Abstract
Endocrine disrupting chemicals (EDC) interfere with the natural hormone balance and may induce epigenetic changes through exposure during sensitive periods of development. In this study, the effects of short-term estradiol-17β (E2) exposure on various tissues of pregnant sows (F0) and on day 10 blastocysts (F1) were assessed. Intergenerational effects were investigated in the liver of 1-year old female offspring (F1). During gestation, sows were orally exposed to two low doses and a high dose of E2 (0.05, 10, and 1000 µg/kg body weight/day). In F0, perturbed tissue specific mRNA expression of cell cycle regulation and tumour suppressor genes was found at low and high dose exposure, being most pronounced in the endometrium and corpus luteum. The liver showed the most significant DNA hypomethylation in three target genes; CDKN2D, PSAT1, and RASSF1. For CDKN2D and PSAT1, differential methylation in blastocysts was similar as observed in the F0 liver. Whereas blastocysts showed hypomethylation, the liver of 1-year old offspring showed subtle, but significant hypermethylation. We show that the level of effect of estrogenic EDC, with the periconceptual period as a sensitive time window, is at much lower concentration than currently presumed and propose epigenetics as a sensitive novel risk assessment parameter.
Similar content being viewed by others
Introduction
The environment can critically influence the complex regulation of the human body. Several environmentally present exogenous compounds are able to interfere with the endocrine system, possibly leading to adverse consequences to human health1,2. These substances are classified as ‘endocrine disrupting chemicals’ (EDC)3. Potent low dose effects of EDC, specifically estrogenic EDC as well as exogenous natural hormones, have been identified4,5. Exposure predominantly occurs through ingestion, inhalation, skin contact, and via the placenta of a pregnant female6. The molecular mechanisms of action involve the mimicking of regulatory pathways of endogenous hormones, such as oestrogens. Surface water has been found to be polluted with natural oestrogens such as estrone (E1), estradiol-17β (E2) and estriol (E3)7. These natural oestrogens, specifically E2, are globally released at varying, but polluting levels from wastewater treatment plants and effluents from livestock feedlots7. E2 is involved in various physiological functions such as growth, development and cell proliferation8,9,10. To explain observed cell proliferation and tumour initiation upon exposure to estrogenic substances, gene expression and epigenetic changes of cell cycle regulators, tumour suppressors, and methylation specific enzymes have been widely studied11,12,13,14,15,16,17.
Epigenetic changes have emerged as possible underlying mechanism through which EDC may exert their effect18. Epigenetics is defined as heritable changes in gene expression that are not due to changes in the DNA sequence19. One of the most widely explored and stable epigenetic mark is DNA methylation, which comprises the addition of a methyl group to the cytosine base of CpN dinucleotides20,21. Epigenetic changes at critical periods of development may permanently alter the epigenome in the germline, that can also be transmitted through generations18. When a pregnant female F0 is exposed, the subsequent two generations, namely the F1 and F2, are also directly exposed in utero or through the germline, respectively, and may exhibit intergenerational epigenetic inheritance. Any observed effects in subsequent generations are known as transgenerational epigenetic inheritance without any direct exposure22.
A far greater concern than direct effects of EDC - with large implications for human health - is that alterations to the epigenome may not cause immediate phenotypic effects, but remain as molecular fingerprints; only leading to a phenotype long after exposure or in subsequent generations23,24. Nonetheless, the current understanding of the link between E2 exposure during critical periods of early development and intergenerational epigenetic changes is limited.
In the present study, we aimed at
-
(1)
Studying the effects of a short-term low dose E2 exposure on tissue specific mRNA expression and DNA methylation in pregnant sows,
-
(2)
Assessing the methylation changes in gestationally exposed blastocysts to identify whether the exposure during early embryo development already induces a fingerprint to these low E2 doses,
-
(3)
Investigating whether changes in DNA methylation as observed in the embryos were also present in the one-year old adult offspring that had been exposed during the entire pregnancy.
To that end, two low doses, namely the acceptable daily intake (ADI) and a dose close to the no observed effect level (NOEL) (0.05 and 10 μg/kg body weight/day, respectively) and a high dose (HIGH) (1000 μg/kg body weight/day) of E2 were orally applied to sows as described previously25 (Fig. 1 and Supplementary Fig. S1 and Supplementary Text). Targeted gene expression analysis of relevant genes was undertaken, followed by the analysis of global DNA methylation changes in different tissues, such as liver, endometrium, corpus luteum, heart, spleen and skeletal muscles of exposed pregnant females (F0). Furthermore, gene specific DNA methylation changes were analysed in target regions of selected genes, namely Cyclin Dependent Kinase Inhibitor 2D (CDKN2D), Phosphoserine Aminotransferase 1 (PSAT1) and Ras Association Domain Family Member 1 (RASSF1). The gene specific DNA methylation was analysed in the liver, endometrium and corpus luteum of F0 sows, day 10 embryos (blastocysts) and the liver of one-year old female offspring (both F1) (Fig. 1). CDKN2D, a member of the INK4 family, encodes for p19INK4d and is involved in the regulation of cell growth26. The promoter region of CDKN2D has previously been shown to be differentially methylated, influencing its transcriptional levels26. PSAT1 is an enzyme that catalyses the serine biosynthesis pathway, with documented evidence of its cell proliferation activity, including cell cycle progression and tumorigenesis27,28. The tumour suppressor gene RASSF1 is known to induce cell cycle arrest and senescence in the G1 phase29. In addition, RASSF1 has been shown to be epigenetically silenced in various tumours and upon E2 exposure16,30.
Summary of the workflow undertaken in this study. The experimental plan indicates the oral exposure to estradiol-17β (E2) (indicated with red bars) at concentrations of 0, 0.05, 10 and 1000 μg/kg body weight/day for the control, ADI – ‘acceptable daily intake level’, the NOEL- ‘no observed effect level’, and a HIGH dose, respectively. Pregnant sows (F0) were exposed to treatments twice a day from conception to birth and again from conception to day 10 post-conception, after which they were slaughtered one hour after the last treatment. Embryos (F1) were flushed from the uterus directly after slaughter. Female offspring (F1) were sampled at one year of age during the diestrous phase of the oestrous cycle.
Our data shows that E2 doses as low as the acceptable daily intake cause changes in both the gene expression and DNA methylation profile of selected targets, underlining that epigenetics could serve as a sensitive novel parameter for risk assessment of estrogenic EDC.
Results
E2 induces tissue specific gene expression changes of cell cycle regulators and tumour suppressor genes in F0 tissues
The gene expression analyses were carried out in F0 liver, endometrium, corpus luteum, heart, spleen, and skeletal muscle using 57 selected estrogen-related target genes (Supplementary Table S1). Overall, 25 genes were found to be differentially expressed in various tissues and treatment groups as compared to the control group (Fig. 2). These included 11 cell cycle regulators (CDKN2D, CDC42EP4, GADD45A, MGMT, GADD45B, CCDC34, CDC42EP3, CDKN1B, CCN1, CDC42BPA, and CDKN2C), 10 tumour suppressor genes (HIC1, BGN, RB1, RASSF1, LATS1, PTEN, P53, ITIH5, PSAT1, and ACTG2), two methylation specific genes (DNMT3a and MeCP2), one gene associated with aberrant expression in cancer (BPGM), and one gene functionally associated with the biosynthesis of sex steroids (HSD17B7). Most differentially expressed genes (DEG) were observed in the corpus luteum followed by the endometrium, heart and skeletal muscle.
Heat map using hierarchical clustering of differentially expressed genes (DEG) in F0 tissues. Left to Right: Corpus luteum, endometrium, heart, skeletal muscle, liver, and spleen, where rows represent genes and columns represent the centred mean log-fold changes per treatment group compared to the control (0.05, 10, and 1000 μg/kg body weight/day represented by ADI, NOEL, and HIGH dose, respectively). *, **, and *** indicate statistical significance with respect to the control group at p < 0.05, p < 0.01, and p < 0.001, respectively (n = 4–6 per group).
Amongst the DEG, the cell cycle regulator CDKN2D was most highly upregulated in the NOEL and HIGH dose group in the endometrium (9.0-fold and 15.2-fold, respectively). CDKN2D also showed a significant upregulation in the HIGH dose group of corpus luteum (9.7-fold), and a dose-dependent increase of its expression in heart with 4.1-fold, 6.2-fold and 10.6-fold for ADI, NOEL, and HIGH, respectively. The most downregulated DEG (12.5-fold) was the tumour suppressor gene hypermethylated in cancer 1 (HIC1), in skeletal muscle in the NOEL dose group. The expression of PSAT1 was significantly downregulated in the HIGH dose in the corpus luteum and endometrium (4.3-fold and 2.4-fold, respectively). RASSF1 was not affected in either groups nor tissues, with exception of the HIGH dose group of the liver (1.3-fold downregulated) (Supplementary Table S2).
E2 does not significantly influence global DNA methylation in F0 tissues
The analysis of the global DNA methylation in the F0 tissues showed a tendency of hypomethylation in the HIGH dose group of spleen (2%), skeletal muscle (2%), heart (1%), and in all dose groups of corpus luteum (1% in ADI, NOEL and HIGH) as compared to controls. None of these differences reached statistical significance (Supplementary Table S4).
E2 induces DNA hypomethylation of CDKN2D, PSAT1, and RASSF1 in F0 liver
Gene specific DNA methylation analysis was carried out on selected CpN sites in the putative promoter region of CDKN2D, the coding region of the first exon of PSAT1, and the 5′ untranslated region (UTR) of the first exon of RASSF1 (Supplementary Table S5).
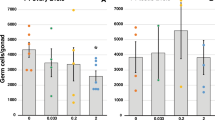
The E2 exposure induced subtle, but significant hypomethylation in several CpN sites analysed for all three genes in the NOEL and HIGH dose groups of F0 liver (Fig. 3). The most pronounced and statistically significant hypomethylation compared to control in each of the three genes analysed was in the HIGH group with a 1.7-fold lower methylation for RASSF1 (CpG −132), followed by a 1.4-fold lower methylation in the NOEL group for RASSF1 (GpC −267) and the HIGH group for PSAT1 (CpG +144).
Heat maps showing gene-specific DNA hypomethylation at CpN sites of NOEL and HIGH dose groups of F0 liver. Each column of the heat maps shown represents a CpN site analysed with its position with respect to the translational start site (indicated by +1), while each row represents the log2 methylation relative to control (0.05, 10, and 1000 μg/kg body weight/day represented by ADI, NOEL, and HIGH, respectively). *, **, and *** indicate statistical significance compared to the control group at p < 0.05, p < 0.01, and p < 0.001 respectively (n = 4–6 per group). (A) Gene diagram of CDKN2D, indicating the CpN sites analysed in the putative promoter region. (B) Gene diagram of PSAT1 indicating the CpN sites analysed in the coding region of the first exon. (C) Gene diagram of RASSF1 indicating the CpN sites analysed in the first exon.
E2-mediated hypomethylation of the CDKN2D promoter region in F0 tissues and F1 embryos, but hypermethylation in adult F1 liver
The differentially methylated CpN sites in the promoter region of CDKN2D primarily showed hypomethylation in exposed versus control animals. This effect was more pronounced in the liver than in any of the reproductive tissues under analysis (Fig. 4). Eventhough the most pronounced DNA methylation differences were observed in the F0 liver, there was no significant correlation to the subtle downregulation in mRNA expression (Supplementary Table S6). Similar to the F0 tissues, the blastocysts showed only hypomethylated CpN sites. Interestingly, an opposite pattern of hypermethylation was revealed in the liver of F1 adult animals. Focusing on CpG −200, a trend towards a dose-dependent decrease was observed for the reproductive tissues, with statistically significant hypomethylation in the corpus luteum of the F0 HIGH dose group. Even though the methylation differences were very subtle, the DNA methylation at this site significantly correlated with the transcription in the corpus luteum (R = 0.56 p = 0.01) (Supplementary Table S6). In addition, the DNA methylation of CpC −242 and CpG −190 significantly correlated with the transcription in the corpus luteum (R = 0.49, p = 0.029 and R = 0.60 p = 0.005, respectively) (Supplementary Table S6). The DNA methylation in the endometrium was only lower in the HIGH dose group at GpG −216 (Fig. 2), whereas mRNA expression was increased in the NOEL and HIGH dose group. The F1 embryos showed statistically significant hypomethylation at CpG −267, CpA −246, CpC −242, and CpG −200 for all doses, and at CpG −190 in the NOEL and HIGH dose. Opposing the methylation pattern in the maternal (F0) liver and the F1 embryos, the adult F1 liver showed statistically significant hypermethylation at CpC −267, CpA −246, CpC −242, CpG −216, CpG −200, and CpG −190.
E2 induced subtle, but consistent DNA hypomethylation in the CDKN2D putative promoter region in F0 liver, endometrium, and corpus luteum, and F1 embryos, but hypermethylation in liver of F1 adult gilts. Bar charts represent mean ± SEM of % DNA methylation per group in different shades of grey (0.05, 10, and 1000 μg/kg body weight/day represented by ADI, NOEL, and HIGH dose, respectively). *, **, and *** indicate statistical significance with respect to the control group at p < 0.05, p < 0.01, and p < 0.001, respectively (n = 4–6 per group for tissue samples, and n = 8–10 for embryos). (A) F0 liver DNA hypomethylation at CpN −270, −267, −249, −242, −216, −200, and −190. (B) F0 endometrium DNA hypomethylation at CpG −216. (C) F0 corpus luteum DNA hypomethylation at CpG −200. (D) F1 embryo DNA hypomethylation at CpN −267, −246, −242, −200, and −190 (E) F1 liver DNA hypermethylation at CpN −267, −246, −242, −216, −200, and −190.
E2-mediated hypomethylation of the coding region of PSAT1 in F0 liver and hypermethylation in F1 liver
All 13 CpN sites in the coding region of the PSAT1 first exon showed statistically significant hypomethylation in the F0 liver in NOEL and/or HIGH dose animals (Fig. 5). CpG +161 was significantly hypomethylated in the F0 endometrium and corpus luteum in the NOEL and HIGH dose animals, and in NOEL dose animals, respectively. Except for CpA +170 (R = −0.47 p = 0.044) in the liver, there was no correlation between methylation and gene expression at any of the sites analysed (Supplementary Table S6). In the F1 blastocysts CpT +121 and CpG +128 showed differential methylation patterns, which were comparable to the F0 liver. Moreover, while CpG +128 was hypomethylated in blastocysts, it was found to be hypermethylated in the 1-year post-partum F1 animals. In contrast to the hypomethylation of all 13 CpN sites in the F0 liver, in the F1 liver all 13 CpN sites were hypermethylated.
E2 induced consistent DNA hypomethylation in the PSAT1 coding region (first exon) in F0 liver, endometrium, and corpus luteum, and F1 embryos but hypermethylation in corresponding CpG sites of the liver of adult F1 animals. Bar charts represent mean ± SEM of % DNA methylation per group in different shades of grey (0.05, 10, and 1000 μg/kg body weight/day represented by ADI, NOEL, and HIGH dose, respectively). *, **, and *** indicate statistical significance with respect to the control group at p < 0.05, p < 0.01, and p < 0.001, respectively (n = 4–6 per group for tissue samples, and n = 8–10 for embryos). (A) F0 liver DNA hypomethylation at all analysed CpN sites. (B) F0 endometrium DNA hypomethylation at CpN +161. (C) F0 corpus luteum DNA hypomethylation at CpG +161. (D) F1 embryo DNA hypomethylation at CpN +121, and +128 (E) F1 liver DNA hypermethylation at all analysed CpN sites.
E2 exposure alters DNA methylation of the 5′ UTR of RASSF1 in F0 liver correlating with RASSF1 gene expression
Eight of the 15 CpG sites analysed in the 5′ untranslated region of RASSF1 first exon showed statistically significant hypomethylation in the F0 liver. The DNA methylation assessed at CpG −134, CpG −132, CpG −72, CpG −65, and CpG −59 significantly correlated with RASSF1 liver gene expression (Fig. 6 and Supplementary Table S6), i.e. higher methylation was correlated with higher expression. Besides the F0 liver, neither endometrium nor corpus luteum of the F0 animals showed differential methylation, and there was no correlation between methylation and gene expression (Supplementary Fig. S4 and Table S6). In F1 blastocysts, the CpG −55 was significantly hypomethylated in ADI and NOEL exposed animals. The F1 liver did not show any differential DNA methylation (Supplementary Fig. S4).
E2 induced DNA hypomethylation in the RASSF1 5′ UTR of the F0 liver. Bar charts represent mean ± SEM of % DNA methylation per group in different shades of grey (0.05, 10, and 1000 μg/kg body weight/day represented by ADI, NOEL, and HIGH dose, respectively). *, **, and *** indicate statistical significance with respect to the control group at p < 0.05, p < 0.01, and p < 0.001, respectively (n = 4–6 per group). (A) Liver DNA methylation showing most pronounced hypomethylation at CpG −134, −132, −116, −113, −85, −72, −67, −65, and −59. (B) Significant correlation of mRNA expression of RASSF1 with its methylation levels at CpG −134, −132, −65, and −59.
Discussion
Our results indicate that continuous oral exposure to a low dose E2 throughout pregnancy, currently considered as safe in the range of human “acceptable daily intake” (ADI) and the “no observed effect level” (NOEL)31, induces aberrant gene expression and DNA methylation. Given the high degree of physiological and anatomical similarity between pigs and humans, specifically the low circulating endogenous oestrogen concentrations during the periconceptional period which opposes the later pronounced synthesis of oestrogens by the placenta during pregnancy, the pig resembles human physiology32. Therefore, the pig was chosen as model organism to investigate the effects of gestational exposure to an additional low-dose estrogenic substance, potentially mimicking intergenerational epigenetic changes in women. The observed molecular changes were apparent in treated mothers, and in both her developing blastocysts and one-year old offspring. These findings are of particular note, as the circulating levels of E2 in animals under both low-dose treatments remained similar to controls25.
The tissue-specific differential gene expression of cell cycle regulators and tumour suppressors highlights the tissue-specific transcriptional E2 responsiveness33. The female reproductive tract is known to be a major target of estrogenic activity due to the high expression of oestrogen receptors34,35. Moreover, recent studies have shown that skeletal muscle is likewise an E2-responsive tissue36. CDKN2D showed the highest upregulation in reproductive tissues and heart, whereas HIC1 was most pronouncedly downregulated in skeletal muscle. In addition, PSAT1 was significantly downregulated in the corpus luteum. As possible explanation for the pronounced upregulation of CDKN2D as cell cycle inhibitor in heart and reproductive tissues we propose a negative feedback mechanism, i.e. repression of cell proliferation, of E2-induced cell hyperproliferation37. In addition, HIC1, the most pronouncedly downregulated DEG, is a known tumour suppressor gene38. Interestingly, the expression of CDKN2D, HIC1, and PSAT1 have been linked to the transcriptional factor of cell cycle regulation E2F127,39,40. The latter, in turn, has been shown to be mediated by E241. Therefore, the results may indicate a complex interplay of molecular factors leading to the tissue specific differential expression of CDKN2D, HIC1, and PSAT1 at high, as well as at low-dose E2 exposure. Lastly, RASSF1 was lower expressed in the liver of the HIGH dose group. Lower expression of RASSF1 has been reported to play a role in tumorigenesis30. In addition to that, silencing of RASSF1 gene expression has been linked to de novo hypermethylation; RASSF1 hypermethylation has previously been found in human tumours, including liver cancer30, indicating the HIGH dose of E2 as potential cancer-inducing risk factor.
The gene specific DNA methylation of selected regions in the CDKN2D, PSAT1 and RASSF1 genes revealed subtle, but particularly interesting changes. In terms of number of CpG sites displaying a significant difference in DNA methylation, the liver was the most affected tissue.
The methylation state of different regions within a gene is known to differentially impact gene expression. In this respect, promoter hypermethylation and gene silencing have been studied most extensively42. A negative correlation between mRNA transcription and DNA methylation of the promoter region of CDKN2D has recently been shown26. Likewise, the analysed sites CpG −200 and CpG −190 in the putative promoter region of CDKN2D in the corpus luteum of F0 sows showed subtle changes in the methylation status that negatively correlated with its gene expression. On the contrary, the methylation status of neither CpG −200 nor CpG −190 in the liver did not. Specific methylation patterns of known transcriptionally silenced tumour suppressor genes in primary tumours have been shown previously; the CDKN2D promotor was differentially methylated in different types of colorectal cancer, but not specifically correlated with gene expression43. Thus, while our results clearly indicate tissue-specific methylation, the methylation independent gene expression which probably underlies the lack of correlation between methylation and gene expression in the liver, remains to be further elucidated.
Besides the promotor methylation as regulator of gene expression, regions close to the transcriptional start site such as first exons, CpG islands and CpG shores have also been shown to influence gene expression44. In both the PSAT1 first exon coding region and the RASSF1 first exon 5′UTR, several CpN sites were significantly hypomethylated in the liver of F0 animals across treatment groups. Despite the consistent hypomethylation of PSAT1, there was no correlation with the gene expression of PSAT1 in the liver of F0 sows. We assume that the location of the PSAT1 target region, i.e. the first exon coding region, might explain the absence of any correlation between methylation and gene expression. The methylation levels of RASSF1 in the NOEL and HIGH dose groups at CpG sites −134, −132, −65, and −59 significantly positively correlated with its gene expression. Our data underline the importance of CpG islands in the promotor region (CDKN2D) and the first exon 5′UTR (PSAT1) as gene expression regulators45,46.
In general, the changes in DNA methylation in our study were very subtle compared to differences found during development, between tissues and in normal versus cancer cells47, raising the question whether methylation status solely resembles a molecular fingerprint of exposure, or that it emerges a precursor for development of disease later in life and/or evokes transgenerational epigenetic effects46. Developmental-stage specific DNA methylation changes might hinder the assessment of local DNA methylation during early embryo development, but it has previously been shown that the embryoblast and trophectoderm have reached relatively stable levels of DNA methylation in day 10 pig embryos48,49. In addition, an increased number of studies have shown that exposure to environmental compounds during pregnancy can affect epigenetic marks in developing offspring, thereby priming individuals for the development of disease later in life50. Previously, we have shown that the low-dose E2 exposure during pregnancy increased the body fat percentage of prepubertal male offspring25, and slightly changed bone density parameters of female offspring5. In addition, we have reported a subtle but significant hypermethylation of the BGN gene in prostate of F1 male offspring upon in utero exposure to HIGH E251. In the current study, we provide evidence that the impact of the low-dose E2 treatment during pregnancy on DNA methylation changes were evident in F0 tissues as well in female offspring and, most importantly, occurred as early as the blastocyst stage. Whether the exposure to E2 directly imposed a molecular fingerprint on the embryo or indirectly by perturbing the uterine environment, which in turn affected the susceptible preimplantation conceptus, remains to be shown. One drawback of the current study is that the sows (F0) analysed for a short-term treatment effect had received E2 twice, once during a whole gestation period and again for 10 days, with a non-treated period of at least 8 months in between. Driven by the 3R (replacement, reduction, and refinement) principle, the endogenous high production of placental E2 during the end of pregnancy, and the fact that lasting effects of E2 exposure are much more likely to occur during the periconceptual period, this potential disadvantage was accepted. We thereby hypothesized that both F1 sampled (1) at 10 days of pregnancy and (2) at one year post parturition experienced the same E2 and/or perturbed uterine milieu due to E2 during the periconceptional period. Our findings clearly, in line with literature, highlight the periconceptual period as sensitive time window for the action of EDC52, and show that the level of effect of estrogenic EDC is at much lower concentration than currently presumed31. This may be of relevance in case of, e.g. oral food contamination, an unexpected conception during ongoing contraceptive treatment, or for women undergoing ovarian stimulation protocols during stimulated cycles in routine ART programs.
Most surprisingly, CDKN2D was hypomethylated at CpG − 200 and CpG − 190 in both liver of F0 sows and blastocysts of F1, whereas these sites were found to be hypermethylated in the liver of F1 offspring at one year of age. Likewise, while CpG +128 PSAT1 was hypomethylated in both liver of F0 sows and blastocysts of F1, this CpG was similarly hypermethylated in the liver of F1 offspring at one year of age. In mice, the exposure to estrogenic EDC such as diethylstilboestrol (DES) and bisphenol-A (BPA) during pregnancy or neonatally has been shown to alter epigenetics marks in the uterus and liver, and has been linked to breast cancer via altered expression of histone methyltransferases53,54,55. One of the few studies investigating methylation changes upon exposure to estrogenic EDC in both F0 and F1 evidences DNA methylation changes of human foetal lung and placenta tissue upon in utero nicotine smoke exposure56. This epigenetic fingerprint is in line with the methylation patterns of CDKN2D and PSAT1 observed in the F0 tissues and the F1 blastocysts. The switch within the F1 during development, namely from hypomethylation in blastocysts to hypermethylation at one year of age remains puzzling. From a regulatory point of view, epigenetic effects are frequently observed as opposing phenotypes, e.g. over- and undergrowth, and resemble a disequilibrium or dysregulation. Thus, if DNA methylation leads to gene expression changes of CDKN2D and PSAT1 over time from foetus to adult, the observed hypermethylation in adult gilts could resemble the consequence of a dysregulated response. Unravelling the physiological DNA methylation changes during foetal and post-partum development over time, including the phase of puberty, may substantiate this assumption.
It seems highly unlikely that CDKN2D and PSAT1 are the only two genes in F1 targeted by the periconeptional E2 treatment. A genome-wide, high resolution DNA methylome screening might unravel further differential DNA methylation to functionally link the molecular state of the blastocysts with the sex-specific metabolic phenotype observed in the adult F15,25. Our data could therefore be indicative of the priming for development of disease later in life50.
In summary, the gestational effect of E2 was measurable as a tissue-specific molecular fingerprint on gene expression and DNA methylation including intergenerational epigenetic changes. The rising prevalence of endocrine related disorders in the recent past cannot be solely attributed to genetic factors and, therefore, our findings are relevant for human health50,51,53,54,55. Future studies should emphasize on studying short-term, intergenerational, as well as transgenerational tissue-specific effects to gain further understanding of the mechanism by which exposure to estrogenic EDC affects DNA methylation. Further, it should be investigated whether the absence of a clear phenotypic change upon the exposure to low-dose estrogenic EDC implies that the observed molecular effects are only transitory and thus non-alarming, or mirror latent fingerprints that may lead to a functional effect later in time. The latter implies that epigenetics could serve as novel sensitive parameter for risk assessment of estrogenic EDC.
Materials and Methods
Animals
The animal trial was performed with German Landrace sows as previously described25. Experimental procedures are described in detail in the Supplementary Text S1. In brief, animals were randomly distributed in four groups receiving different doses of E2 (0.05, 10 and 1000 µg/kg body weight/day, and a control ethanol carrier). The doses of E2 were orally applied to sows (n = 4–6 per treatment group) twice a day, to guarantee lasting elevated circulating estrogen concentrations, during the whole period of pregnancy. On day 7 of pregnancy, the average weight ± standard deviation of the sows was 218 ± 41, 239 ± 44, 239 ± 40, and 221 ± 40 kg for control, ADI, NOEL, and HIGH, respectively. On day 107 of pregnancy, the average weight ± standard deviation was 273 ± 29, 293 ± 34, 295 ± 31, and 286 ± 30 kg for for control, ADI, NOEL, and HIGH, respectively. The average age ± standard deviation of the sows at delivery was 29 ± 9, 37 ± 10, 31 ± 6, and 27 ± 3 months for control, ADI, NOEL, and HIGH, respectively. The average number of living offspring ± standard deviation was 8 ± 4, 10 ± 2, 10 ± 4, and 10 ± 3 piglets for for control, ADI, NOEL, and HIGH, respectively. The average birth weight of the offspring ± standard deviation was 1.8 ± 0.7, 2.0 ± 0.5, 1.7 ± 0.6, and 1.5 ± 0.4 kg for for control, ADI, NOEL, and HIGH, respectively. Piglets were weaned at 21 days of age and followed up to 1 year of age. Sows were subjected to an additional similar treatment during a following conception and slaughtered one hour after the last treatment on day 10 of this pregnancy, whereby each sow received the same treatment dose in both pregnancies. Embryos were recovered by uterine flushing upon slaughter. For the subsequent analysis, embryos with a minimum DNA content of 125 ng and an average DNA content of 18.1 ± 1.5 ng/µl (mean ± SEM) for the local DNA methylation analysis were selected randomly, and distributed as follows: control, ADI, NOEL, and HIGH dose, 9 embryos (4 females, 5 males), 9 embryos (5 females, 4 males), 10 embryos (4 females, 6 males), and 8 embryos (4 females, 4 males), respectively. The female offspring were slaughtered at one year of age (n = 6 per treatment group) during the luteal phase (10–13 days following estrus behaviour) after at least three ostroes cycles after onset of pubery. All experiments and samplings were conducted in accordance with accepted standards of humane animal care and were approved by the District Government of Upper Bavaria, reference # 55.2-1-54-2531-68-09.
DNA/RNA extraction, quantification and quality assessment
Tissue samples and embryos for nucleic acid extraction were collected after slaughter, snap frozen in liquid nitrogen and stored at −80 °C until further analysis. Total DNA and RNA of tissues was extracted with the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. Due to high fiber content of heart and skeletal muscle, total DNA and RNA was extracted using a specialized AllPrep Universal DNA/RNA/microRNA kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. After extraction, RNA and DNA was quantified and the quality was assessed as explained previously51. Total DNA and RNA of embryos was extracted with the AllPrep DNA/RNA Micro Kit (Qiagen, Hilden, Germany), according to manufacturer’s instructions with slight modifications (SI Text). Purity and quantity was assessed spectrophotometrically using the NanoDrop 1000 (peqLab, Erlangen, Germany). Additionally, RNA quantity of embryos was determined using the Qubit (Invitrogen) with the Qubit™ RNA BR Assay.
Global DNA methylation analysis
Global DNA methylation was assessed by combining enzyme digestion with pyrosequencing using the LUminometric Methylation Assay (LUMA) as described previously57, with minor modifications due to the PyroMark Q48 Autoprep System (Qiagen): the complete reaction digest was transferred to each well of a PyroMark Q48 Autoprep Disc, and 6.5 µl of PyroMark Annealing Buffer was added automatically by the instrument. The nucleotides were added to the cartridge chambers as follows: 50 μl dATPαS and 50 μl water to “A”, 50 μl dTTP and 50 μl water to “T”, 50 μl dCTP and 50 μl dGTP to “C”, and 100 μl water to “G”. Pyrosequencing was performed with the nucleotide dispensation order 5′-ACTCGA-3′.
Gene expression analysis
The cDNA was synthesized as described previously51. Target gene specific primers were designed using the publicly available NCBI Primer Blast software and purchased from Microsynth (Balgach, Switzerland). Target genes were selected based on their previously reported E2-mediated effects. The PubMed database was used to search papers with the following keywords: ‘estradiol-17β’, ‘transcriptomics’, ‘gene expression’, ‘pregnancy’, ‘cancer’, and ‘tumor suppressors’. All primers were tested for specificity by melt-curve analysis and gel electrophoresis. Primer sequences for all target genes can be found in (Supplementary Table S1). To determine gene expression, a high throughput gene expression platform was used based on Dynamic Array™ microfluidic chips (Fluidigm 96.96 Dynamic Array 15 IFC, BioMark™ Systems). The mRNA expression of selected target genes and four reference genes was measured at an annealing temperature of 60 °C and according to the Fluidigm Advanced Development Protocol 14 (SI Text). Cq values were obtained using a single threshold, a linearity range of 7 to 21 Cq was selected for further analysis, and relative quantification of the mRNA levels was performed with the 2−ΔΔCq method58. The geometrical mean of four reference genes (HPRT1, TBP, RPL4 and ActB) was used for normalization (Supplementary Table S2).
Design of DNA methylation assays
The region of interest for the DNA methylation assay of CDKN2D was selected based on previous evidence of a putative promoter region in Homo sapiens lying within the region −774 bp to +18 bp from the translational start site26. This region was aligned with the CDKN2D sequence of Sus scrofa with the NCBI Blastn tool revealing 77% identity. The pyrosequencing was performed on CpN sites −269 to −182 for methylation analysis (Supplementary Figs S2 and 3). The region of interest for the DNA methylation assay of RASSF1 was also selected based on previously documented evidence16. Similar to Homo sapiens, the RASSF1 gene in Sus scrofa revealed a large CpG island spanning −304 bp to +349 bp from the translational start site (Supplementary Fig. S3). The CpG islands indicated in the figure were identified using a publicly available software (Meth Primer)59, with the following criteria (Island size > 100, GC % > 50.0, Obs/Exp > 0.6). Pyrosequencing was performed on CpG sites −137 to −55 (Fig. 3) in the 5′UTR. For PSAT1, thirteen CpN sites from +119 to +175 in the coding region of the first exon were selected for methylation analysis (Fig. 3).
Bisulfite pyrosequencing
The bisulfite pyrosequencing was performed as explained earlier51, with slight modifications (SI Text). The primers for the PyroMark assays were designed using the PyroMark Assay Design Software 2.0 (Qiagen, Hilden, Germany) (Supplementary Table S3). The annealing temperatures were 56 °C, 58 °C, and 56 °C for the CDKN2D, PSAT1, and RASSF1 assays, respectively. The product specificity was validated by gel electrophoresis and CpN methylation was quantified using the PyroMark Q48 Autoprep System (Qiagen) and the PyroMark Q48 Advanced CpG Reagents (Qiagen). Methylation values [%] were calculated with the PyroMark Q48 Autoprep 2.4.1 Software (Qiagen).
Statistical analysis
All statistical analyses were performed using the IBM SPSS Statistics Software version 22 (IBM, 295 Böblingen, Germany). In order to determine the effects of E2 on gene expression, global DNA methylation and gene specific DNA methylation, a one-way ANOVA was used to detect significant differences followed by Dunnett’s post hoc test. To detect site-specific changes, and as reported previously60,61, each individual CpN site has been analysed by a one-way ANOVA followed by Dunnett’s post hoc test. The correlation analyses were performed using Pearson correlation. Statistically significance is displayed in the figures by *, **, and *** indicating statistical significance with respect to the control group at p < 0.05, p < 0.01, and p < 0.001, respectively.
References
Rosenmai, A. K. et al. An effect-directed strategy for characterizing emerging chemicals in food contact materials made from paper and board. Food and chemical toxicology: an international journal published for the British Industrial Biological Research Association 106, 250–259, https://doi.org/10.1016/j.fct.2017.05.061 (2017).
Johansson, H. K. L., Svingen, T., Fowler, P. A., Vinggaard, A. M. & Boberg, J. Environmental influences on ovarian dysgenesis - developmental windows sensitive to chemical exposures. Nat Rev Endocrinol, https://doi.org/10.1038/nrendo.2017.36 (2017).
Solecki, R. et al. Scientific principles for the identification of endocrine-disrupting chemicals: a consensus statement. Archives of Toxicology 91, 1001–1006, https://doi.org/10.1007/s00204-016-1866-9 (2017).
Soto, A. M. & Sonnenschein, C. Environmental causes of cancer: endocrine disruptors as carcinogens. Nat Rev Endocrinol 6, 363–370, https://doi.org/10.1038/nrendo.2010.87 (2010).
Floter, V. L. et al. Sex-specific effects of low-dose gestational estradiol-17beta exposure on bone development in porcine offspring. Toxicology 366-367, 60–67, https://doi.org/10.1016/j.tox.2016.07.012 (2016).
Gore, A. C. et al. EDC-2: The Endocrine Society’s Second Scientific Statement on Endocrine-Disrupting Chemicals. Endocrine reviews 36, E1–e150, https://doi.org/10.1210/er.2015-1010 (2015).
Adeel, M., Song, X., Wang, Y., Francis, D. & Yang, Y. Environmental impact of estrogens on human, animal and plant life: A critical review. Environment International 99, 107–119, https://doi.org/10.1016/j.envint.2016.12.010 (2017).
Leung, K. C., Johannsson, G., Leong, G. M. & Ho, K. K. Estrogen regulation of growth hormone action. Endocrine reviews 25, 693–721, https://doi.org/10.1210/er.2003-0035 (2004).
Chalbos, D., Vignon, F., Keydar, I. & Rochefort, H. Estrogens stimulate cell proliferation and induce secretory proteins in a human breast cancer cell line (T47D). The Journal of clinical endocrinology and metabolism 55, 276–283, https://doi.org/10.1210/jcem-55-2-276 (1982).
Tong, W. & Pollard, J. W. Progesterone inhibits estrogen-induced cyclin D1 and cdk4 nuclear translocation, cyclin E- and cyclin A-cdk2 kinase activation, and cell proliferation in uterine epithelial cells in mice. Mol Cell Biol 19, 2251–2264 (1999).
Jadhav, R. R. et al. Genome-wide DNA methylation analysis reveals estrogen-mediated epigenetic repression of metallothionein-1 gene cluster in breast cancer. Clinical epigenetics 7, 13, https://doi.org/10.1186/s13148-015-0045-9 (2015).
Sato, K. et al. Neonatal exposure to diethylstilbestrol alters expression of DNA methyltransferases and methylation of genomic DNA in the mouse uterus. Endocrine journal 56, 131–139 (2009).
Laing, L. V. et al. Bisphenol A causes reproductive toxicity, decreases dnmt1 transcription, and reduces global DNA methylation in breeding zebrafish (Danio rerio). Epigenetics 11, 526–538, https://doi.org/10.1080/15592294.2016.1182272 (2016).
Fernandez, S. V. et al. Expression and DNA methylation changes in human breast epithelial cells after bisphenol A exposure. International journal of oncology 41, 369–377, https://doi.org/10.3892/ijo.2012.1444 (2012).
Rodriguez, B. A. et al. Estrogen-mediated epigenetic repression of the imprinted gene cyclin-dependent kinase inhibitor 1C in breast cancer cells. Carcinogenesis 32, 812–821, https://doi.org/10.1093/carcin/bgr017 (2011).
Starlard-Davenport, A. et al. Mechanisms of epigenetic silencing of the Rassf1a gene during estrogen-induced breast carcinogenesis in ACI rats. Carcinogenesis 31, 376–381, https://doi.org/10.1093/carcin/bgp304 (2010).
Sunami, E. et al. Estrogen receptor and HER2/neu status affect epigenetic differences of tumor-related genes in primary breast tumors. Breast cancer research: BCR 10, R46, https://doi.org/10.1186/bcr2098 (2008).
Bommarito, P. A., Martin, E. & Fry, R. C. Effects of prenatal exposure to endocrine disruptors and toxic metals on the fetal epigenome. Epigenomics 9, 333–350, https://doi.org/10.2217/epi-2016-0112 (2017).
Inbar-Feigenberg, M., Choufani, S., Butcher, D. T., Roifman, M. & Weksberg, R. Basic concepts of epigenetics. Fertility and sterility 99, 607–615, https://doi.org/10.1016/j.fertnstert.2013.01.117 (2013).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322, https://doi.org/10.1038/nature08514 (2009).
He, Y. & Ecker, J. R. Non-CG Methylation in the Human Genome. Annual review of genomics and human genetics 16, 55–77, https://doi.org/10.1146/annurev-genom-090413-025437 (2015).
Heard, E. & Martienssen, R. A. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157, 95–109, https://doi.org/10.1016/j.cell.2014.02.045 (2014).
Vaiserman, A. Early-life Exposure to Endocrine Disrupting Chemicals and Later-life Health Outcomes: An Epigenetic Bridge? Aging and disease 5, 419–429, https://doi.org/10.14336/ad.2014.0500419 (2014).
Ho, S. M. et al. Environmental epigenetics and its implication on disease risk and health outcomes. ILAR journal 53, 289–305, https://doi.org/10.1093/ilar.53.3-4.289 (2012).
Furst, R. W. et al. Maternal low-dose estradiol-17beta exposure during pregnancy impairs postnatal progeny weight development and body composition. Toxicology and applied pharmacology 263, 338–344, https://doi.org/10.1016/j.taap.2012.07.006 (2012).
Zhu, W. G. et al. Increased expression of unmethylated CDKN2D by 5-aza-2′-deoxycytidine in human lung cancer cells. Oncogene 20, 7787–7796, https://doi.org/10.1038/sj.onc.1204970 (2001).
Yang, Y. et al. PSAT1 regulates cyclin D1 degradation and sustains proliferation of non-small cell lung cancer cells. International journal of cancer 136, E39–50, https://doi.org/10.1002/ijc.29150 (2015).
Vie, N. et al. Overexpression of phosphoserine aminotransferase PSAT1 stimulates cell growth and increases chemoresistance of colon cancer cells. Molecular cancer 7, 14, https://doi.org/10.1186/1476-4598-7-14 (2008).
Thaler, S., Hahnel, P. S., Schad, A., Dammann, R. & Schuler, M. RASSF1A mediates p21Cip1/Waf1-dependent cell cycle arrest and senescence through modulation of the Raf-MEK-ERK pathway and inhibition of Akt. Cancer research 69, 1748–1757, https://doi.org/10.1158/0008-5472.can-08-1377 (2009).
Pfeifer, G. P. & Dammann, R. Methylation of the tumor suppressor gene RASSF1A in human tumors. Biochemistry. Biokhimiia 70, 576–583 (2005).
WHO, J. F. W. E. C. o. F. A. In (52: 1999.02.02.-11: Rome) (eds Food and Agriculture Organization of the United Nations & World Health Organization) (World Health Organization, 1999).
Strauss, J. F. 3rd, Martinez, F. & Kiriakidou, M. Placental steroid hormone synthesis: unique features and unanswered questions. Biology of reproduction 54, 303–311 (1996).
Gordon, F. K. et al. Research resource: Aorta- and liver-specific ERalpha-binding patterns and gene regulation by estrogen. Molecular endocrinology (Baltimore, Md.) 28, 1337–1351, https://doi.org/10.1210/me.2013-1395 (2014).
Diamanti-Kandarakis, E. et al. Endocrine-disrupting chemicals: an Endocrine Society scientific statement. Endocrine reviews 30, 293–342, https://doi.org/10.1210/er.2009-0002 (2009).
Crain, D. A. et al. Female reproductive disorders: the roles of endocrine-disrupting compounds and developmental timing. Fertility and sterility 90, 911–940, https://doi.org/10.1016/j.fertnstert.2008.08.067 (2008).
Lowe, D. A., Baltgalvis, K. A. & Greising, S. M. Mechanisms behind estrogen’s beneficial effect on muscle strength in females. Exercise and sport sciences reviews 38, 61–67, https://doi.org/10.1097/JES.0b013e3181d496bc (2010).
Mokarram, P. et al. Interconnection of Estrogen/Testosterone Metabolism and Mevalonate Pathway in Breast and Prostate Cancers. Current molecular pharmacology 10, 86–114, https://doi.org/10.2174/1874467209666160112125631 (2017).
Wales, M. M. et al. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med 1, 570–577 (1995).
Carcagno, A. L. et al. E2F1-mediated upregulation of p19INK4d determines its periodic expression during cell cycle and regulates cellular proliferation. PloS one 6, e21938, https://doi.org/10.1371/journal.pone.0021938 (2011).
Zheng, J. et al. Signification of Hypermethylated in Cancer 1 (HIC1) as Tumor Suppressor Gene in Tumor Progression. Cancer microenvironment: official journal of the International Cancer Microenvironment Society 5, 285–293, https://doi.org/10.1007/s12307-012-0103-1 (2012).
Wang, W., Dong, L., Saville, B. & Safe, S. Transcriptional activation of E2F1 gene expression by 17beta-estradiol in MCF-7 cells is regulated by NF-Y-Sp1/estrogen receptor interactions. Molecular endocrinology (Baltimore, Md.) 13, 1373–1387, https://doi.org/10.1210/mend.13.8.0323 (1999).
Siegfried, Z. & Simon, I. DNA methylation and gene expression. Wiley interdisciplinary reviews. Systems biology and medicine 2, 362–371, https://doi.org/10.1002/wsbm.64 (2010).
Cheng, Y. W., Shawber, C., Notterman, D., Paty, P. & Barany, F. Multiplexed profiling of candidate genes for CpG island methylation status using a flexible PCR/LDR/Universal Array assay. Genome research 16, 282–289, https://doi.org/10.1101/gr.4181406 (2006).
Zhang, W., Spector, T. D., Deloukas, P., Bell, J. T. & Engelhardt, B. E. Predicting genome-wide DNA methylation using methylation marks, genomic position, and DNA regulatory elements. Genome biology 16, 14, https://doi.org/10.1186/s13059-015-0581-9 (2015).
Jones, P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews. Genetics 13, 484–492, https://doi.org/10.1038/nrg3230 (2012).
Leenen, F. A., Muller, C. P. & Turner, J. D. DNA methylation: conducting the orchestra from exposure to phenotype? Clinical epigenetics 8, 92, https://doi.org/10.1186/s13148-016-0256-8 (2016).
Clark, S. J. & Melki, J. DNA methylation and gene silencing in cancer: which is the guilty party? Oncogene 21, 5380–5387, https://doi.org/10.1038/sj.onc.1205598 (2002).
Fulka, J., Fulka, H., Slavik, T., Okada, K. & Fulka, J. Jr. DNA methylation pattern in pig in vivo produced embryos. Histochemistry and cell biology 126, 213–217, https://doi.org/10.1007/s00418-006-0153-x (2006).
Reik, W., Dean, W. & Walter, J. Epigenetic reprogramming in mammalian development. Science 293, 1089–1093, https://doi.org/10.1126/science.1063443 (2001).
Martos, S. N., Tang, W. Y. & Wang, Z. Elusive inheritance: Transgenerational effects and epigenetic inheritance in human environmental disease. Progress in biophysics and molecular biology 118, 44–54, https://doi.org/10.1016/j.pbiomolbio.2015.02.011 (2015).
Kradolfer, D. et al. Epigenetic effects of prenatal estradiol-17beta exposure on the reproductive system of pigs. Molecular and cellular endocrinology 430, 125–137, https://doi.org/10.1016/j.mce.2016.04.005 (2016).
Van Soom, A. & Fazeli, A. Epigenetics and periconception environment: an introduction. Reproduction, fertility, and development 27, iii–v, https://doi.org/10.1071/RDv27n5_IN (2015).
Jefferson, W. N. et al. Persistently altered epigenetic marks in the mouse uterus after neonatal estrogen exposure. Molecular endocrinology (Baltimore, Md.) 27, 1666–1677, https://doi.org/10.1210/me.2013-1211 (2013).
Doherty, L. F., Bromer, J. G., Zhou, Y., Aldad, T. S. & Taylor, H. S. In utero exposure to diethylstilbestrol (DES) or bisphenol-A (BPA) increases EZH2 expression in the mammary gland: an epigenetic mechanism linking endocrine disruptors to breast cancer. Hormones & cancer 1, 146–155, https://doi.org/10.1007/s12672-010-0015-9 (2010).
Kim, J. H. et al. Perinatal bisphenol A exposure promotes dose-dependent alterations of the mouse methylome. BMC genomics 15, 30, https://doi.org/10.1186/1471-2164-15-30 (2014).
Chhabra, D. et al. Fetal lung and placental methylation is associated with in utero nicotine exposure. Epigenetics 9, 1473–1484, https://doi.org/10.4161/15592294.2014.971593 (2014).
Furst, R. W., Meyer, H. H., Schweizer, G. & Ulbrich, S. E. Is DNA methylation an epigenetic contribution to transcriptional regulation of the bovine endometrium during the estrous cycle and early pregnancy? Molecular and cellular endocrinology 348, 67–77, https://doi.org/10.1016/j.mce.2011.07.032 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif.) 25, 402–408, https://doi.org/10.1006/meth.2001.1262 (2001).
Li, L. C. & Dahiya, R. MethPrimer: designing primers for methylation PCRs. Bioinformatics (Oxford, England) 18, 1427–1431 (2002).
Hsieh, H. Y. et al. Epigenetic silencing of the dual-role signal mediator, ANGPTL4 in tumor tissues and its overexpression in the urothelial carcinoma microenvironment. Oncogene 37, 673–686, https://doi.org/10.1038/onc.2017.375 (2018).
Mao, W. et al. Pyrosequencing analysis of methylation levels of clock genes in leukocytes from Parkinson’s disease patients. Neuroscience letters 668, 115–119, https://doi.org/10.1016/j.neulet.2018.01.027 (2018).
Acknowledgements
The knowledgable contribution of Myriam Reichenbach and of several members of both Physiology Weihenstephan, TUM and Animal Physiology, ETH is highly acknowledged. Gene expression data were generated in collaboration with the Genetic Diversity Centre (GDC), ETH Zurich. The authors are active members of the European Union COST actions FA1201 EPICONCEPT and CA16119 CELLFIT. This article is dedicated to Heinrich HD Meyer†. The ZIEL PhD Graduate School “Nutritional Adaptation”, Technische Universität München, and the Swiss National Science Foundation SNSF (IZCOZ0_177141) partially funded the study. They were neither involved in decisions regarding the study design and conductance nor in any further analysis.
Author information
Authors and Affiliations
Contributions
V.v.d.W. performed the analytical experiments, analysed the data and wrote the manuscript. V.L.F. conceived of the study, performed the animal experiments, wrote the manuscript. S.E.U. conceived of the study, supervised data analysis, and critically reviewed the manuscript. All authors reviewed the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
van der Weijden, V.A., Flöter, V.L. & Ulbrich, S.E. Gestational oral low-dose estradiol-17β induces altered DNA methylation of CDKN2D and PSAT1 in embryos and adult offspring. Sci Rep 8, 7494 (2018). https://doi.org/10.1038/s41598-018-25831-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-25831-9
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.