Abstract
Skeletal muscle in the forelimb develops during embryonic and fetal development and perinatally. While much is known regarding the molecules involved in forelimb myogenesis, little is known about the specific mechanisms and interactions. Migrating skeletal muscle precursor cells express Pax3 as they migrate into the forelimb from the dermomyotome. To compare gene expression profiles of the same cell population over time, we isolated lineage-traced Pax3+ cells (Pax3EGFP) from forelimbs at different embryonic days. We performed whole transcriptome profiling via RNA-Seq of Pax3+ cells to construct gene networks involved in different stages of embryonic and fetal development. With this, we identified genes involved in the skeletal, muscular, vascular, nervous and immune systems. Expression of genes related to the immune, skeletal and vascular systems showed prominent increases over time, suggesting a non-skeletal myogenic context of Pax3-derived cells. Using co-expression analysis, we observed an immune-related gene subnetwork active during fetal myogenesis, further implying that Pax3-derived cells are not a strictly myogenic lineage, and are involved in patterning and three-dimensional formation of the forelimb through multiple systems.
Similar content being viewed by others
Introduction
Skeletal muscle formation in the forelimb during embryogenesis is a tightly regulated and controlled process. Forelimb muscles derive from paraxial mesoderm-derived anatomical structures called somites. Somites segment themselves into the myotome, sclerotome, and dermomyotome. The dermomyotome is divided into epaxial and hypaxial layers, the latter of which is the origin of all skeletal muscle of the trunk and back1,2. Pax3 is a homeodomain sequence-specific transcription factor (SSTF) that marks all somite-derived skeletal muscles in the forelimb. Pax3 is expressed starting at embryonic day (E) 10 in embryonic myogenic progenitor cells (EMPCs), which triggers migration and delamination of EMPCs from the ventrolateral lip of the hypaxial dermomyotome into the limb bud3,4,5,6. In Pax3 knockout (KO) mutant mice, myogenic progenitor cells fail to migrate and delaminate from the somite, which ultimately leads to a forelimb deficient of skeletal muscle7,8.
After EMPCs colonize the limb bud, skeletal muscle forms in distinct, successive stages9. Between E10 and E12, embryonic myoblasts fuse into embryonic myotubes. Between E12 and E16, fetal myoblasts fuse with both each other and embryonic myotubes to form fetal myofibers that serve as the foundation for future skeletal muscle. During this process, significant changes occur in gene expression10 and the underlying gene regulatory networks11,12, but little information is known regarding specifics that drive the molecular processes. Many of the mechanisms that take place during myogenesis are re-activated during skeletal muscle regeneration in adults, including the activation of skeletal muscle-specific SSTFs13, making it possible to translate any insights gained between systems. Since all known forelimb skeletal muscles derive from Pax3+ progenitor cells, the Pax3EGFP lineage offers a genetic tool to uncover the molecular processes that determine forelimb myogenesis and organogenesis. By observing the gene expression profiles of Pax3EGFP cells across the developmental time course as they migrate from the dermomyotome into forelimb, we can identify the molecular players coincident with muscle stages as they are formed and maintained in coordination with other cell lineages in the developing limb structure.
Network analysis is a quantitative paradigm for analyzing biological systems as individual parts working and interacting together14,15,16. Technological advances combined with reduced prices in next-generation sequencing have resulted in development of advanced techniques for network analysis of cell specific changes in organ development and disease17. Graphical representation via network analysis of gene expression data enables the visualization of complex interactions in large data sets in an intuitive format. In such a representation, nodes represent genes that are then connected to each other via edges that represent interactions. A specific type of network, co-expression networks, are created from transcriptomics data to reveal patterns of gene expression in dynamic systems18,19,20, and have been used to identify cell-type specific patterns of gene expression during development and the changes in regulatory interactions responsible for cell-state phenotypes21,22, among other uses.
Applying co-expression analysis to Pax3EGFP lineage-traced myoblasts provides a model system to decode the mechanisms behind embryonic and fetal myogenesis in the forelimb. In this study, we used next generation RNA sequencing of lineage-traced cells isolated through fluorescent-activated cell sorting (FACS-Seq) to perform differential expression and co-expression analysis during distinct stages of embryonic development. We discovered that the Pax3EGFP lineage harbors several cell populations not previously defined, including cells that will likely populate the immune and hematopoietic systems parallel to the already known skeletal muscle, smooth muscle, and neuronal systems. Development of these diverse systems is tightly orchestrated as cells migrate from the dermomyotome, enter the forelimb space, and receive signals from the highly plastic environment. SSTFs integrate external signals during patterning with shifting gene expression networks that coordinate the migration, proliferation, differentiation, and integration of cell types into fully functioning organs and multi-system limb structures. For example, homeodomain SSTFs in combination of Shh, Fgf and Wnt signaling dominate the early patterning events in embryonic forelimb myogenesis, followed by the rise in importance of zinc-finger and helix-turn-helix SSTFs in fetal states. In this study, we observed that Pax3-derived cells contribute more fully to the three-dimensional formation of the forelimb than previously thought, and give rise to cells with characteristics of the skeletal, vascular, nervous, hemolymphoid and immune systems in addition to muscle. Thus, the dermomyotome might give rise to more many cell populations than originally thought.
Results and Discussion
Isolation of Pax3-derived embryonic forelimb cells
To trace the genes involved in myogenesis in the forelimb in real time, we used a transgenic mouse model genetically composed of a Pax3Cre driver23 combined with a ROSA26EGFP tracer24. When both genotypes are combined into one mouse, all cells that at any point ever expressed Pax3 will also express EGFP, including any and all daughter cells (lineage tracer). This system enables the tracking of the same cell population in the mouse forelimb over time as it develops and differentiates. We chose E11, E12, E13, and E14 as time points for analysis to trace development from the beginning of embryonic myogenesis, when the Pax3+ dermomyotome-derived cells enter the myogenic lineage, to the onset of fetal myogenesis, when the myoblasts/myotubes start to form myofibers. Mouse embryos at each stage show strong EGFP expression, especially in the forelimbs (Fig. 1a). As the forelimb develops, individual digits and muscle groups develop too, seen clearly at E14. FACS25 was used to isolate EGFP expressing cells (Pax3EGFP) at each stage. Density-based scatter plots that represent EGFP fluorescence intensity vs. cell size show two distinct cell populations in each stage (Fig. 1b), EGFP-positive and EGFP-negative cells. A histogram representation gives a more clear image of the two distinct cell populations (Fig. 1c). Pax3EGFP cells comprise 92% of the whole cell population of the forelimb at E11 and E12 (Fig. 1b) in agreement with strong EGFP-fluorescence seen by microscopy (Fig. 1a). At E13, the Pax3EGFP cell population was reduced to 68% (and was further reduced at E14) due to reduced efficiency of our tissue disaggregation/cellular dissociation procedure (Fig. 1a,b). The onset of fetal myogenesis occurs between E12 and E13, when embryonic myofibers fuse with fetal myoblasts/myotubes to form fetal myofibers. The cytoskeletal rearrangements that occur among cells at E12-E13 generates a larger extracellular matrix which imparts resistance to our enzo-mechanical dissociation process (see Materials and Methods), and many cells were filtered out as clumps including dense tissue that failed to dissociate. The exact genes and molecular mechanisms involved in this process remain elusive and would be interesting to study.
Pax3EGFP expression in mouse embryonic forelimbs. (a) Fluorescent microscopy showing Pax3EGFP expression based on a Pax3Cre|Rosa26EGFP driver at E11, E12, E13, and E14 forelimbs. (b) Scatter plots from FACS showing EGFP intensity on the x-axis, and forward scatter (FSC) on the y-axis. Gate R5 shows 92%, 92%, 68%, and 61% EFGP-positive cells in forelimbs at E11, E12, E13, and E14, respectively. (c) Histograms depict EGFP intensity on the x-axis vs cell number (count) on the y-axis. Green peaks represent EFGP-positive populations based on gating from R5.
Gene expression profiling of Pax3-derived embryonic forelimb cells
After sorting, total RNA from each sample was extracted and tested by the Bioanalyzer for quality control. Only high-quality samples with an RNA Integrity Number (RIN) above 7.0 were retained for library preparation and processed for sequencing. Upon aligning the mapped sequence reads to the published murine mm10 genome and calculating the differentially expressed (DE) genes between each time point, a quality control step was performed via principal component analysis (PCA)26. The PCA plot (Fig. 2a) shows the variability between biological replicates in our system, and emphasizes the value of ample biological replicates for a study like this. The PCA plot also shows a distinct clustering of samples by stage with greater biological variation between stages that among samples from the same stage. Interestingly, the clustering of samples by stages appears to follow a developmental trajectory. Samples from E11 cluster in the bottom left and follow a horizontal parabola-like trajectory through E14, suggesting that time of conception is a significant factor in our analysis, as would be expected. Vaginal plug checking to determine timing of pregnancy was performed only once per day and therefore litters could be up to 12 hours apart in age but still marked as the same embryonic day for analysis. Variability may be accentuated at early stages when developmental changes are more dramatic.
Differential expression (DE) and Gene Ontology (GO) term analysis of RNA-Seq data from sorted, Pax3EGFP cells. (a) Principal component analysis (PCA) and plot of all 28 samples. PCA shows good clustering of samples by biological time point, with variation between samples in the same group. Samples appear to follow the developmental trajectory. (b) Heatmap of signed difference ratio (SDR) based on all 4,481 DE genes between any two consecutive developmental states. Columns represent samples, and each row represents one DE gene. Yellow indicates high expression and blue indicates low expression, relative to the average expression of each gene between all samples. Red, green, blue, and purple bars on the left indicate clusters of DE genes expressed at E11, E11, E12, E13 and E14, respectively.
Gene expression levels were calculated for each sample. DE genes were determined and a heatmap was generated based on the signed difference ratio (SDR) from log2-normalized reads (Fig. 2b). Distinct clusters of genes form at each stage based on their expression patterns. Clusters of interest are graphically delineated with red, green, blue, and purple boxes to the left of the heatmap.
Genes segregated with the red cluster were expressed specifically at E11, implying that they are early embryonic myogenesis markers. Gene ontology (GO) term enrichment analysis of the red cluster revealed an overrepresentation of genes associated with pattern specification processes (false discovery rate, FDR = 6.49 × 10−5), neuron differentiation (FDR = 0.008), and appendage morphogenesis (FDR = 2.03 × 10−4). Example genes in these categories were primarily homeodomain SSTFs represented by the Hox family, in agreement with previous reports that associated the Hox genes with regulation of patterning and digit formation in the embryonic limb27,28,29, including the Hoxc and Hoxd family genes expressed at E1110.
Genes segregated within the green cluster were expressed during E11 and E12 (Fig. 2b), and marked embryonic myogenesis. GO term functional annotation enrichment analysis indicated that “green” genes are over-represented with those involved in epithelial tube morphogenesis (FDR = 0.009), central nervous system (CNS) development (FDR = 2.24 × 10−4), mesenchyme development (FDR = 9.66 × 10−2), and neuron fate commitment (FDR = 4.71 × 10−4), among others, suggesting that formation of the CNS is taking place during E11–E12. Since Pax3EGFP cells are known to mark all cells in the skeletal muscle lineage in the forelimb, detection of many so genes that are not usually expressed in myoblasts was surprising.
Genes clustered in the blue box (Fig. 2b; Fig. S1) showed high expression levels at E13 and E14 which coincides with the onset of fetal development, and they were involved in angiogenesis (FDR = 2.61 × 10−13), negative regulation of cell proliferation (FDR = 2.46 × 10−8), and differentiation (FDR = 1.60 × 10−8). Example genes in the “blue” cluster included the angiogenesis markers Angpt2 and Anpep, and negative markers of cell proliferation such as Ar and Dpt. Their expression suggests that in fetal states cells of the Pax3EGFP lineage stop proliferating, exit the cell cycle and possibly enter the smooth muscle cell lineage. Angiogenesis and myogenesis are highly interrelated and co-dependent during forelimb development. Expression of certain angiogenesis-related genes can increase the rate of muscle regeneration in adult skeletal muscle30,31. Additionally, colonization of vascular cells in the developing forelimb is required for migration of Pax3+ myoblasts into the limb bud32, implying communication between muscle and vascular systems during development.
Genes in the purple cluster (Fig. 2b; Fig. S1) were expressed explicitly at E14 and were associated with immune response regulation (FDR = 9.49 × 10−39) and processes (FDR = 9.51 × 10−54). This cluster included interleukin receptors and the CD antigens Ccl6, Cd44, Il20rb, and Ciita. There is little information on the interaction between skeletal muscle and immune systems during fetal development, so the inclusion of immune system-related genes in our analysis of Pax3+ cells was a bit surprising. All cells were sorted to a final purity of 97–99% (data not shown), so genes detected in our analysis were unlikely to have originated in non-green cells. As confirmation, we compared our DE genes with those from a similar study by Biressi et al.10, and found a similar list of immune-related genes such as Anxa1, Cd44, and Myb among others. Previous studies have shown that macrophage infiltration and inflammation occur during satellite cell-mediated skeletal muscle regeneration33,34 in adults. Although not a developmental process, many developmental mechanisms are reactivated during adult regeneration of skeletal muscle. It should be noted that these genes were specifically expressed at the latest stage of development that we sampled, E14, after the onset of fetal development, and unlikey to mark angiogenesis.
Pax3 expression in non-myogenic embryonic forelimb cells
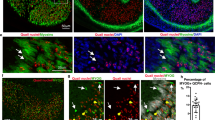
To further investigate the gene expression profiling findings, we performed double labelled immunohistochemistry for Pax3 and the myogenic markers Myf5 and Myog (Figs. 3a–d1) in forelimbs from E11 and E12 mice. Myf5 marks skeletal muscle cells and brown and white adipocytes35, while Myog marks cells committed to the myogenic lineage36. Pax3+ cells largely overlap with the Myf5+ in skeletal muscle at E11 (Fig. 3a,a1) and both brown fat and subcutaneous white fat37. At E12, three distinct cell populations were detected: Pax3+, Myf5+ and Pax3+Myf5+ double positive (Fig. 3b,b1). At E11, only a small Pax3+Myog+ cell population was detected (Fig. 3c,c1). By E12, cells are committed to the muscle cell lineage and the Pax3+Myog+ cell population was enlarged (Fig. 3d,d1). Similarly, triple labeling immunohistochemistry on E11 Pax3EGFP forelimbs using antibodies against EGFP (to mark the Pax3-derived cells), Myog and Pitx2 (to mark the skeletal muscle cells) was performed38 (Fig. 3e,e1). Several cell populations were detected besides the Pax3+Myog+ and Pax3+Pitx2+ populations, suggesting the presence of non-myogenic cell types within the Pax3EGFP lineage. Immunohistochemistry on E12 Splotch (Pax3Sp) mice, a natural mutation of the Pax3 locus that results in ablation of skeletal muscle in the forelimbs, also indicated the presence of a small population of Pax3+Pitx2− cells in the forelimb (Fig. 3f,g), further supporting the observations that Pax3-derived cells populate other lineages. These observations were in accord with previous studies showing that a subset of Pax3EGFP cells in the forelimb differentiate into vascular epithelial cells39. In limb formation, the vascular and nervous systems develop in parallel to the skeletal system40,41, and the Pax3 lineage is likely to give rise to cells that will populate different systems42,43.
Myogenic and non-myogenic cells populate the Pax3EGFP lineage in the forelimb. (a–d) Immunohistochemistry of E11 (a,a1,c,c1) and E12 (b,b1,d,d1) forelimb frontal sections of wild type mice for Pax3, Myf5 and Myog. (a1–d1) higher magnification of the a–d images. (e) Immunohistochemistry of E11 forelimb frontal sections of Pax3EGFP mice for EGFP (Pax3), Myog and Pitx2. (e1) higher magnification of the e image. (f,g) Immunohistochemistry of E12 forelimb frontal sections of Pitx2Z/+ (f) and Pitx2Z/+|Pax3Sp/Sp (g) mouse for ß-gal(Pitx2) and Pax3.
Construction of co-expression network during forelimb development
To observe the biological network underlying forelimb development we performed a co-expression analysis, using differentially expressed (DE) genes. A single co-expression network was constructed from pairwise correlation coefficients between each of 4,481 DE genes, using all samples. We opted to construct a single network for all biopsies, rather than state-specific individual networks, to increase the power of our analysis. We focused only on genes that were DE between consecutive developmental states to highlight the genes of most biological relevance, and to decrease computational time. Upon calculating Pearson correlation coefficients (PCC) in a pairwise manner, we determined a FDR cutoff for significant correlation using the following rationale. The p-value choice reflected the condition that the node-degree distribution of biological networks closely follows a scale-free distribution14. We plotted the p-value cutoff vs. the R2 of a best-fit power line for the resulting node-degree distribution and observed that our co-expression network fits a scale-free topology well (Fig. S2a), allowing the choice of a p-value cutoff of 1E-16 resulting in an R2 cutoff of 0.88. The resulting network had a scale-free degree distribution (Fig. S2b) and a giant component comprising the vast majority (97%) of the nodes, consistent with previous studies of gene regulatory networks44. Ultimately, a network with 682 nodes and 3,655 edges was generated, with an average node degree of 10.7.
When the network was graphically visualized with Cytoscape software45, we observed a single network composed of two mostly independent subnetworks, with smaller individual networks present (Fig. 4). Each node (circle) represents a gene transcript, and edges represent significant correlation between the transcripts. GO term enrichment analysis revealed an overrepresentation of cytoskeletal, skeletal and neuronal system-related genes (skeletal system, neuronal system), and immune response-related genes (immune system, hematopoietic system). The strong presence of the immune and skeletal system-related genes implied that two different transcriptional co-expression networks co-exist during forelimb development with little interaction between them. To identify modules, which are clusters of highly interconnected nodes that together perform a specific biological function16, we used the MCL package in R statistical software and performed Markov clustering46. Markov clustering identifies modules by simulating flow in networks, and determining the clusters in which the most flow accumulates. However, the weakness of this method was that it assigned each gene to only a single module, which rarely reflects the true underlying biology. Using a module size cutoff of eight, markov clustering identified seven modules, marked by number and color. Two modules (blue6, purple5) comprise one subnetwork, and two (pink7, teal3) comprised another subnetwork. The three smaller modules (red4, green1, yellow2) were mostly independent of either subnetwork (Fig. 4).
Co-expression network and module identification of the Pax3EGFP lineage in the forelimb. The generated co-expression network was visualized in Cytoscape software. Nodes (transcripts) are shown as circles, with size proportional to the degree of the node (i.e., the number of neighbor nodes to which it is connected in the network). Seven modules with at least nine nodes were identified via markov clustering, and are color-coded accordingly. The full co-expression network is comprised of two, mostly-distinct subnetworks.
GO term enrichment identified significant overrepresentation of collagen fibril organization (FDR = 6.38 × 10−11), extracellular matrix organization (7.56 × 10−11), and skeletal system morphogenesis (FDR = 8.69 × 10−04) related genes in the teal3 module, but no significant enrichment in the pink7 module. The teal3 module represented the muscular and skeletal systems during development, and identification of such a module was expected. Most components of the skeletal system were lumped into one module rather than multiple separate modules with more specific functions, suggesting that the skeletal and muscular systems are intertwined and co-dependent during development, and/or share common mechanisms. Immune and defense-related GO terms such as immune system processes (FDR = 3.61 × 10−5) and defense response (FDR = 9.52 × 10−6) were overrepresented in the blue6 module, while T-cell related GO terms such as mast cell activation (FDR = 2.29 × 10−9) and T-cell proliferation (FDR = 5.26 × 10−8) were overrepresented in the purple5 module. Unlike the teal3 module, both immune-related modules were expressed most strongly at E14, implying that immune-related genes are expressed most highly later in fetal myogenesis, rather than at the onset.
Of the smaller modules, the yellow2 module showed enrichment in genes related to vasculogenesis (FDR = 1.38 × 10−2) and angiogenesis (3.49 × 10−4). Unlike the immune system-related modules, the yellow2 module was not connected to the main network (Fig. 4). Since it was such a small module, angiogenesis and vascularization are likely a relatively small part of the Pax3EGFP lineage in the mouse forelimb. It should also be noted that the network was constructed to represent all states of development. If co-expression of myogenesis and angiogenesis-related genes is strongest during late embryonic or early fetal myogenesis, it would not be detected in the current network analysis. The red4 module showed no significant enrichment in specific GO terms, making its function difficult to assess. Observing the individual genes in the red4 network, the presence of the SSTF Gata2 points to a possible function related to the hematopoietic system, since Gata2 has been shown to be a marker of hematopoietic cells in early development47. This argument was strengthened by the proximity and weak connection of the red4 module to the purple5 module, rather than the teal3 module, because of the interdependence and co-regulation between the hematopoietic and immune systems48. The only module that showed high overall expression during the early developmental states is the pink7 module, which contained the embryonic myoblast-marker genes Crip1, Lhx9, Mecom, Phf6, Prdm16, and Sox11. Also, it needs to be noted that these are RNA-based measurements, and that protein abundances may not correlate exactly with RNA abundances. This is a topic of great interest that could lead to the discovery of unknown and/or novel cell states.
An alternate explanation for the presence of immune-related genes in Pax3+ cells during fetal development could be that the GO term enrichment analysis is inherently biased to some degree because it only takes into account the known and annotated functions of genes. Because of the prevalence of pleiotropy in humans as well as the rich hierarchy of functional annotations in the gene ontology, most genes have multiple, if not dozens of annotated biological functions that can be context-dependent based on tissue type or other variables. Genes that have only been studied in only one system are likely annotated with incomplete information in regard to functions in our contexts. Additionally, certain GO terms such as “immune response” are semantically broad and thus somewhat loosely defined. Taken together, a GO term enrichment analysis could include the wrong context of one or more genes, and bias the results in a way that does not reflect the true underlying biology. More stringent biological validation such as immunohistochemistry with known lineage markers, or transgenic mouse KO studies is required to truly determine whether immune-related genes are expressed during fetal myogenesis in Pax3+ cells.
Among the questions raised by this analysis, taking into account the limitations described above, were whether the enrichment in immune-related genes during fetal embryogenesis was caused by non-myogenic Pax3EGFP subpopulations, or whether the skeletal muscle cells were expressing these genes. The former possibility seems plausible, insofar as it is already known that the Pax3EGFP lineage gives rise to a small population of vascular endothelial cells. Since samples were sorted to 97–99% purity, gene expression was unlikely to be caused by impurities. Co-expression network analysis revealed that the immune-related subnetwork genes were expressed at E11 (Fig. 4), but overall expression did not peak until E14 (Fig. S2C). One possible explanation is that an immune-related subpopulation of the Pax3EGFP exists at E11, but these cells do not expand in number until significantly until later in development. Another possibility was that there were two separate networks expressed in the same cell type. These questions reveal the power of co-expression analysis, which can detect gene expression patterns even at low levels of expression49.
Gene expression profiling of SSTFs in the Pax3-derived cells in developing forelimbs
To identify the genes involved in early forelimb development we cross referenced the 4,481 DE genes with a list of known SSTFs, and identified SSTFs as stage-specific depending on the averaged SDR values. SDR values for all SSTFs in all samples were calculated as mentioned previously, and SDR values were averaged by time point. SSTFs were classified as specific to each developmental state if the average SDR value for a stage was at least 0.4, and was at least 0.3 greater (additively) than the average for any other state. Similarly, SSTFs were classified as specific to two embryonic states if the SDR for each state was at least 0.3, the average of both SDR values was at least 0.4, and the average of both SDR values was at least 0.3 greater (additively) than the average for any other state. Additionally, the mouse genome informatics (MGI) batch tool was used to determine the known expression of stage-specific SSTFs (Table 1).
SSTFs expressed specifically at E11 belonged to homeodomain (HD) and/or zinc-finger (ZNF) transcription factor families, were primarily expressed in the CNS during early-mid gestation (Table 1) and are nodes of the network (Fig. 4) with few expressed specifically in skeletal muscle. This further supports the previous observations that multiple Pax3+ non-myogenic cells exist within the Pax3EGFP lineage. SSTFs expressed at both E11 and E12 follow the same trends, except were composed mostly of homeodomain SSTFs. No SSTFs expressed specifically at E12 or E13 were present in the co-expression network, implying that they may perform stage-specific roles. Expression of immune-related genes occurs later in fetal embryogenesis. SSTFs expressed specifically at E13 and E14 belonged to the ZNF family, with only Zeb2 and Pou2f2 possessing a homeodomain. While most of the SSTFs expressed at E13 and E14 are expressed in the CNS, they are also expressed in other tissue types such as the vascular system (VS) and hemolymphoid systems (HLS), among others, and were present in the co-expression network in skeletal system, nervous system, and immune system subnetworks. SSTFs with established immunogenic functions included Bcl6, Ikzf1, and Zbtb7b among others. Bcl6 is also part of the skeletal and nervous system subnetwork, meaning it may not have an immune-specific role in this context.
Gene expression profiling of signaling molecules in the Pax3-derived cells in developing forelimbs
During development, signaling molecules convey information to cells about their direction, behavior and specification by activating transcriptional programs. Cell shapes and cytoskeletal changes regulate cell lineages and organ formation. Notch, Hedgehog (Hh), Wingless/Wnt, Bmp (bone morphogenetic proteins), Egf (epidermal growth factor), and Fgf (fibroblast growth factor) signaling can generate morphogen gradients across varying distances that pattern cells in a concentration-dependent manner.
Notch functions in organ formation during development, including somitogenesis, as well as in adult homeostasis by determining cell fate and maintaining pluripotency50. Members of the Notch pathway were highly expressed in the Pax3EGFP cells in the embryonic forelimb (E11, Fig. 5). The elevated Shh levels in the Pax3EGFP lineage at E11 were in accord with its expression at the posterior margin of limb buds between E9–E1251. Shh is produced by cells located in the zone of polarizing activity (ZPA) in the mesenchyme and regulates patterning along the anterior-posterior axis. Shh signaling is also involved in timing myogenic differentiation, promoting slow muscle differentiation, and controlling migration into the distal part of the limb52,53. Wnts are secreted proteins that control a multitude of diverse developmental processes. At the onset of limb development, the limb buds form as a result of an interplay between Fgf and Wnt signaling (E11, Fig. 5). Wnt proteins control the morphogenesis of specific tissues in the limb such as musculature, synovial joints, cartilage, and bone. Wnt6 from the limb ectoderm promotes limb myogenesis via Pax3 and Myf554. Wnt7a maintains the expression of N-cadherin, which is essential for myogenic migration and chondrogenesis55. Wnt3a induces the apical ectodermal ridge (AER) formation and Fgf8 expression through the canonical Wnt pathway56. Wnt4 and Wnt11 are expressed in the mesenchyme surrounding the developing cartilage elements which form at the onset of fetal development (E13, E14, Fig. 5). Fgfs , produced by the AER (Fgf2, Fgf4, Fgf8, Fgf9) and in the underlying mesenchyme (Fgf2, Fgf10), are required for proximal-distal outgrowth57. Specification of the vascular and hematopoietic systems is a characteristic of fetal development, following the patterning of the skeletal system characterized by expression of members of the Egf signaling pathway (E12–E14, Fig. 5). These data collectively suggest that interactions of muscle, bone, cartilage, tendon, and ligament are critical for the correct assembly of the musculoskeletal system during development, which is further enhanced by the development of the immune and hematopoietic systems.
Relative expression of known signaling ligands. Relative expression of known signaling ligands relevant in forelimb development was visualized as a heatmap using SDR values, with columns representing samples ordered by stage, and rows representing ligands. Yellow indicated high gene expression relative to the average of all samples, and blue indicates low expression.
Conclusion
FACS-Seq analysis of embryonic and fetal Pax3EGFP cells identified many transcripts outside the myogenic lineage. Pax3-derived cells populated the musculoskeletal, vascular, neuronal, immune and hematopoietic lineages. These findings suggest that the dermomyotome Pax3-derived cells may have the characteristics of a stem cell niche that can give rise to several lineages to form a functional forelimb, and provides a framework for future single cell sequencing within the forelimb during development.
Materials and Methods
All methods were carried out in accordance with relevant guidelines and regulations. All experimental protocols were approved by the Environmental Health and Safety Committee at Oregon State University. All animal experiments were performed in accordance to institutional and National Health and Medical Research Council guidelines. The experimental protocol was approved by the Institutional Animal Care and Use Committee at Oregon State University.
Fluorescence assisted cell sorting (FACS) of embryonic mouse forelimb cells
Mice were fed the standard PicoLab Rodent Diet 20, 5053*, a managed formulation delivers constant nutrition. Female ICR mice were plugged on consecutive days by male Pax3Cre|RosaEGFP mice. At 11, 12, 13, and 14 days post vaginal plug, female mice were euthanized, and embryos collected in PBS over ice and embryos were genotyped under a fluorescent microscope. Forelimbs were dissected between the caudal edge of the shoulder and the lumbar region. Isolated forelimbs from each litter were pooled in Dulbecco’s Modified Eagle Medium (DMEM) with 4.5 g/L glucose, based on Pax3Cre|RosaEGFP positive (green, G) and negative (white, W) genotypes. Dissociation of embryonic forelimbs was carried out as described previously58 with the following modifications. DMEM was removed, and dissociation buffer (HBSS without CaCl2, MgCl2, MgSO4 (Gibco), 2 mg/mL Type I Collagenase (Worthington Biochem), 5 mM EDTA was added, ~6 forelimbs per 1 mL buffer for E11 and E12, and ~2 forelimbs per 1 mL buffer at E13 and E14. Forelimbs were incubated for 3 minutes at 37 °C, and pipetted 10 times through a 1 mL pipette tip to promote dissociation. Forelimbs were incubated and pippeted once more at E11, E12, and E13, and twice more at E14. After the final dissociation step, each pooled sample was centrifuged at 5,000 rpm for one minute. The media was aspirated, cells were resuspended in PBS by pipetting 15 times, to a final concentration between 1 × 106 and 1 × 107 cells/mL. Cell suspensions were passed through a 35 µm nitex filter again before they were sorted. Cell suspensions were sorted using a Sony SH800 cell sorter (Sony Inc). EGFP+ (G) cells were sorted directly into PBS. Once the full samples have been sorted, each tube (G) was spun at 3800 rpm for 15 minutes at 4 °C. PBS was aspirated off the cell pellets, and cell pellets were lysed with 350 µL Buffer RLT with added ß-mercaptothanol (Qiagen). Lysates were kept over ice until all samples were sorted.
RNA preparation, sequencing and analysis
RNA was extracted using RNAeasy mini kit (Qiagen) following the manufacturer’s protocol. RNA was tested for quality and degradation using the AATI Fragment Analyzer (ATI). RNA libraries were sequenced on a 100 bp single-end run on the Illumina Hiseq. 4000 (Illumina, San Diego, CA). Library preparation was done by trained technicians at the GC3F core facility using the Kapa Biosystems Stranded mRNA-Seq Kit (Kapa). Libraries were created and sequenced, corresponding to six (E11.5), seven (E12.5), nine (E13.5), and six (E14.5) biological replicates. Primary Illumina data image analysis, base calling, and read-quality filtering were done using the Casava pipeline version 1.8.2 (Illumina). Each sample was processed and analyzed with the same methods. After filtering low quality reads TopHat version 2.1.0 was used to align all reads to the mm10 genome with default parameters and to identify splice junctions59,60. HTseq was used to create count tables from tophat2 aligned reads61. DESeq2 was used to calculate differential gene expression between time points62 using an FDR adjusted cutoff of p ≤ 0.05, with a fold change ≥1.5, between any two consecutive time points. Principal component analysis was performed using the prcomp function in R software63. Heatmaps were generated using the pheatmap package in R software64. Signed difference ratios (SDR) were calculated similar to65, except the average for each gene across all samples was subtracted from each sample. Fastq sequences were deposited to the NCBI gene expression omnibus (GEO) sequence read archive (SRA) under the accession SRP126903.
Co-expression Network Construction and Analysis
Co-expression networks were constructed as previously described18. Pairwise correlation coefficients were calculated between each of 4,481 identified DEGs, in all samples, sing an adjusted FDR cutoff of p ≤ 1e-16. The co-expression network was visualized in Cytoscape45, and modules were identified via markov clustering66 using the package MCL in R software. GO term enrichment in modules was determined by Panther GO67,68. R software custom code used for co-expression analysis is available in supplemental code file.
Immunohistochemistry
Immunohistochemistry was performed as previously described69.
References
Christ, B. & Ordahl, C. P. Early stages of chick somite development. Anat. Embryol. 191, 381–396 (1995).
Burke, A. C. & Nowicki, J. L. A new view of patterning domains in the vertebrate mesoderm. Dev. Cell 4, 159–165 (2003).
Goulding, M., Lumsden, A. & Paquette, A. J. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development 120, 957–971 (1994).
Bladt, F., Riethmacher, D., Isenmann, S., Aguzzi, A. & Birchmeier, C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 376, 376768a0 (1995).
Dietrich, S. et al. The role of SF/HGF and c-Met in the development of skeletal muscle. Development 126, 1621–1629 (1999).
Hayashi, K. & Ozawa, E. Myogenic cell migration from somites is induced by tissue contact with medial region of the presumptive limb mesoderm in chick embryos. Development 121, 661–669 (1995).
Bober, E., Franz, T., Arnold, H. H., Gruss, P. & Tremblay, P. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development 120, 603–612 (1994).
Daston, G., Lamar, E., Olivier, M. & Goulding, M. Pax-3 is necessary for migration but not differentiation of limb muscle precursors in the mouse. Development 122, 1017–1027 (1996).
Tajbakhsh, S. Skeletal muscle stem and progenitor cells: reconciling genetics and lineage. Exp. Cell Res. 306, 364–372 (2005).
Biressi, S. et al. Intrinsic phenotypic diversity of embryonic and fetal myoblasts is revealed by genome-wide gene expression analysis on purified cells. Dev. Biol. 304, 633–651 (2007).
Buckingham, M. & Rigby, P. W. J. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell 28, 225–238 (2014).
Messina, G. et al. Nfix regulates fetal-specific transcription in developing skeletal muscle. Cell 140, 554–566 (2010).
Yusuf, F. & Brand-Saberi, B. Myogenesis and muscle regeneration. Histochem. Cell Biol. 138, 187–199 (2012).
Barabási, A.-L. & Albert, R. Emergence of Scaling in Random Networks. Science 286, 509–512 (1999).
Kirschner, M. W. The meaning of systems biology. Cell 121, 503–504 (2005).
Barabasi, A.-L. & Oltvai, Z. N. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 5, 101–113 (2004).
Singh, A. J., Ramsey, S. A., Filtz, T. M. & Kioussi, C. Differential gene regulatory networks in development and disease. Cell. Mol. Life Sci. https://doi.org/10.1007/s00018-017-2679-6 (2017).
Dong, X. et al. Reverse enGENEering of Regulatory Networks from Big Data: A Roadmap for Biologists. Bioinform. Biol. Insights 9, 61–74 (2015).
Kioussi, C., Shih, H.-P., Loflin, J. & Gross, M. K. Prediction of active nodes in the transcriptional network of neural tube patterning. Proc. Natl. Acad. Sci. USA 103, 18621–18626 (2006).
Kioussi, C. & Gross, M. K. How to Build Transcriptional Network Models of Mammalian Pattern Formation. PLoS One 3, e2179 (2008).
Hsiao, T.-H. et al. Differential network analysis reveals the genome-wide landscape of estrogen receptor modulation in hormonal cancers. Sci. Rep. 6, 23035 (2016).
Földy, C. et al. Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc. Natl. Acad. Sci. USA 113, E5222–31 (2016).
Engleka, K. A. et al. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev. Biol. 280, 396–406 (2005).
Mao, X., Fujiwara, Y., Chapdelaine, A., Yang, H. & Orkin, S. H. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood 97, 324–326 (2001).
Herzenberg, L. A., Sweet, R. G. & Herzenberg, L. A. Fluorescence-activated cell sorting. Sci. Am. 234, 108–117 (1976).
Ringnér, M. What is principal component analysis? Nat. Biotechnol. 26, 303–304 (2008).
Pineault, K. M. & Wellik, D. M. Hox genes and limb musculoskeletal development. Curr. Osteoporos. Rep. 12, 420–427 (2014).
Raines, A. M., Magella, B., Adam, M. & Potter, S. S. Key pathways regulated by HoxA9,10,11/HoxD9,10,11 during limb development. BMC Dev. Biol. 15, 28 (2015).
Martin, P. Tissue patterning in the developing mouse limb. Int. J. Dev. Biol. 34, 323–336 (1990).
Mofarrahi, M. et al. Angiopoietin-1 enhances skeletal muscle regeneration in mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 308, R576–89 (2015).
Borselli, C. et al. Functional muscle regeneration with combined delivery of angiogenesis and myogenesis factors. Proc. Natl. Acad. Sci. USA 107, 3287–3292 (2010).
Young, K. et al. Endoglin is required in Pax3-derived cells for embryonic blood vessel formation. Dev. Biol. 409, 95–105 (2016).
Saclier, M., Cuvellier, S., Magnan, M., Mounier, R. & Chazaud, B. Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J. 280, 4118–4130 (2013).
Costamagna, D., Costelli, P., Sampaolesi, M. & Penna, F. Role of Inflammation in Muscle Homeostasis and Myogenesis. Mediators Inflamm. 2015, 805172 (2015).
Sanchez-Gurmaches, J. et al. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 16, 348–362 (2012).
Grounds, M. D., Garrett, K. L., Lai, M. C., Wright, W. E. & Beilharz, M. W. Identification of skeletal muscle precursor cells in vivo by use of MyoD1 and myogenin probes. Cell Tissue Res. 267, 99–104 (1992).
Sanchez-Gurmaches, J. & Guertin, D. A. Adipocyte lineages: tracing back the origins of fat. Biochim. Biophys. Acta 1842, 340–351 (2014).
Shih, H. P., Gross, M. K. & Kioussi, C. Expression pattern of the homeodomain transcription factor Pitx2 during muscle development. Gene Expr. Patterns 7, 441–451 (2007).
Lagha, M. et al. Pax3:Foxc2 reciprocal repression in the somite modulates muscular versus vascular cell fate choice in multipotent progenitors. Dev. Cell 17, 892–899 (2009).
Jostes, B., Walther, C. & Gruss, P. The murine paired box gene, Pax7, is expressed specifically during the development of the nervous and muscular system. Mech. Dev. 33, 27–37 (1990).
Deries, M. & Thorsteinsdóttir, S. Axial and limb muscle development: dialogue with the neighbourhood. Cell. Mol. Life Sci. 73, 4415–4431 (2016).
Mansouri, A., Pla, P., Larue, L. & Gruss, P. Pax3 acts cell autonomously in the neural tube and somites by controlling cell surface properties. Development 128, 1995–2005 (2001).
Kubic, J. D., Little, E. C., Kaiser, R. S., Young, K. P. & Lang, D. FOXD3 Promotes PAX3 Expression in Melanoma Cells. J. Cell. Biochem. 117, 533–541 (2016).
van Noort, V., Snel, B. & Huynen, M. A. The yeast coexpression network has a small-world, scale-free architecture and can be explained by a simple model. EMBO Rep. 5, 280–284 (2004).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Van Dongen, S. M. Graph clustering by flow simulation, Graduate Dissertation (2001).
Tsai, F. Y. et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371, 221–226 (1994).
Riether, C., Schürch, C. M. & Ochsenbein, A. F. Regulation of hematopoietic and leukemic stem cells by the immune system. Cell Death Differ. 22, 187–198 (2015).
Ideker, T. & Krogan, N. J. Differential network biology. Mol. Syst. Biol. 8, 565 (2012).
Carrieri, F. A. & Dale, J. K. Turn It Down a Notch. Front Cell Dev Biol 4, 151 (2016).
Zhu, J. et al. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev. Cell 14, 624–632 (2008).
Hu, J. K.-H., McGlinn, E., Harfe, B. D., Kardon, G. & Tabin, C. J. Autonomous and nonautonomous roles of Hedgehog signaling in regulating limb muscle formation. Genes Dev. 26, 2088–2102 (2012).
Anderson, C. et al. Sonic hedgehog acts cell-autonomously on muscle precursor cells to generate limb muscle diversity. Genes Dev. 26, 2103–2117 (2012).
Geetha-Loganathan, P. et al. Ectodermal Wnt-6 promotes Myf5-dependent avian limb myogenesis. Dev. Biol. 288, 221–233 (2005).
Tufan, A. C. & Tuan, R. S. Wnt regulation of limb mesenchymal chondrogenesis is accompanied by altered N-cadherin-related functions. FASEB J. 15, 1436–1438 (2001).
Kengaku, M. et al. Distinct WNT pathways regulating AER formation and dorsoventral polarity in the chick limb bud. Science 280, 1274–1277 (1998).
Min, H. et al. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 12, 3156–3161 (1998).
Campbell, A. L., Eng, D., Gross, M. K. & Kioussi, C. Prediction of gene network models in limb muscle precursors. Gene 509, 16–23 (2012).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 15, 550 (2014).
Ihaka, R. & Gentleman, R. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Stat. 5, 299–314 (1996).
Kolde, R. Pheatmap: pretty heatmaps. R package version 61 (2012).
Ramsey, S. A. et al. Uncovering a Macrophage Transcriptional Program by Integrating Evidence from Motif Scanning and Expression Dynamics. PLoS Comput. Biol. 4, e1000021 (2008).
Enright, A. J., Van Dongen, S. & Ouzounis, C. A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002).
Mi, H., Muruganujan, A., Casagrande, J. T. & Thomas, P. D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 (2013).
Mi, H. et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189 (2017).
Ma, H.-Y., Xu, J., Eng, D., Gross, M. K. & Kioussi, C. Pitx2-mediated cardiac outflow tract remodeling. Dev. Dyn. 242, 456–468 (2013).
Beverdam, A., Brouwer, A., Reijnen, M., Korving, J. & Meijlink, F. Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development 128, 3975–3986 (2001).
Visel, A., Thaller, C. & Eichele, G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 32, D552–6 (2004).
Gray, P. A. et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 306, 2255–2257 (2004).
Chotteau-Lelièvre, A., Desbiens, X., Pelczar, H., Defossez, P. A. & de Launoit, Y. Differential expression patterns of the PEA3 group transcription factors through murine embryonic development. Oncogene 15, 937–952 (1997).
Dush, M. K. & Martin, G. R. Analysis of mouse Evx genes: Evx-1 displays graded expression in the primitive streak. Dev. Biol. 151, 273–287 (1992).
Vlaeminck-Guillem, V. et al. The Ets family member Erg gene is expressed in mesodermal tissues and neural crests at fundamental steps during mouse embryogenesis. Mech. Dev. 91, 331–335 (2000).
Reymond, A. et al. Human chromosome 21 gene expression atlas in the mouse. Nature 420, 582–586 (2002).
Ruest, L.-B. et al. dHAND-Cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Dev. Biol. 257, 263–277 (2003).
Leimeister, C., Schumacher, N. & Gessler, M. Expression of Notch pathway genes in the embryonic mouse metanephros suggests a role in proximal tubule development. Gene Expr. Patterns 3, 595–598 (2003).
Nakagawa, O., Nakagawa, M., Richardson, J. A., Olson, E. N. & Srivastava, D. HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev. Biol. 216, 72–84 (1999).
Coré, N. et al. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development 124, 721–729 (1997).
Burke, A. C., Nelson, C. E., Morgan, B. A. & Tabin, C. Hox genes and the evolution of vertebrate axial morphology. Development 121, 333–346 (1995).
Gaunt, S. J., Krumlauf, R. & Duboule, D. Mouse homeo-genes within a subfamily, Hox-1.4, −2.6 and −5.1, display similar anteroposterior domains of expression in the embryo, but show stage- and tissue-dependent differences in their regulation. Development 107, 131–141 (1989).
Kania, A., Johnson, R. L. & Jessell, T. M. Coordinate roles for LIM homeobox genes in directing the dorsoventral trajectory of motor axons in the vertebrate limb. Cell 102, 161–173 (2000).
Stanton, B. R., Perkins, A. S., Tessarollo, L., Sassoon, D. A. & Parada, L. F. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 6, 2235–2247 (1992).
Blackshaw, S. et al. Genomic analysis of mouse retinal development. PLoS Biol. 2, E247 (2004).
Neubüser, A., Koseki, H. & Balling, R. Characterization and developmental expression of Pax9, a paired-box-containing gene related to Pax1. Dev. Biol. 170, 701–716 (1995).
Voss, A. K. et al. Protein and gene expression analysis of Phf6, the gene mutated in the Börjeson-Forssman-Lehmann Syndrome of intellectual disability and obesity. Gene Expr. Patterns 7, 858–871 (2007).
Diez-Roux, G. et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 9, e1000582 (2011).
Fedtsova, N. G. & Turner, E. E. Brn-3.0 expression identifies early post-mitotic CNS neurons and sensory neural precursors. Mech. Dev. 53, 291–304 (1995).
McKee, A. E. et al. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev. Biol. 5, 14 (2005).
Carmona, M. C. et al. Mitochondrial biogenesis and thyroid status maturation in brown fat require CCAAT/enhancer-binding protein alpha. J. Biol. Chem. 277, 21489–21498 (2002).
Visel, A. et al. Regulatory pathway analysis by high-throughput in situ hybridization. PLoS Genet. 3, 1867–1883 (2007).
Lewandowski, J. P. et al. Spatiotemporal regulation of GLI target genes in the mammalian limb bud. Dev. Biol. 406, 92–103 (2015).
Rovescalli, A. C., Asoh, S. & Nirenberg, M. Cloning and characterization of four murine homeobox genes. Proc. Natl. Acad. Sci. USA 93, 10691–10696 (1996).
Kunath, M., Lüdecke, H.-J. & Vortkamp, A. Expression of Trps1 during mouse embryonic development. Mech. Dev. 119(Suppl 1), S117–20 (2002).
Fantauzzo, K. A., Bazzi, H., Jahoda, C. A. B. & Christiano, A. M. Dynamic expression of the zinc-finger transcription factor Trps1 during hair follicle morphogenesis and cycling. Gene Expr. Patterns 8, 51–57 (2008).
Mansouri, A. et al. Paired-related murine homeobox gene expressed in the developing sclerotome, kidney, and nervous system. Dev. Dyn. 210, 53–65 (1997).
Purandare, S. M. et al. A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development 129, 2293–2302 (2002).
Liu, J., Schiltz, J. F., Shah, P. C., Benson, K. F. & Chada, K. K. Genomic structure and expression of the murine Hmgi(y) gene. Gene 246, 197–207 (2000).
Kessel, M. & Gruss, P. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell 67, 89–104 (1991).
Dony, C. & Gruss, P. Specific expression of the Hox 1.3 homeo box gene in murine embryonic structures originating from or induced by the mesoderm. EMBO J. 6, 2965–2975 (1987).
Larochelle, C., Tremblay, M., Bernier, D., Aubin, J. & Jeannotte, L. Multiple cis-acting regulatory regions are required for restricted spatio-temporal Hoxa5 gene expression. Dev. Dyn. 214, 127–140 (1999).
Kostic, D. & Capecchi, M. R. Targeted disruptions of the murine Hoxa-4 and Hoxa-6 genes result in homeotic transformations of components of the vertebral column. Mech. Dev. 46, 231–247 (1994).
Evans, S. M. & O’Brien, T. X. Expression of the helix-loop-helix factor Id during mouse embryonic development. Dev. Biol. 159, 485–499 (1993).
Jen, Y., Manova, K. & Benezra, R. Expression patterns of Id1, Id2, and Id3 are highly related but distinct from that of Id4 during mouse embryogenesis. Dev. Dyn. 207, 235–252 (1996).
Duncan, M., DiCicco-Bloom, E. M., Xiang, X., Benezra, R. & Chada, K. The gene for the helix-loop-helix protein, Id, is specifically expressed in neural precursors. Dev. Biol. 154, 1–10 (1992).
Rétaux, S., Rogard, M., Bach, I., Failli, V. & Besson, M. J. Lhx9: a novel LIM-homeodomain gene expressed in the developing forebrain. J. Neurosci. 19, 783–793 (1999).
Jonk, L. J. et al. Cloning and expression during development of three murine members of the COUP family of nuclear orphan receptors. Mech. Dev. 47, 81–97 (1994).
Hjalt, T. A., Semina, E. V., Amendt, B. A. & Murray, J. C. The Pitx2 protein in mouse development. Dev. Dyn. 218, 195–200 (2000).
Holmberg, J., Ingner, G., Johansson, C., Leander, P. & Hjalt, T. A. PITX2 gain-of-function induced defects in mouse forelimb development. BMC Dev. Biol. 8, 25 (2008).
Haller, K., Rambaldi, I., Kovács, E. N., Daniels, E. & Featherstone, M. Prep2: cloning and expression of a new prep family member. Dev. Dyn. 225, 358–364 (2002).
Laclef, C., Souil, E., Demignon, J. & Maire, P. Thymus, kidney and craniofacial abnormalities in Six 1 deficient mice. Mech. Dev. 120, 669–679 (2003).
Xu, P.-X. et al. Six1 is required for the early organogenesis of mammalian kidney. Development 130, 3085–3094 (2003).
Fogelgren, B. et al. Misexpression of Six2 is associated with heritable frontonasal dysplasia and renal hypoplasia in 3H1 Br mice. Dev. Dyn. 237, 1767–1779 (2008).
Hargrave, M. et al. Expression of the Sox11 gene in mouse embryos suggests roles in neuronal maturation and epithelio-mesenchymal induction. Dev. Dyn. 210, 79–86 (1997).
Spitz, F. et al. A t(2;8) balanced translocation with breakpoints near the human HOXD complex causes mesomelic dysplasia and vertebral defects. Genomics 79, 493–498 (2002).
Alaynick, W. A. et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 6, 13–24 (2007).
Hermans-Borgmeyer, I., Süsens, U. & Borgmeyer, U. Developmental expression of the estrogen receptor-related receptor gamma in the nervous system during mouse embryogenesis. Mech. Dev. 97, 197–199 (2000).
Süsens, U., Hermans-Borgmeyer, I. & Borgmeyer, U. Alternative splicing and expression of the mouse estrogen receptor-related receptor gamma. Biochem. Biophys. Res. Commun. 267, 532–535 (2000).
Monaghan, A. P., Kaestner, K. H., Grau, E. & Schütz, G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development 119, 567–578 (1993).
Gaunt, S. J., Coletta, P. L., Pravtcheva, D. & Sharpe, P. T. Mouse Hox-3.4: homeobox sequence and embryonic expression patterns compared with other members of the Hox gene network. Development 109, 329–339 (1990).
Candia, A. F. et al. Mox-1 and Mox-2 define a novel homeobox gene subfamily and are differentially expressed during early mesodermal patterning in mouse embryos. Development 116, 1123–1136 (1992).
Jacquemin, P. et al. Cloning and embryonic expression pattern of the mouse Onecut transcription factor OC-2. Gene Expr. Patterns 3, 639–644 (2003).
Armstrong, J. F., Pritchard-Jones, K., Bickmore, W. A., Hastie, N. D. & Bard, J. B. The expression of the Wilms’ tumour gene, WT1, in the developing mammalian embryo. Mech. Dev. 40, 85–97 (1993).
Ith, B., Wei, J., Yet, S.-F., Perrella, M. A. & Layne, M. D. Aortic carboxypeptidase-like protein is expressed in collagen-rich tissues during mouse embryonic development. Gene Expr. Patterns 5, 533–537 (2005).
Layne, M. D. et al. Impaired abdominal wall development and deficient wound healing in mice lacking aortic carboxypeptidase-like protein. Mol. Cell. Biol. 21, 5256–5261 (2001).
Abbott, B. D., Birnbaum, L. S. & Perdew, G. H. Developmental expression of two members of a new class of transcription factors: I. Expression of aryl hydrocarbon receptor in the C57BL/6N mouse embryo. Dev. Dyn. 204, 133–143 (1995).
Crocoll, A., Zhu, C. C., Cato, A. C. & Blum, M. Expression of androgen receptor mRNA during mouse embryogenesis. Mech. Dev. 72, 175–178 (1998).
Fukuda, T. et al. The murine BCL6 gene is induced in activated lymphocytes as an immediate early gene. Oncogene 11, 1657–1663 (1995).
Ormestad, M., Astorga, J. & Carlsson, P. Differences in the embryonic expression patterns of mouse Foxf1 and −2 match their distinct mutant phenotypes. Dev. Dyn. 229, 328–333 (2004).
Hjerling-Leffler, J. et al. The boundary cap: a source of neural crest stem cells that generate multiple sensory neuron subtypes. Development 132, 2623–2632 (2005).
Izpisùa-Belmonte, J. C. et al. Primary structure and embryonic expression pattern of the mouse Hox-4.3 homeobox gene. Development 110, 733–745 (1990).
Jin, C. et al. Identification of mouse Jun dimerization protein 2 as a novel repressor of ATF-2. FEBS Lett. 489, 34–41 (2001).
Koscielny, G. et al. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 42, D802–9 (2014).
Hodgkinson, C. A. et al. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74, 395–404 (1993).
Chaudhry, A. Z., Lyons, G. E. & Gronostajski, R. M. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev. Dyn. 208, 313–325 (1997).
Lau, E. C., Li, Z. Q., Santos, V. & Slavkin, H. C. Messenger RNA phenotyping for semi-quantitative comparison of glucocorticoid receptor transcript levels in the developing embryonic mouse palate. J. Steroid Biochem. Mol. Biol. 46, 751–758 (1993).
Alam, S., Zinyk, D., Ma, L. & Schuurmans, C. Members of the Plag gene family are expressed in complementary and overlapping regions in the developing murine nervous system. Dev. Dyn. 234, 772–782 (2005).
Tsuda, T. et al. Zinc finger protein Zac1 is expressed in chondrogenic sites of the mouse. Dev. Dyn. 229, 340–348 (2004).
Piras, G. et al. Zac1 (Lot1), a potential tumor suppressor gene, and the gene for epsilon-sarcoglycan are maternally imprinted genes: identification by a subtractive screen of novel uniparental fibroblast lines. Mol. Cell. Biol. 20, 3308–3315 (2000).
Hong, K. Y. et al. Perilipin+ embryonic preadipocytes actively proliferate along growing vasculatures for adipose expansion. Development 142, 2623–2632 (2015).
Li, M. et al. Expression of murine ELL-associated factor 2 (Eaf2) is developmentally regulated. Dev. Dyn. 228, 273–280 (2003).
Silver, L. & Palis, J. Initiation of murine embryonic erythropoiesis: a spatial analysis. Blood 89, 1154–1164 (1997).
Nardelli, J., Thiesson, D., Fujiwara, Y., Tsai, F. Y. & Orkin, S. H. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev. Biol. 210, 305–321 (1999).
Erselius, J. R., Goulding, M. D. & Gruss, P. Structure and expression pattern of the murine Hox-3.2 gene. Development 110, 629–642 (1990).
Yeo, J.-C. et al. Klf2 is an essential factor that sustains ground state pluripotency. Cell Stem Cell 14, 864–872 (2014).
Anderson, K. P., Kern, C. B., Crable, S. C. & Lingrel, J. B. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Krüppel-like factor: identification of a new multigene family. Mol. Cell. Biol. 15, 5957–5965 (1995).
Tamplin, O. J., Cox, B. J. & Rossant, J. Integrated microarray and ChIP analysis identifies multiple Foxa2 dependent target genes in the notochord. Dev. Biol. 360, 415–425 (2011).
Stoykova, A. S., Sterrer, S., Erselius, J. R., Hatzopoulos, A. K. & Gruss, P. Mini-Oct and Oct-2c: two novel, functionally diverse murine Oct-2 gene products are differentially expressed in the CNS. Neuron 8, 541–558 (1992).
Tanaka, T., Yoshida, N., Kishimoto, T. & Akira, S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J. 16, 7432–7443 (1997).
Chen, S. S., Chen, J. F., Johnson, P. F., Muppala, V. & Lee, Y. H. C/EBPbeta, when expressed from the C/ebpalpha gene locus, can functionally replace C/EBPalpha in liver but not in adipose tissue. Mol. Cell. Biol. 20, 7292–7299 (2000).
Elefanty, A. G., Begley, C. G., Hartley, L., Papaevangeliou, B. & Robb, L. SCL expression in the mouse embryo detected with a targeted lacZ reporter gene demonstrates its localization to hematopoietic, vascular, and neural tissues. Blood 94, 3754–3763 (1999).
Kallianpur, A. R., Jordan, J. E. & Brandt, S. J. The SCL/TAL-1 gene is expressed in progenitors of both the hematopoietic and vascular systems during embryogenesis. Blood 83, 1200–1208 (1994).
Acknowledgements
The authors thank Maggie Weitzman for library preparations and RNAseq. The work was supported by the College of Pharmacy (AJS, TMF and CK), and Oregon State University (CK).
Author information
Authors and Affiliations
Contributions
C.K. designed the experiments. A.J.S., C.N.C. and H.Y.M. performed the experiments. C.K., A.J.S., S.A.R. and T.M.F. analyzed the data. A.J.S. and C.K. prepared the figures. C.K., A.J.S., S.A.R. and T.M.F. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Singh, A.J., Chang, CN., Ma, HY. et al. FACS-Seq analysis of Pax3-derived cells identifies non-myogenic lineages in the embryonic forelimb. Sci Rep 8, 7670 (2018). https://doi.org/10.1038/s41598-018-25998-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-25998-1
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.