between malignant cancer cells and the surrounding nonmalignant stroma is a good viewpoint to investigate cell-cell interactions. Here, we developed two stable isotope labeling of amino acids in cell culture (SILAC)-based mass spectrometry (MS)/MS approaches termed spike-in SILAC and triple-SILAC to quantify changes of protein secretion level in a cell co-cultured system. Within the co-culture system of CT26 and Ana-1 cells, the spike-in SILAC and triple-SILAC MS approaches are sensitive to quantitatively measure protein secretion changes. Three representative quantified proteins (Galectin-1, Cathepsin L1 and Thrombospondin-1) by two SILAC-based MS methods were further validated by Western blotting, and the coming result matched well with SILACs’. We further applied these two SILACs to human cell lines, NCM460 and HT29 co-culture system, for evaluating the feasibility, which confirmed the spike-in and triple SILAC were capable of monitoring the changed secreted proteins of human cell lines. Considering these two strategies in time consuming, sample complexity and proteome coverage, the triple-SILAC way shows more efficiency and economy for real-time recording secreted protein levels in tumor microenvironment.
Similar content being viewed by others
Introduction
Tumor microenvironment, a complex system of many cell types including tumor, stroma, endothelial and immune system cells1,2, was recognized as the product of a developing crosstalk with secreted proteins elements. The secreted proteins in extracellular matrix (ECM), which produced by various cells in cell microenvironment, play very important roles in their growth and progression2,3. Proteins that are secreted into ECM, taken as a promising source of biomarkers3,4, also are responsible for signaling and communication in tumor microenvironment4,5. It provides new sights in cancer biology to pursue the dynamic changes of low-abundance secreted proteins in cancer microenvironment gives6.
The secretory pattern of tumor cells and their neighboring stromal cells is dynamically changed in tumor microenvironment. The secreted proteins are often present with high numbers and low abundance, and interfered by serum proteins in culture medium and contaminations of intracellular proteins released by cell lysis during sample preparation7,8,9. In order to sensitively detect active cell-cell interactions mediated by secreted proteins in tumor microenvironment, the SILAC (stable isotope labeling of amino acids in cell culture) based quantitative proteomic strategy showed us the good confidence due to its sensitivity and accuracy6,10. The SILAC-combined mass spectrometry (MS) has been widely used in varied studies, including screening disease biomarkers9,10,11,12,13, drug targets14,15,16, monitoring changes in post-translational modifications17,18,19 and finding key factors in the complex signal pathway10,20,21.
In classical SILAC-MS, two cell populations are respectively labeling with light, heavy isotope amino acids10,13, then cell lysates were combined to analyze together by LC-MS/MS. In the MS spectra, each isotope labeling peptide appears as a doublet with distinct mass differences. The differential protein abundances between two samples are calculated directly by comparing the intensity differences of the pair of isotope labeling peaks in MS12,15. So far, the typical SILAC-MS is only applicable for cell protein isotope labeling and quantification.
Lately, multiple SILAC-derived technical modifications have been developed to enlarge its practicability in protein quantification. For instance, the spike-in SILAC22, is developed in which the labeling is separated or ‘decoupled’ from the biological experiment. The non-labeled samples are combined with the SILAC standard, and each of these combined samples is analyzed separately by LC-MS/MS. The difference between the experimental samples is calculated as the ‘ratio of ratios’, where the ratio of one sample relative to the standard is divided by the ratio of the other relative to the standard. Actually, we previously have extended the SILAC-MS approach to tissue proteome comparisons, in which the heavy isotope labeling cells serve as the spike-in standards to compare the proteome changes of two states of tissues20.
Another similar strategy named super-SILAC method has been broadened five SILAC-labeling cell lines to serve as the internal standards for tissue proteome quantification23. Later, the triple-SILAC, namely SILAC with three isotope labeling states24, has been employed to characterize dynamic interaction partners in signal pathway. Generally, the multiple SILAC-based MS technique progresses greatly widen classic SILAC application in the study of clinical samples, secretome, post-translational modifications and organelle proteomes25.
Due to the complexity of multiple cell-involved tumor microenvironment, we aim to develop the spike-in SILAC or triple-SILAC approaches to real-time record protein-mediated cell-cell interactions. Based on our established SILAC methods6,26, we applied two cell lines in a co-culture system to simulate the tumor microenvironment, and adopted both the spike-in SILAC and triple-SILAC strategies to identify and quantify secreted protein changes. By comparing the feasibilities of these two methods, the triple-SILAC way shows more efficiency, economy for real-time recording secreted proteins in tumor microenvironment.
Results
Quantitative MS strategy for triple-SILAC and spike-in SILAC
For triple-SILAC, cell culture supernatant collected from each state, including unlabeling (Light, L), labeling with 13C6-Lys (Heavy, H) or 2H3-Leu (Medium, M) was combined for a simultaneous comparison of protein abundances. The relative amount of proteins from different culture media (CM) can be calculated directly by comparing the intensities of the certain labeling peptide(s) peaks in MS. The SILAC ratio (Ratio T) was defined as the ratio of co-cultured system versus mono-cultured one. A certain secreted protein’s Ratio T equaled to the unlabeling peptide(s) intensity divided by the sum of 13C6-Lys and 2H3-Leu labeling one(s) (Fig. 1A).
The strategies of ‘triple-SILAC’ (A) and ‘spike-in SILAC’ (B) approaches. In triple-SILAC, we combined the CMs from none labeled (L) co-cultured system, 13C6-Lys (H) labeled mono-cultured CT26 and 2H3-Leu (M) labeled Ana-1, then simultaneously analyzed by LC-MS/MS. The final ratio (Ratio T) of co-cultured system versus mono-cultured ones can be calculated directly by comparing the intensities of peaks with the certain molecular weight shifts in mass spectrum. In spike-in SILAC, CMs from 13C6-Lys (H) labeled Ana-1 and CT26 were combined separately with co-cultured one (L). To compare the peaks intensity with heavy (H) and light (L) labeled, Ratio1 and 2 were identified. The Ratio S is the inverse of the sum of. Ratio1 and Ratio2. Light: non-labeled. Medium: 2H3-Leu labeled. Heavy: 13C6-lysine labeled. Ratio T and Ratio S: Final ratio of a protein for co-cultured system versus mono-cultured in triple and spike-in SILAC. Ratio 1: the protein ratio of 13C6-Lys-labeling mono-cultured CT26 versus co-cultured cells. Ratio 2: the protein ratio of 13C6-Lys-labeling mono-cultured Ana-1versus co-cultured cells.
For spike-in SILAC, each 13C6-Lys labeling mono-cultured sample is separately combined with the co-cultured SILAC standard (none-label), which are then processed and analyzed together. Each of these two combined samples is analyzed separately by LC-MS/MS. The difference (Ratio S) between the experimental samples is calculated as the ‘ratio of ratios’, where the ratio of one sample relative to the standard is divided by the ratio of the other relative to the standard (Fig. 1B). The SILAC ratio1 was shown as the relative concentration of a secreted protein of 13C6-Lys-labeling mono-cultured cells versus co-cultured cells. Similarly, SILAC ratio2 was represented the relative protein amount of 13C6-Lys-labeling mono-cultured Ana-1 versus co-cultured cells.
In both SILAC-based strategies, it should be averaged when several peptides were used to quantify a protein. Due to uncertainty of the exact cell number we add in, the final ratios of both SILAC approaches should be normalized by total protein concentration in mono-and co-cultured CM.
MS quantification analysis for secreted proteins
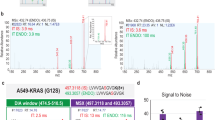
CT26 and Ana-1 cells were co-cultured in a transwell system with 1:1 cell number ratio. To compare efficiency of triple-SILAC method with spike-in SILAC way, three identified proteins were representatively shown the quantitative results. For triple-SILAC, Cathepsin L1 was upregulated by 4.5 fold, and Thrombospondin-1 was down regulated by 0.13 fold in co-cultured system. The abundance was no change for Galectin-1. For spike-in SILAC, Cathepsin L1 increased by 2.86 fold (Ratio S ), which was calculated by inversing the sum of Ratio1 and 2 (1/(0.17 + 0.18)) (Fig. 2B). Thrombospondin-1 was decreased by 0.18 fold (Ratio S = 1/ (Ratio1 + Ratio2) = 1/ (1.60 + 4.08)) (Fig. 2C). The unchanged protein, Galectin-1, had a ratio at 1.07 (1/ (0.22 + 0.71) (Fig. 2A). In both strategies, these three proteins showed the same change trend (Table 1).
Quantitative MS peaks of three proteins identified by spike-in SILAC and Triple-SILAC in mouse Ana-1 and CT26 co-culture system. In triple-SILAC way, the MS spectra showed the peak intensity of the quantitative peptides with none (L), 2H3-Leu (M) and 13C6-Lys (H) labels. In spike-in SILAC, the quantitative peptide containing 2H3-Leu (M) and 13C6-Lys (H) labels were compared to none labeling (L) separately. In both ways, the monoisotopic peaks with highest intensity were taken to be compared. (A) The MS spectra of the quantitative peptide“R.LNMEAINYMAADGDFK.I” for Galectin-1. (B)The quantitative peptide “K.ENGGLDSEESYPYEAK.D” for Cathepsin L1. (C) The quantitative peptide “K.AGTLDLSLSLPGK.Q” for Thrombospondin-1.
Western blotting validation for SILAC-MS approaches
To validate the SILAC-MS quantification results, the three identified proteins were further detected by Western blotting in Ana-1/CT26 system (Fig. 3). The protein abundances were compared the band density using the Imagine J software (version 1.8.0)27. Firstly, to confirm the same loading amount of samples, we plotted the total proteins of 3 samples by Coomassie blue staining (Fig. 3A). Before immunoblotting, we also provided Ponceau S staining of the PVDF membrane to evaluate the efficiency of protein transfer from the SDS gel to membrane (Fig. 3B). The Gel analysis results showed Cathepsin L1 was upregulated to 3.29 fold (14372/4375). Thrombospondin-1 had lower intensity in co-cultured system versus mono-culture with a ratio of 0.21 (2043/9517). And Galectin-1 showed no obvious change with a ratio of 0.93 (10994/11813) in Western blotting analysis (Fig. 3C). The Western blotting results were highly consistent with the SILAC-MS quantification.
In-gel validation for three representative proteins. (A)The total proteins of loaded samples. (B) The total proteins after transferring to PVDF membrane. (C) The three MS- quantified proteins were verified by Western blot. Line 1–3: Conditional media (CM) collected from mono-cultured Ana-1 (1), CT26 (2) and co-cultured system (3). Line 4: the mixture CMs from mono-cultured Ana-1 and CT26 that have been incubated for 48 h respectively (4). Data showed the band intensity. The unprocessed original scans of blots (Fig. 3) are shown in Supplementary Fig. 1.
The extensive application of SILACs in human cell line co-culture system
After successfully applying the two (SILAC)-based MS/MS approaches in CT26 and Ana-1 co-culture system, two human cell lines, NCM460 and HT29, were further chosen to validate the feasibility of analyzing secreted proteins in human colorectal carcinoma in vitro.
Similarly, we quantified two changed proteins in NCM460 and HT29 co-culture system using the above two (SILAC)-based MS/MS approaches. Two representative proteins, Myosin-9 and Prosaposin, were shown the quantification data (Fig. 4). Myosin-9 was respectively decreased by 0.45, 0.41 fold in triple and spike-in SILAC MS analysis for the co-culture system (Fig. 4A). Prosaposin (Psap), was identified to increase by the ratio of 2.23(Ratio T) in triple SILAC and 1.45 (Ratio S = 1/ (Ratio1 + Ratio2) = 1/ (0.29 + 0.4)) in spike-in SILAC MS analysis (Fig. 4B). In both strategies, these two proteins showed the same trend (Table 2).
Discussion
Each SILAC-MS method has individual strength and shortcoming (Table 3). For the triple-SILAC way, three samples are differentially labeled and combined before MS identification. The three-in-one sample just needs to process once, so the quantification deviation introduced between multiple unparalleled sample preparations can be eliminated. This also means we can apply as much steps to enrich proteins or peptides as we want to meet experiment demands. With a spike-in standard, each sample is measured separately, and therefore the measuring time is two to threefold longer than that for triple-SILAC experiments. In spike-in SILAC, the samples are processed and analyzed separately, the results are based on the ratios between the samples, in which it is dependent on the ratio of ratios, leading to an increase in the quantification variation. This effect will also take place when two triple-SILAC experiments are combined. The relative statistical error of a ratio of ratios is the square root of the sum of the squares of individual relative errors. Secondly, the biological issue and the system studied would be considered for Spike-in SILAC analysis. The makeup of the SILAC standard will vary depending on the complexity of the system of interest, ranging from single proteins to a complex mixture of thousands of proteins, and the approach is dependent on the variability of the samples. When there are more than three samples to be compared, triple-SILAC is out of our options.
Owing to the partial stochastic nature of shotgun proteomics, when the proteome coverage is low, the overlap between runs is also low, and it is difficult to compare samples. Naturally, the aim should always be to achieve high proteome coverage. This problem is irrelevant in triple-SILAC experiments, as all the peptides are directly compared in a single analysis. The complexity of the MS spectra is 1.5-fold higher in triple-labeled samples than that in spike-in standard experiments. High complexity may lead to lower numbers of identified proteins due to re-sequencing of peptides in the different SILAC states. But it is not observed in our co-cultured system which includes two cell lines.
The CT26 and Ana-1 both are mouse cell lines. The CT26 is a mouse colon fibroblast carcinoma cell line, and Ana-1 is a normal murine macrophage cell line. We chose these two cell lines for recording protein-mediated cell-cell interactions due to the following reasons. 1) Mouse cell lines are the mostly used in biological and medical research due to similarity with human cells in gene expression, ageing, and immune responses to infection and disease. 2) The secreted proteins in ECM are produced by various cells in cell microenvironment, which is dynamic to regulate cell growth and progression2. Tumor-associated macrophages are present in the stroma of many tumors, which are frequently associated with the progression of several types of cancer including colon cancer6. So CT26 and Ana-1 cell lines were selected to simulate colorectal carcinoma tumor microenvironment for developing and evaluating the power of SILAC-based MS approaches we applied in this paper.
In Ana-1 and CT26 system, totally 221 secreted proteins were quantified through spike-in SILAC-MS analysis, and 203 proteins were identified by triple-SILAC MS. The upregulated protein Cathepsin L1 in our co-cultured cells, is a lysosomal cysteine protease, which is overexpressed to secret into serum in several cancers, including ovarian cancer, pancreatic neuroendocrine cancer, glioma and others28,29,30. The overexpression of Cathepsin L1 is involved in tumor invasion, metastasis and chemotherapy resistance31,32. Thrombospondin-1 is a large glycoprotein secreted by platelets and synthesized by many cell types, including endothelial and tumor cells. Thrombospondin-1 may potentiate tumor progression, promotes tumor cell adhesion, migration and invasion33. The two changed secreted proteins are further explored their molecular functions between tumor cells and macrophages.
We further applied two (SILAC)-based MS/MS approaches to validate the feasibility of analyzing secreted proteins in human NCM460 and HT29 system in vitro, which confirmed the spike-in and triple SILAC were capable of monitoring the changed secreted proteins of human cell lines (Fig. 4).
The original SILAC labeling is limited to cell culture or microorganisms. To extend this powerful technique to higher organisms, Hayter and colleagues demonstrated chicken can be partially labeled at the amino acid level by feeding them with a diet containing stable isotope-labeled valine34. Drosophila can be labeled by feeding the flies with 13C6-Lys labeled yeast. Krueger et.al achieved the 13C6-Lys labeling laboratory mouse35. Until now, this so-called ‘SILAC mouse’ is the only multicellular organism that has been completely labeled with the SILAC approach and partial labeling was recently achieved in newts34,36.
SILAC-labeled mice are generated by feeding them with 13C6-Lys containing diet. Although labeling rates reach 95% which is required to perform comparative and quantitative proteomics by MS after two generations. This is both costly and time-consuming and as a result it hampers repeated generation of labeled organisms for each experimental condition, each strain or transgenic organism. Different cell types and tissues in the same organism have different SILAC label incorporation, and extending the labeling time cannot perform complete labeling due to the recycling of internal amino acid sources35. This may lead to the quantification uncertainty.
It is recommended using the SILAC model organisms just as standards rather than as the experimental system themselves, since the SILAC food might have metabolic effects35. Moreover, with the spike-in standard approach the same SILAC organism can be used for experiments with strains of various genetic backgrounds. In our co-culture system, we use the unlabeled co-culture cells as the spike-in standard.
For triple SILAC MS application on animal models, we need to build two different labeled model animals, which is time consuming and much more cost-expensive. Besides lysine, there are limited peptides are suited for the whole organism labeling now. Arginine is also problematic since it is converted into proline in some cell lines37. Leucine is one of the most used amino acid for SILAC labeling, but it is not the tryptic-digested peptide end, which is not optimal for global or large-scale SILAC quantitation12.
15N is also used to label entire rats, particularly for quantitative brain proteomics38. Despite its usefulness, 15N labeling has also several disadvantages. Since most peptides contain dozens of nitrogen atoms, labeling with highly enriched 15N still results in only partial peptide labeling and therefore complex isotope clusters. In addition, the mass shift between the labeled (i.e. heavy) and unlabeled (i.e. light) form of a peptide depends on the number of nitrogen atoms and therefore varies depending on the peptide sequence. This leads to an increase in the number of candidate masses that need to be considered and therefore complicates peptide identification by search algorithms. Both problems result in smaller identification rates and less accurate quantification that can partially overcome by computational correction39.
Conclusion
The SILAC-based quantitative proteomic techniques, including the spike-in SILAC and triple-SILAC, are successfully applied to identify secreted protein changes in mimic tumor microenvironment in vitro. Considering the availability of these two methods, the triple-SILAC MS shows more efficiency, economy for real-time recording secreted protein levels in tumor microenvironment.
Methods
Chemicals and reagents
Fetal bovine serum (FBS) (AUS-01N, CELL-BOX), RPMI 1640 media (Gibco Gaithersburg), Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco, Gaithersburg) 13C6-lysine (98%; 89988, Thermos), and 2H3-leucine (DLM-1259-1, 99%; Cambridge Isotope Laboratories). Coomassie blue and Ponceau S (GOLDBIO). The other reagents for MS analysis were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated.
Cell culture
CT-26, a mouse colon fibroblast carcinoma cell line, and a normal murine macrophage cell line Ana-1 were both ordered from American Type Culture Collection (ATCC). Cells were cultured in the RPMI-1640 Medium, supplemented with 10% FBS at 37 °C in humidified atmosphere with 5% (v/v) CO2.
The NCM460, a normal human colon mucosal epithelial cell line, was ordered from Incell Corporation, LLC (www.incell.com). HT29, a colon adenocarcinoma cell line, was ordered from ATCC. These cells were stored in our laboratory40. Cells were routinely maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco, Gaithersburg, MD) supplemented with 10% FBS at 37 °C in humidified atmosphere with 5% (v/v) CO2.
Stable isotope labeling
CT-26 cells were cultured in RPMI 1640 media with 0.1 mg/ml 13C6- Lys replacing normal Lys. Meanwhile Ana-1 cells were cultured in RPMI 1640 media with 0.1 mg/ml 13C6- Lys or 2H3-Leu to respectively replace normal Lys or Leu.
NCM460 cells were cultured in DMEM with 0.1 mg/ml 13C6- Lys replacing normal Lys. Meanwhile HT29 cells were cultured in DMEM media with 0.1 mg/ml 13C6- Lys or 2H3-Leu to respectively replace normal Lys or Leu.
All Cell lines were cultured and routinely maintained at 37 °C in humidified air containing 5% CO2. The culture media were changed every 2–3 days. Cells were kept in ‘labeled’ medium for at least 5 cell passages to achieve >95% labeling.
Co-culture conditions
To modulate the interactions between the two kinds of cell lines, Ana-1 and CT26 cells were co-cultured on a transwell system (3419, Corning Inc., Corning, NY) with 1:1 cell number ratio. In co-culture system, 5 × 106 unlabeling Ana-1 cells were seeded into the bottom chamber of a 75mm-transwell plate and incubated in 8 ml serum-free RPMI 1640, meanwhile the same amounts of unlabeling CT26 cells were also cultured in 8 ml serum-free RPMI 1640 onto the top chamber of the transwell plate. Cells were co-cultured for 48 h.
For NCM460 and HT29 cells, the co-cultured system is just as same as Ana-1 and CT26 in 8 ml serum-free DMEM, with NCM460 cells in the bottom chamber and HT29 in the top one.
In triple-SILAC way, the same amount of 2H3-Leu labeled cells (Ana-1 or HT29) and 13C6-Lys cells (CT26 or NCM460), as the mono-culture system, were cultured in 8 ml serum-free 2H3-Leu and 13C6-Lys added-in RPMI 1640 or DMEM for 48 h respectively. For spike-in, 13C6-Lys labeled cells (Ana-1, CT26, NCM460 and HT29) were cultured in 13C6-Lys containing RPMI 1640 or DMEM.
Secreted protein preparation
Secreted protein preparation was performed according to our previous method6. Briefly the conditioned medium (CM), which separately derived from co- and mono- culture system in triple-SILAC and spike-in SILAC strategies, was collected by centrifugation at 1500 × g for 3 min to remove cell debris following with a sterile filtration (0.22μm, Millipore). With an addition of proteinase inhibitor (cocktail,P-8340,sigma), the CM was concentrated by ultrafiltration using a centrifugal filter device “AmiconUltra-15” (UFC900324, Millipore) with centrifugation at 4000 × g for 2 h. Proteins were precipitated with cold acetone at −20 °C overnight and collected by centrifugation at 13,000 g for 20 min at 4 °C.The protein pellet was air-dried at room temperature, and resolved in 40 μL sample buffer (60 mM Tris-HCL pH 6.8, 2% SDS, 10% glycerol, 5% DTT, 0.01% bromophenol blue). The protein concentration was determined by Bradford protein assay (P0011, Beyotime).
Sample combination
The CMs were mixed from the different cultured cell system. In triple-SILAC mode, we mix all three CMs into one part, which contains non-labeled (light), 2H3-Leu (medium) and 13C6-Lys (heavy) labeled proteins. For spike-in, the different labeled mono-cultured CM was combined with co-cultured one respectively.
Proteins in-gel digestion
Proteins were separated by 12%SDS-PAGE and stained with coomassie to visualize gel bands. The gel lanes were excised to cut into 1 mm3 slices and destained twice with 100 mM NH4HCO3/50% acetonitrile (ACN) at 37 °C for 30 min. After being dehydrated and dried, gel slices were digested with 10–20 μl MS-grade trypsin (V5280, Trypsin Gold, Promega, USA) at 37 °C overnight. Peptides were extracted by ultrasonic with extraction solution (50%ACN/5% trifluoroacetic acid) for twice. The combined extraction was dried in a vacuum concentrator at room temperature. Samples were then subjected to LC-MS/MS analysis.
LC-MS/MS analysis
Proteins were identified by nanoLC-MS/MS with LTQ-Orbitrap (Thermo Finnigan, San Jose, CA). Peptide samples were separated on a C18 column (150 μm, 150 mm length, Column Technology Inc.) after being desalted on a trap column (Zorbax 300 SB C18, Agilent Technologies, Palo Alto, CA). The mobile phase A was 0.1% formic acid in HPLC-grade water, and 0.1% formic acid in ACN for the mobile phase B. The peptide mixture was separated at 2 μl/min with a linear gradient of 4%–50% B for 110 min, following by 50%–100% B from 110 min to 115 min, then maintained at 100% B for 5 min. Data dependent MS/MS mode was acquired, in which each scan cycle consisted of one full MS scan in profile mode followed by seven MS/MS scans in centroid mode.
Mass data search
The Mass data were processed by Maxquant software 1.6.0.1 plugging Andromeda 1.5.6.0 as the database search engine. The parameters for database searching were set as following: database, Swiss-Prot databases (16523 proteins (Mouse, Aug 30, 2017) and 43943 proteins (Human, Oct 13, 2017)); enzyme, trypsin; two missed cleavages are allowed. Carbamidomethyl (C) was selected as a fixed modification, and oxidation (M), acetyl (protein N-term), 13C6-Lys and 2H3-Leu labeling (only for triple-SILAC) were set as the variable modification. Initial peptide mass tolerance was set to 7 ppm and fragment mass tolerance was 0.5 Da. + 2 as default charge state of each peptide. The false discovery rates (FDRs) of peptide and protein were both set to 0.01, which ensure that protein identifications with FDRs ≤ 1% (95% confidence). At least one unique peptide of a protein successfully detected was considered to be acceptable.
Coomassie blue and Ponceau S staining
The SDS-PAGE gel was fixed in solution (50% methanol and 10% glacial acetic acid) for 2 hours with gentle agitation. It was stained in staining solution (0.1% Coomassie Brilliant blue R-250, 50% methanol and 10% glacial acetic acid) for 2–3 hours with gentle agitation. After destaining in solution (40% methanol and 10% glacial acetic acid) for several times until no unspecific background, the gel was stored in 5% glacial acetic acid.
The blotted membrane was immersed in a sufficient amount of Ponceau S staining solution (0.1% (x/v) Ponceau S in 1% (v/v) acetic acid) to stain for 5 minutes. Then membrane was immersed in an aqueous solution containing 5% acetic acid (v/v) for 5 minutes to evaluate the efficiency of protein transfer from the SDS gel to PVDF membrane.
Western blot
We choose three typical secreted proteins (Cathepsin L1, Galectin-1 and Thrombospondin-1) to validate by Western blot analysis. Besides three identified proteins in SILACs changed in different direction: no change, up-and down regulation.
It was totally same in the way of cell culturing and protein extraction just as described above with the only difference that all culture system was made of non-labeling serum-free RPMI1640 medium, no 2H3-Leu or 13C6-lys. The protein concentration was determined with the Bradford protein method. 40 μg protein was separated on a 12% SDS-PAGE gel and transferred onto a PVDF membrane at 100 V for 1 h. Subsequently the membrane was incubated in TBST buffer (20 mM Tris–HCl, pH 7.6, 150 mM NaCl, 0.1%Tween-20) with 5% non-fat milk at room temperature for 2 h. The specific primary antibodies were diluted in TBST (50 mM Tris–HCl, with 150 mM NaCl,0.1%Tween-20, pH7.4) buffer to incubate PVDF membrane at 4 °C overnight. Primary antibodies included mouse anti-Galectin-1 (1:200, sc373717, Santa-Cruz, CA), mouse anti-Thrombospondin-1 (1:200, sc59886, Santa-Cruz, CA), and mouse anti-Cathepsin L1 antibodies (1:200, sc13094, Santa-Cruz, CA). The corresponding mouse derived secondary antibody conjugated horseradish peroxidase was subsequently incubated with the PVDF membrane for 60 min at room temperature. Signal detection was performed with ECL reagent (Amersham Biosciences, Piscataway, New Jersey, USA).
Data availability statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Clementz, A. G. et al. Collagen XV: exploring its structure and role within the tumor microenvironment. Mol Cancer Res. 11, 1481–1486 (2013).
Sangaletti, S. et al. The good and bad of targeting cancer-associated extracellular matrix. Curr Opin Pharmacol. 35, 75–82 (2017).
Slany, A. et al. Targeting breast cancer-associated fibroblasts to improve anti-cancer therapy. Breast. 24, 532–538 (2015).
Giussani, M. et al. Tumor-extracellular matrix interactions: Identification of tools associated with breast cancer progression. Semin Cancer Biol. 35, 3–10 (2015).
Kostourou, V. et al. Non-collagenous ECM proteins in blood vessel morphogenesis and cancer. Biochim Biophys Acta. 1840, 2403–2413 (2014).
Zeng, X. et al. Quantitative secretome analysis reveals the interactions between epithelia and tumor cells by in vitro modulating colon cancer microenvironment. J Proteomics. 89, 51–70 (2013).
Zhong, L. et al. Identification of secreted proteins that mediate cell-cell interactions in an in vitro model of the lung cancer microenvironment. Cancer Res. 68, 7237–7245 (2008).
Li, M. et al. Intercellular transfer of proteins as identified by stable isotope labeling of amino acids in cell culture. J Biol Chem. 285, 6285–6297 (2010).
Li, X. et al. Quantitative proteomic analysis and comparison of two bone marrow stromal cell lines using the SILAC method. Exp Hematol. 44, 1059–1071 (2016).
Liang, S. et al. Affinity purification combined with mass spectrometry-based proteomics to study mammalian protein complex and protein-protein interactions. Curr Proteomics. 6, 25–31 (2009).
Kashyap, M. K. et al. SILAC-based quantitative proteomic approach to identify potential biomarkers from the esophageal squamous cell carcinoma secretome. Cancer Biol Ther. 10, 796–810 (2010).
Zhang, D. et al. Downregulation of ATP1A1 promotes cancer development in renal cell carcinoma. Clin Proteomics. 14, 15 (2017).
Liang, S. et al. Quantitative proteomics for cancer biomarker discovery. Comb Chem High Throughput Screen. 15, 221–231 (2012).
Chen, B. et al. Proteomic progresses in microbial physiology and clinical antimicrobial therapy. Eur J Clin Microbiol Infect Dis. 36, 403–413 (2017).
Liang, S. et al. Honokiol inhibits cell migration via IQGAP1 downregulation discovered bya quantitative pharmaceutical proteomic analysis. Proteomics. 10, 1474–1483 (2010).
Ma, W. et al. Antibacterial mechanism of daptomycin antibiotic against Staphylococcus aureus based on a quantitative bacterial proteome analysis. J Proteomics. 150, 242–251 (2017).
Yang, Y. et al. Protein SUMOylation modification and its associations with disease. Open Biol. 7, 170167 (2017).
Cuomo, A. et al. SILAC-based proteomic analysis to dissect the “histone modification signature” of human breast cancer cells. Amino Acids. 41, 387–399 (2011).
Liang, Z. et al. SUMOylation of IQGAP1 promotes the development of colorectal cancer. Cancer Lett. 411, 90–99 (2017).
Tatyana, A. P. et al. Stable isotope labeling by amino acids in cell culture (SILAC) and quantitative comparison of the membrane proteomes of self-renewing and differentiating human embryonic stem cells. Mol Cell Proteomics. 8, 959–970 (2009).
Liang, S. et al. Analysis of the protein complex associated with 14-3-3 epsilon bya deuterated-leucine labeling quantitative proteomics strategy. J. Chromatography B. 877, 627–634 (2009).
Geiger, T. et al. Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat Protoc. 6, 147–157 (2011).
Tamar, G. et al. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat methods. 7, 383–387 (2010).
Hilger, M. et al. Triple SILAC to determine stimulus specific interactions in the Wnt pathway. J Proteome Res. 11, 982–994 (2012).
Anjana, S. et al. Super-SILAC: current trends and future perspectives. Expert Rev Proteomics. 12, 13–19 (2014).
Xu, Y. et al. Application of the SILAC (stable isotope labelling with amino acids in cell culture)technique in quantitative comparisons for tissue proteome expression. Biotechnol Appl Biochem. 54, 11–20 (2009).
Schneider, C. A. et al. NIH Image to ImageJ: 25 years of image analysis. Nat methods. 9, 671–675 (2012).
Zhang, W. et al. Overexpression of cysteine Cathepsin L is a marker of invasion and metastasis in ovarian cancer. Oncol Rep. 31, 1334–1342 (2014).
Brindle, N. R. et al. Deficiency for the cysteine protease Cathepsin L impairs Myc-induced tumorigenesis in a mouse model of pancreatic neuroendocrine cancer. PLoS One. 10, e0120348 (2015).
Fei, Y. et al. Cathepsin L knockdown enhances curcumin-mediated inhibition of growth, migration, and invasion of glioma cells. Brain Res. 1646, 580–588 (2016).
Zhang, L. et al. Cathepsin L is involved in proliferation and invasion of ovarian cancer cells. Mol Med Rep. 11, 468–474 (2015).
Zhang, H. et al. Knockdown of Cathepsin L sensitizes ovarian cancer cells to chemotherapy. Oncol Lett. 11, 4235–4239 (2016).
Tuszynski, G. P. et al. The role of thrombospondin-1 in tumor progression and angiogenesis. Bioessays. 18, 71–76 (1996).
Hayter, J. R. et al. The subunit structure and dynamics of the 20 S proteasome in chicken skeletal muscle. Mol Cell Proteomics. 4, 1370–1381 (2005).
Krüger, M. et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 134, 353–364 (2008).
Looso, M. et al. Advanced identification of proteins in uncharacterized proteomes by pulsed in vivo SILAC. Mol Cell Proteomics. 9, 1157–1166 (2010).
Ong, S. E. et al. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat Protoc. 1, 2650–2660 (2006).
Liao, L. et al. Quantitative analysis of brain nuclear phosphoproteins identifies developmentally regulated phosphorylation events. J Proteome Res. 7, 4743–4755 (2008).
Gouw, J. W. et al. Optimizing identification and quantitation of 15N-labeled proteins in comparative proteomics. Anal Chem. 80, 7796–7803 (2008).
Chen, B. et al. STC2 promotes the epithelial-mesenchymal transition of colorectal cancer cells through AKT-ERK signaling pathways. Oncotarget. 7, 71400–71416 (2016).
Acknowledgements
This work was financially supported by the grants from the National Key Basic Research Program of China (2013CB911303, 2011CB910703), National 863 High Tech Foundation (2014AA020608), National Natural Sciences Foundation of China (31470810), the Science & Technology Department of Sichuan Province (2017JY0232), and the Health and Family Planning Commission of Sichuan Province (17ZD045).
Author information
Authors and Affiliations
Contributions
Wang X. performed experiments and wrote paper draft; Deng S, Zhao X., He Y., Ye Y., performed experiments; He G., Zhu H., Xu N. analyzed data. Liang S. conceived, instructed experiments and revised paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, X., He, Y., Ye, Y. et al. SILAC–based quantitative MS approach for real-time recording protein-mediated cell-cell interactions. Sci Rep 8, 8441 (2018). https://doi.org/10.1038/s41598-018-26262-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-26262-2
This article is cited by
-
SUMOylation of annexin A6 retards cell migration and tumor growth by suppressing RHOU/AKT1–involved EMT in hepatocellular carcinoma
Cell Communication and Signaling (2024)
-
SUMOylation of AnxA6 facilitates EGFR-PKCα complex formation to suppress epithelial cancer growth
Cell Communication and Signaling (2023)
-
SAE1 promotes human glioma progression through activating AKT SUMOylation-mediated signaling pathways
Cell Communication and Signaling (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.