Abstract
Indoor wet cells serve as an environmental reservoir for a wide diversity of melanized fungi. A total of 313 melanized fungi were isolated at five locations in Guangzhou, China. Internal transcribed spacer (rDNA ITS) sequencing showed a preponderance of 27 species belonging to 10 genera; 64.22% (n = 201) were known as human opportunists in the orders Chaetothyriales and Venturiales, potentially causing cutaneous and sometimes deep infections. Knufia epidermidis was the most frequently encountered species in bathrooms (n = 26), while in kitchens Ochroconis musae (n = 14), Phialophora oxyspora (n = 12) and P. europaea (n = 10) were prevalent. Since the majority of species isolated are common agents of cutaneous infections and are rarely encountered in the natural environment, it is hypothesized that indoor facilities explain the previously enigmatic sources of infection by these organisms.
Similar content being viewed by others
Introduction
Black yeast-like and other melanized fungi are frequently isolated from clinical specimens and are known as etiologic agents of a gamut of opportunistic infections, but for many species their natural habitat is unknown and hence the source and route of transmission remain enigmatic. The majority of clinically relevant black yeast-like fungi belong to the order Chaetothyriales, while some belong to the Venturiales. Propagules are mostly hydrophilic1 and reluctantly dispersed by air, infections mostly being of traumatic origin. Members of the group are associated with mild implantation diseases of skin and nails or chronic mutilating infections such as chromoblastomycosis and phaeohyphomycosis, but some show neurotropic dissemination. All infections may occur in immunocompetent individuals. Since special methods are required for their isolation from nature, a low competitive ability towards fast-growing contaminants has been hypothesized2.
In the environment, black yeast-like fungi have been recovered from unexpected, rather hostile (micro)environments, including rock, creosote-treated wood, hydrocarbon-polluted soil, and lichens3,4,5. Recent studies have demonstrated that black yeast-like fungi of Chaetothyriales, particularly Exophiala and Cladophialophora species are common colonizers in indoor wet cells6,7. The thermotolerant species Exophiala dermatitidis is frequent in steambaths, house baths and dishwashers8,9. Southeast Asia countries have a similar (sub)tropical climate, hot moisture sustaining thermophilic fungal growth during a large part of the year, although with marked geographical differences. However, disseminated Exophiala dermatitidis infections are particularly observed in East Asia countries10, e.g. Thailand, while the neurotropic species Cladophialophora bantiana occurs relatively often in India11. In order to understand the origin of infections, screening of households for fungi other than the common airborne fraction mainly involved in allergy (Aspergillus, Alternaria, Cladosporium) seems overdue. Seveal selective techniques have been developed enabling recovery the black yeast-like fungi2,7. In the present study we focused on black fungi in Sourthern China, where chromoblastomycosis and phaeohyphomycosis are endemic12,13,14,15,16. Our aim was to identify the spectum of black yeast-like fungi in indoor wet cells, and clarify whether these facilities harbour potential etiologic agents of disease.
Methods
Sampling and isolation
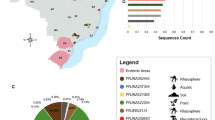
Environmental samples were collected from 53 families living in five regions (Yue Xiu, Li Wan, Bai Yun, Tian He, Hai Zhu) of Guangzhou city, during July to September, 2015. Black biofilms were sampled from bathrooms (water ladle, wall, soap box, washbasin, door, sprinklers, toothbrush cup, floor, brush, mirror), kitchen (water tank, chopping board, sterilizing cabinet, wall, cooking bench), refrigerator (rubber seal), washing machine (rubber seal, water tap) and water dispenser (water tank) using sterile cotton swabs and immediately contained in sterile tubes. The sampling tubes were stored for max. 24 h at room temperature before processing.
For the inoculation and culture of suspect fungus, 500 gram of Potato Dextrose Agar (PDA) (Oxoid, Thermo Fisher Scientific, Basingstoke, UK) powder was solved in 1.28 liter demi water, and autoclaved under 121 °C, 105 kPa for 30 min. Each 250 mg of Chloramphenicol (Sigma Aldrich, St. Louis, MO.USA) and cycloheximide (Sigma Aldrich, St. Louis, MO.USA) were added into the PDA medium when it reach to about 60 °C, and mixted sufficiently before plating. Sterile cotton swabs contained the suspect fungus specimens were rubbed over the surface of PDA medium plate with 200 mg/L chloramphenicol and 200 mg/L cycloheximide, and incubated at 28 °C for 2 weeks. The workflow of sampling and isolation are shown in Fig. 1.
The workflow of sampling and isolation. The black biofilms samples from particular areas of bathroom, kitchen, refrigerator rubber seal, and washing machine were taken by using sterile cotton swabs; the black biofilm materials were cultured on PDA medium (200 mg/L chloramphenicol and 200 mg/L cycloheximide) for 2 weeks. Susceptive black yeast like single colony strains were taken for further culture under same condition as above.
Morphology
After 2 weeks culturing, the suspected slow-growing, blackish-brown colonies in the same plate were viewed and compared. The predominant colony type in each plate was considered as target colony, and one was selected and transferred to new growth media on PDA using a loop for further culture. Preliminary identification at the generic level was carried out by colony appearance and morphology by microscopy using slide cultures on Sabouraud’s glucose agar (SDA) (Oxoid, Thermo Fisher Scientific, Basingstoke, UK) after 2 weeks culturing at room temperature.
DNA extraction
Methods of DNA extraction were those of Sun et al.13. Briefly, about 1 cm2 mycelium or yeasts of 15-d-old cultures was transferred to a 2 mL Eppendorf tube containing 300 μL CTAB (cetyltrimethylammonium bromide) buffer [CTAB 2% (w/v), NaCl 1.4 M, Tris-HCl 100 mM, pH 8.0; EDTA 20 mM, b-mercaptoethanol 0.2% (v/v)] and about 80 mg of a silica mixture (silica gel H, Merck 7736, Darmstadt, Germany/Kieselguhr Celite 545, Machery, Düren, Germany, 2:1, w/w). Cells were disrupted with sterile glass beads (Sigma, St. Louis, MO, USA) for approximately 5 min. Subsequently 200 μL CTAB buffer was added, the mixture was vortexed and incubated for 10 min at 65 °C. After addition of 500 μL chloroform, the solution was mixed and centrifuged for 5 min at 13, 000 rpm and the supernatant transferred to a new tube with 2 vols of ice cold 96% ethanol. DNA was allowed to precipitate for 30 min at −20 °C and then centrifuged again for 5 min at 13, 000 rpm. Subsequently the pellet was washed with cold 70% ethanol. After drying at room temperature it was resuspended in 97.5 μL TE-buffer plus 2.5 μL RNAse 20 U.mL−1 and incubated for 5 min at 37 °C, before storage at −20 °C.
Sequencing and molecular identification
Ribosomal DNA (rDNA, codes for ribosomal RNA) Internal Transcribed Spacer (ITS) was amplified using primers V9G and LS266 and sequenced with ITS1 and ITS42. Amplicons were cleaned with GFX PCR DNA purification kit (GE Healthcare, U.K.). Sequencing was performed on an ABI 3730XL automatic sequencer. Sequences were edited using the Seqman package (DNAStar, Madison, U.S.A.) and aligned using BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium). Sequences were compared in a research data database of black fungi maintained at Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands, and validated by ex-type strains of known species. In addition, all the ITS sequences were deposited to GenBank database, and conducted Blastn analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) as default parameters settings. The most hits sequence of known species was considered as the target species showed in Supplementary (Table S1).
Statistical analysis
Descriptive statistics were carried out with the statistical software package Statistical Product and Service Solutions (SPSS, 17.0) (IBM SPSS, Chicago, USA). Mann-Whitney U test for unparametric variables was used for comparison of strains among five different regions. The strains isolated in each family and location were also analyzed using one way ANOVA parametric test, followed with Turkey analysis. For all statistical tests, p-value of <0.05 was considered statistically significant.
Results
A total of 509 samples were taken from 53 families at five regions of Guangzhou, China, of which 399 samples showed positive growth of melanized fungi (Table 1). Subsequently, 313 (61.49%) strains of black yeast-like fungi were isolated from 399 positive samples (Table 2). Eighty-six suspected colonies could not successfully be isolated during subsequent subculturing. Although the percentages of positive isolation per region ranged from 59.48% to 71.88%, no significant differences of total isolates were found between five regions investigated either based on unparametric Mann-Whitney U test (p = 0.427) or one way ANOVA parametric test (p = 0.671), followed by Turkey analysis (p = 0.714–0.999) (Table 2).
Initial identification of suspected isolates based on colony appearance and slide culture micromorphology was approximate. Most isolates were identified at genus level due to similar colony and morphology appearance under microscopy, e.g. the genera Exophiala, Cladophialaphora, Phialophora, as well as Ochroconis in the order Venturiales. Therefore, the molecular based identification became essential for classification of extremely similar species in morphology. The rDNA ITS sequences of all isolates were amplified and sequenced successfully. Local blast in a research database on black yeast-like fungi at Westerdijk Fungal Biodiversity Institute, identified 259 strains with 27 species distributed in 10 genera, including Exophiala (n = 77), Phialophora (n = 44), Ochroconis (n = 38), Knufia (n = 37), Cladosporium (n = 37), Rhinocladiella (n = 7), Cyphellophora (n = 7), Hortaea (n = 7), Cladophialophora (n = 4), and Veronaea (n = 1) (Table 3). The most frequently isolated species were K. epidermidis (n = 37, 11.82%), O. musae (n = 32, 10.22%), P. oxyspora (n = 27, 7.99%), Cladosporium halotolerans (n = 21, 6.93%), E. lecanii-corni (n = 18, 5.94%), P. europaea (n = 18, 5.75%), E. alcalophila (n = 13, 4.15%), Cladosporium irritans (n = 13, 4.15%) and E. oligosperma (n = 10, 3.19%). Apparently undescribed species were detected in the genera Exophiala, Cyphellophora, Ochroconis, Hortaea and Cladophialophora (Table 3). Fifty-four isolates were identified as unknown species based on either morphology or on rDNA ITS sequences (17.25%) (Table 3).
To assist species identification of all 313 strains, the rDNA ITS sequences were compared by Blast analysis against archived sequences in GenBank, which is a widely used approach for fungal identification. Unfortunately, only 14/27 species were corrected identified compared to the quality-controlled database at Westerdijk Fungal Biodiversity Institute (Table S1). Six species were misidentified including Cladosporium irritans (0.7% difference) as Toxicocladosporium banksiae, Cyphellophora fusarioides (1% difference) as Cyphellophora laciniata, E. cancerae as E. salmonis, O. humicola (0.2% difference) as O. mirabilis, O. musae as O. humicola and P. verrucosa as P. americana. Eight species (H. werneckii with 8% difference included) were identified at the genus or order level, including C. oxysporum/tenuissimum as Cladosporium sp., E. equina (11% difference) as Chaetothyriales sp., V. japonica (6% difference) as Chaetothyriales sp., H. werneckii (8% difference) as Acremonium sp., as well as E. equina, E. aquamarina (10% difference), E. nishimurae (5% difference), E. jeanselmei (5% difference) as Exophiala sp. The remaining Exophiala sp. was misidentified as E. alcalophila. Interestingly, 44/54 isolates previously identified as unknown species hit 4 known species on GenBank with variable coverage (66–99%) and identity (86–98%), although with different morphology. Four of fifty-four isolates identified as Chaetothyriales sp. with coverage (95–99%) and identity (90–93%). The remaining 6/54 isolates were identified as Herpotrichiellaceae sp. (n = 3), Exophiala sp. (n = 2) and Phialaphora sp. (n = 1), respectively.
An association analysis between isolation frequency and sampling areas indicated that the bathroom (n = 147, 46.96%) was the most common isolation site, followed by kitchen (n = 116, 37.06%), refrigerator (n = 32, 10.22%), washing machine (n = 17, 5.43%) and water dispenser (n = 1, 0.32%) (Table 4). The distribution of isolates was significantly variable in each location, which is proven by either unparametric Mann-Whitney U test (p = 0.015) of total isolates between four locations (water dispensers were not included in this test due to small data point) or one way ANOVA parametric test (p = 0.033), but rejected by a successive Turkey analysis among four locations (p = 0.064–0.995).
The analysis between isolation frequency of each species and sampling areas showed that in bathrooms, the most frequently encountered species were K. epidermidis (n = 26), E. lecanii-corni (n = 14), P. oxyspora (n = 12), O. musae (n = 12) and E. alcalophila (n = 10), while in kitchens, the most frequently isolated species were O. musae (n = 14), P. oxyspora (n = 12), and P. europaea (n = 10) (Table 5). In refrigerators, Cladosporium species such as C. halotolerans were commonly encountered (n = 9). In washing machines and water dispensers, only few black fungal isolates were found other than K. epidermidis (n = 6) and O. musae (n = 5) (Table 5).
Discussion
Implantation of contaminated thorns or wooden splinters has been hypothesized to be a main infection route of chromoblastomycosis or phaeohyphomycosis, and therefore the agents of both diseases probably originate from the natural environment2. Systemic occurrence of black yeasts probably has a pulmonary route of infection, for which hot indoor wet cells have been suggested, such as steambaths10. We establish the prevalence of black yeast-like fungi in the wet cell environment, which thus far has been neglected in indoor studies. Most studies of indoor fungi focus on airborne fungi (Alternaria, Cladosporium, Penicillium, Aspergillus, Rhizopus and Mucor). Those fungi grow very fast compared with black yeast-like fungi, which make it difficult to recover black yeast by direct isolation. The fungal community described in this study is very different from that of studies focus on indoor airborne fungi. Only few studies focus on the black yeast and melanzied fungi in indoor wet cell environments are available. Clarification of the prevalence of melanized fungi in the wet cell environment is significant to estimate potential transmission routes of melanized fungi. In this study, 399 of 509 samples taken in moist indoor sampling sites of human residences in Guangzhou, Southern China proved to be positive for black yeast-like and other melanized fungi. The remaining 110 samples showed no growth of black yeast-like fungi or growth of airborne contaminent fungi, e.g. Aspergillus and Penicillium species. The black yeast genus Exophiala, containing species that reproduce by budding with slimy conidia, was common (n = 77), followed by Knufia (n = 37), and Phialophora (n = 44), while also many strains of Rhinocladiella, Cyphellophora, and Cladophialophora were encountered. All these genera belong to a single fungal order, Chaetothyriales. The genus Ochroconis, which is unrelated (order Venturiales) but has similar ecology as Exophiala was also common (n = 38). Except for E. dermatitidis, the majority of isolated fungi have been listed by de Hoog et al.17 as potential agents of mild skin and nail infections, confirming Lian and de Hoog7 who noticed that bathrooms harbour a remarkably large number of skin fungi that were until then not known from environmental sources. Lian and de Hoog7 hypothesized that human skin that is softened during bathing might be more vulnerable to fungal infection. Our study underlines that indoor wet areas serve as reservoir for fungi known to be involved in human infections18. Temperatures of our sampling sites usually do not exceed 37 °C, which might be a reason that only species were encountered that infect human skin and nails, rather than species causing systemic infections. In contrast, hot moist indoor environments such as steambaths10 and dishwashers19 are regularly colonized by thermophilic systemic opportunists, such as the black yeast Exophiala dermatitidis and the white yeasts Candida parapsilosis and Magnusiomyces capitatus.
Exophiala species are often isolated from indoor water sources, such as sinks, drainpipes, swimming pools, and bathing facilities, and also occur in municipal drinking water enabling biofilm formation20,21. Chaetothyrialean species of Table 3 are involved in mild cutaneous and nail mycoses22. Our isolation of E. dermatitidis from a kitchen (n = 4) and from the rubber seal of a refrigerator (n = 1) may be coincidental. The fungus can occasionally be found in sites with lower temperature, such as sinks and humidifiers, but is very abundant in steambaths10.
Knufia epidermidis was a common species in the bathroom. It was first described from a superficial skin lesion with blackish discoloration in an 80-yr-old Chinese patient23. Subsequent studies detected this species in human toes, skin and nails22,24.
Phialophora europaea, currently classified in Cyphellophora25, is among the fungi causing mild skin and nail infections22,26,27. The species thus far has rarely been found outside the human host, but we encountered it at high isolation rates in bathrooms and kitchens. The genus Cyphellophora, along with some species formerly classified in Phialophora phylogenetically separate from the main group of human opportunists in Herpotrichiellaceae. The two Cyphellophora species identified in this study suggest wide spread in indoor wet cells. Cladophialophora boppii seems to have a similar ecology, occasionally being found as a colonizer on human skin and nails28, as well as in bathrooms22. Rhinocladiella similis was originally described as the sympodial counterpart of Exophiala jeanselmei, and has occasionally been reported from human infections29. We isolated several strains from bathroom facilities, confirming data of Matos et al.30.
Ochroconis musae (order Venturiales) was the most frequently encountered species in kitchens and bathrooms, confirming findings of Lian and de Hoog7. The species was originally described as O. mirabilis from strains from humans and moist sources31, but Lu et al.32 had found a strain of the same species on a banana leaf and introduced the O. musae one month earlier. However, it is not a plant-associated fungus but is regularly associated with low-nutrient waters31. It occasionally causes skin and nail infections of immunocompetent patients33,34, and is particularly common as an invader of cold-blooded, waterborne vertebrates such as frogs and fish31. Yew et al.35 found that melanin production, osmoregulation, expanded gene family encoding taurine catabolism dioxygenase (TauD/TdfA domain), glutathione-S-transferase domains and RTA1-like protein families enabled the fungus to thrive under hostile oligotrophic conditions, such as low-nutrient and moist environments.
Hortaea werneckii (order Capnodiales) is a halophilic black yeast36. In humans it is known from superficial colonization the hand called tinea nigra37,38 which is explained by salty cutaneous conditions e.g. during beach holidays. Our consistent finding of the species in indoor environments demonstrates that the halotolerant amplitude of the fungus is very wide and includes oligotrophic habitats. The fungus has no opportunistic potential.
Cladosporium species (order Capnodiales) are airborne fungi, considered as biofilm formers and contaminants with no clinical significance. Cladosporium halotolerans was recently segregated from C. sphaerospermum as an osmotolerant fungus commonly isolated from hypersaline water of salterns and other environments with low water activity such as peanut shells39; it is regularly encountered on bathroom walls40. In humans Cladosporium species are isolated from the respiratory tract (54.5%), followed by superficial (28.4%) and deep tissues and fluids (14.7%)41, but very few unambiguously proven clinical cases exist. The most common isolation sites of Cladosporium species such as C. irritans were rubber seals of refrigerators and bathrooms, suggesting temperature-independent oligotrophism. Cladosporium oxysporum/tenuissimum was isolated from the same sites as C. halotolerans, indicating similar ecology. Cladosporium irritans otherwise has been reported from plant leaves and other environmental sources42,43 and from respiratory specimens without signs of invasion41. Cladosporium is preponderant in the airborne mycobiota and considered as important allergenic fungi44.
In conclusion, we noticed that most of the black fungi that were recovered from indoor wet cells mostly belong to the order Chaetothyriales and the family Herpotrichiellaceae within this order it otherwise known to contain numerous agents of superficial skin and nail infections. Maximum growth temperatures of most species are around 40 °C. Notably, the related fungus Exophiala dermatitidis, able to grow above 40 °C and found at higher temperatures in the indoor environment, viz. in steambaths and dishwashers, potentially causes disseminated, potential fatal human infections. Exophiala species with maximum growth temperatures around 33 °C are particularly found infecting waterborne cold-blooded animals45. This suggests that thermotolerance is an important virulence factor in black fungi, determining that the fungi which are not capable of growing under human conditions (33–37 °C) are not able to infect humans.
In addition to the above known black yeast-like fungi, a high rate of unknown strains (n = 54, 17.25%) was recovered in our dataset. The most interesting finding is that 44/54 isolates, which previously identified as unknown species against the black yeast database at Westerdijk Fungal Biodiversity Institute, hit 4 known species on GenBank. The remained 10 isolates were also identified in order or genus level. It suggests the stochastic blast analysis on a massive, multifarious nucleotides database, such as GenBank, may guide the further identification of novel species. However, when all 259 known strains sequences were blasted on this database, only 14/27 species hit the correct nucleotides sequences archived in GenBank. The remained 13 species were identified in genus level or completely misidentified. One reasonable explanation is that Genbank is a stock center for nucleotides sequences which deposited by researcher worldwide. The blast analysis only depend on the nucleotides sequences similarity, not refer to either morphology or other test. Another reason is that frequently nomenclature change of dispute species, thus this results suggest that well maintained and organized database will be the preference for novel isolates identification, e.g. the database at Westerdijk Fungal Biodiversity Institute for black yeast. Nevertheless, further studies are needed to describe these strains as novel taxa.
References
Satow, M. M., Attili-Angelis, D., de Hoog, G. S., Angelis, D. F. & Vicente, V. A. Selective factors involved in oil flotation isolation of black yeasts from the environment. Stud Mycol. 61, 157–163 (2008).
Vicente, V. A. et al. Environmental isolation of black yeast-like fungi involved in human infection. Stud Mycol. 61, 137–144 (2008).
Gumral, R. et al. Black yeast diversity on creosoted railway sleepers changes with ambient climatic conditions. Microb Ecol. 68, 699–707 (2014).
Muggia, L., Kocourkova, J. & Knudsen, K. Disentangling the complex of Lichenothelia species from rock communities in the desert. Mycologia. 107, 1233–1253 (2015).
Lutzoni, F., Pagel, M. & Reeb, V. Major fungal lineages are derived from lichen symbiotic ancestors. Nature. 411, 937–940 (2001).
Figel, I. C. et al. Black yeasts-like fungi isolated from dialysis water in hemodialysis units. Mycopathologia 175, 413–420 (2012).
Lian, X. & de Hoog, G. S. Indoor wet cells harbour melanized agents of cutaneous infection. Med Mycol. 48, 622–628 (2010).
Zalar, P., Novak, M., de Hoog, G. S. & Gunde-Cimerman, N. Dishwashers–a man-made ecological niche accommodating human opportunistic fungal pathogens. Fungal Biol. 115, 997–1007 (2011).
Nishimura, K., Miyaji, M., Taguchi, H. & Tanaka, R. Fungi in bathwater and sludge of bathroom drainpipes. 1. Frequent isolation of Exophiala species. Mycopathologia. 97, 17–23 (1987).
Sudhadham, M. et al. The neurotropic black yeast Exophiala dermatitidis has a possible origin in the tropical rain forest. Stud Mycol 61, 145–155 (2008).
Suri, P., Chhina, D. K., Kaushal, V., Kaushal, R. K. & Singh, J. Cerebral Phaeohyphomycosis due to Cladophialophora bantiana - A Case Report and Review of Literature from India. J Clin Diagn Res. 8, DD01–05 (2014).
Xi, L. et al. Molecular diversity of Fonsecaea (Chaetothyriales) causing chromoblastomycosis in southern China. Med Mycol. 47, 27–33 (2009).
Sun, J. et al. Molecular characterization of pathogenic members of the genus Fonsecaea using multilocus analysis. PLoS One. 7, e41512 (2012).
Lu, S. et al. Chromoblastomycosis in Mainland China: a systematic review on clinical characteristics. Mycopathologia. 175, 489–495 (2013).
Li, X. Q. et al. The role of melanin pathways in extremotolerance and virulence of Fonsecaea revealed by de novo assembly transcriptomics using illumina paired-end sequencing. Stud Mycol. 83, 1–18 (2016).
Yang, Y. P., Li, W., Huang, W. M., Zhou, Y. & Fan, Y. M. Chromoblastomycosis caused by Fonsecaea: clinicopathology, susceptibility and molecular identification of seven consecutive cases in Southern China. Clin Microbiol Infect. 19, 1023–1028 (2013).
Moussa, T. A. et al. Nomenclatural notes on Nadsoniella and the human opportunist black yeast genus. Exophiala. Mycoses. 60, 358–365 (2017).
Hilmarsdottir, I., Haraldsson, H., Sigurdardottir, A. & Sigurgeirsson, B. Dermatophytes in a swimming pool facility: difference in dermatophyte load in men’s and women’s dressing rooms. Acta Derm Venereol. 85, 267–268 (2005).
Dogen, A., Kaplan, E., Ilkit, M. & de Hoog, G. S. Massive contamination of Exophiala dermatitidis and E. phaeomuriformis in railway stations in subtropical Turkey. Mycopathologia. 175, 381–386 (2013).
Heinrichs, G., Hubner, I., Schmidt, C. K., de Hoog, G. S. & Haase, G. Analysis of black fungal biofilms occurring at domestic water taps. I: compositional analysis using Tag-Encoded FLX Amplicon Pyrosequencing. Mycopathologia. 175, 387–397 (2013).
Heinrichs, G., Hubner, I., Schmidt, C. K., de Hoog, G. S. & Haase, G. Analysis of black fungal biofilms occurring at domestic water taps. II: potential routes of entry. Mycopathologia. 175, 399–412 (2013).
Saunte, D. M., Tarazooie, B., Arendrup, M. C. & de Hoog, G. S. Black yeast-like fungi in skin and nail: it probably matters. Mycoses. 55, 161–167 (2012).
Li, D. M., de Hoog, G. S., Saunte, D. M., van den Ende, A. H. & Chen, X. R. Coniosporium epidermidis sp. nov., a new species from human skin. Stud Mycol. 61, 131–136 (2008).
Li, D. M. & Chen, X. R. A new superficial fungal infection caused by Coniosporium epidermidis. J Am Acad Dermatol. 63, 725–727 (2010).
Reblova, M., Untereiner, W. A. & Reblova, K. Novel evolutionary lineages revealed in the Chaetothyriales (fungi) based on multigene phylogenetic analyses and comparison of its secondary structure. PLoS One. 8, e63547 (2013).
Decock, C., Delgado-Rodriguez, G., Buchet, S. & Seng, J. M. A new species and three new combinations in Cyphellophora, with a note on the taxonomic affinities of the genus, and its relation to Kumbhamaya and Pseudomicrodochium. Antonie Van Leeuwenhoek. 84, 209–216 (2003).
Feng, P. et al. In vitro activities of nine antifungal drugs against 81 Phialophora and Cyphellophora isolates. Antimicrob Agents Chemother. 56, 6044–6047 (2012).
Brasch, J. et al. Toenail infection by Cladophialophora boppii. Med Mycol. 49, 190–193 (2011).
de Hoog, G. S. et al. Species diversity and polymorphism in the Exophiala spinifera clade containing opportunistic black yeast-like fungi. J Clin Microbiol. 41, 4767–4778 (2003).
Matos, T., de Hoog, G. S., de Boer, A. G., de Crom, I. & Haase, G. High prevalence of the neurotrope Exophiala dermatitidis and related oligotrophic black yeasts in sauna facilities. Mycoses. 45, 373–377 (2002).
Samerpitak, K., Gerrits van den Ende, B. H., Stielow, J. B., Menken, S. B. & de Hoog, G. S. Barcoding and species recognition of opportunistic pathogens in Ochroconis and Verruconis. Fungal Biol. 120, 219–230 (2016).
Lu, H. et al. A new species of Scolecobasidium associated with the sooty blotch and flyspeck complex on banana from China. Mycological Progress. 12, 489–495 (2013).
Kaur, H., Rudramurthy, S. M., Mohindra, S., Gupta, S. & Chakrabarti, A. Ochroconis humicola coexisting with esthesioneuroblastoma: an incidental coloniser or allergen? Mycopathologia. 178, 79–83 (2014).
Shi, D. et al. Subcutaneous infection by Ochroconis mirabilis in an immunocompetent patient. Med Mycol Case Rep. 11, 44–47 (2016).
Yew, S. M. et al. The genome of newly classified Ochroconis mirabilis: Insights into fungal adaptation to different living conditions. BMC Genomics. 17, 91 (2016).
Plemenitas, A., Vaupotic, T., Lenassi, M., Kogej, T. & Gunde-Cimerman, N. Adaptation of extremely halotolerant black yeast Hortaea werneckii to increased osmolarity: a molecular perspective at a glance. Stud Mycol. 61, 67–75 (2008).
de Hoog, G. S. & Gerrits van den Ende AH. Nutritional pattern and eco-physiology of Hortaea werneckii, agent of human tinea nigra. Antonie Van Leeuwenhoek. 62, 321–329 (1992).
Bonifaz, A. et al. Tinea nigra by Hortaea werneckii, a report of 22 cases from Mexico. Stud Mycol. 61, 77–82 (2008).
Zalar, P. et al. Phylogeny and ecology of the ubiquitous saprobe Cladosporium sphaerospermum, with descriptions of seven new species from hypersaline environments. Stud Mycol. 58, 157–183 (2007).
Segers, F. J. et al. The Indoor Fungus Cladosporium halotolerans Survives Humidity Dynamics Markedly Better than Aspergillus niger and Penicillium rubens despite Less Growth at Lowered Steady-State Water Activity. Appl Environ Microbiol. 82, 5089–5098 (2016).
Sandoval-Denis, M. et al. Cladosporium Species Recovered from Clinical Samples in the United States. J Clin Microbiol. 53, 2990–3000 (2015).
Crous, P. W., Braun, U., Schubert, K. & Groenewald, J. Z. Delimiting Cladosporium from morphologically similar genera. Stud Mycol. 58, 33–56 (2007).
Crous, P. W. & Groenewald, J. Z. Why everlastings don’t last. Persoonia. 26, 70–84 (2011).
de Ana, S. G., Torres-Rodriguez, J. M., Ramirez, E. A., Garcia, S. M. & Belmonte-Soler, J. Seasonal distribution of Alternaria, Aspergillus, Cladosporium and Penicillium species isolated in homes of fungal allergic patients. J Investig Allergol Clin Immunol. 16, 357–363 (2006).
de Hoog, G. S. et al. Waterborne Exophiala species causing disease in cold-blooded animals. Persoonia. 27, 46–72 (2011).
Acknowledgements
This work was supported in part by National Natural Science Foundation of China (81301411, 81571970, 81671992 and 81403197). The Nature Science Foundation of Guangdong Province, China (S2012010008614, S2013010013175).
Author information
Authors and Affiliations
Contributions
X.F.W., performed the experiments and wrote the manuscript; W.Y.C., v.d.E.A.H., J.M.Z. and T.X., helped collect the samples and performed statistical analysis; X.Q.L. and J.F.S., degsined the experiment; G.S.d.H. and L.Y.X. helped review and modified the manuscript. All authors have reviewed and approved the fnal content.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, X., Cai, W., van den Ende, A.H.G.G. et al. Indoor wet cells as a habitat for melanized fungi, opportunistic pathogens on humans and other vertebrates. Sci Rep 8, 7685 (2018). https://doi.org/10.1038/s41598-018-26071-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-26071-7
This article is cited by
-
Fungal Colonization of the Airways of Patients with Cystic Fibrosis: the Role of the Environmental Reservoirs
Mycopathologia (2024)
-
Sugarcane: an unexpected habitat for black yeasts in Chaetothyriales
IMA Fungus (2023)
-
A re-evaluation of the Chaetothyriales using criteria of comparative biology
Fungal Diversity (2020)
-
Microorganisms populating the water-related indoor biome
Applied Microbiology and Biotechnology (2020)
-
Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi
Fungal Diversity (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.