Abstract
Triggering receptor expressed on myeloid cells 2 (TREM2) is known for its anti-inflammatory properties during the immune response, and influences negatively on TNF-α expression levels. Genetic epidemiology studies have identified polymorphisms located in the TREM2 gene associated with neurodegenerative and chronic inflammatory diseases. TREM2 levels have been observed to affect plasma levels of TNF-α and plaque stability in symptomatic and asymptomatic patients with carotid stenosis. In this study, we investigated polymorphisms located in the TREM2 gene region and association with TNF-α levels and the intima media thickness of the femoral artery. The discovery population from the STANISLAS Family Study comprised of 809 individuals, whereas the replication population utilized an independent cohort of French origin (n = 916). Our results suggest that the minor allele (T) of SNP rs6918289 is positively associated with elevated plasma levels of TNF-α in discovery and replication populations (P = 0.0026, SE = 0.04 and P = 0.023, SE = 0.09, respectively), including femoral artery thickness in the discovery cohort (P = 0.026, SE = 0.009). Results indicate that rs6918289 may be considered as a risk factor for inflammatory diseases and could be used in stratified medicine with patients diagnosed with chronic inflammatory-related conditions, such as atherosclerosis.
Similar content being viewed by others
Introduction
The triggering receptors expressed on myeloid cells (TREM) family molecules are members of the immunoglobulin superfamily of receptors. All five genes from the TREM family (Table 1) are situated in the 6p21.1 region of the chromosome1 and mediate signaling in immune cells, thus playing critical roles in inflammatory responses2. The region 6p21.1 is in proximity to the MHC/HLA region of the genome, which is implicated in the immune response, autoimmunity and risk of autoimmune diseases3,4. Specifically, the TREM2 molecule is primarily expressed on the cell surface of macrophages and dendritic cells derived from monocytes, as well as in microglia and osteoclasts. TREM2 binds to the DAP12 trans-membrane molecule and is responsible for a series of tyrosine phosphorylation reactions that regulate various inflammatory responses2,5. TREM2 portrays anti-inflammatory properties during the immune response6,7, including: stimulation of phagocytosis and suppression of cytokine production, e.g. TNF-α8,9, one of the most important molecules for the regulation of inflammation, and reflects the degree of inflammatory response.
It is evident that TREM2 acts as a protective molecule in chronic inflammatory diseases. For example, studies in transgenic mice have demonstrated that deficiency of TREM2 protein may accelerate the aging process, reduce microglial activity and result in neuroinflammation, which plays a major role in all neurodegenerative diseases10. Furthermore, atherosclerosis is a chronic cardiovascular inflammatory disease, caused by activation of the immune system mediating the chronic inflammatory process of the arterial wall. Interestingly, TREM2 has also been shown to play an important role in the stability of atherosclerotic plaques11.
Polymorphisms located in the TREM2 gene have been correlated with neurodegenerative and chronic inflammatory diseases, such as Alzheimer’s disease12,13, frontotemporal dementia14,15, Parkinson’s disease16,17, inflammatory bowel disease18 and stroke19. However, no genetic determinants have been identified in the TREM2 locus affecting plasma levels of TNF-α and/or intima media thickness.
Due to the role of TREM2 in the inflammatory response and stability of atherosclerotic plaques, we hypothesize that polymorphisms in the TREM2 gene region may influence the inflammatory process and subsequent atherosclerotic plaque formation. In this investigation, we have studied the association of variants located in the TREM2 gene regionwith plasma levels of TNF-α and intima media thickness of the femoral artery (IMT-F).
Results
Information on the genotyped polymorphisms as well as the demographic and clinical characteristics of the studied populations is shown in Tables 2 and 3, respectively. All SNPs analyzed within this investigation were in agreement with Hardy-Weinberg equilibrium (P > 0.001).
Genetic association of SNPs in TREM2 gene region with TNF-α concentration
Firstly, association analysis was performed in 139 children from the STANISLAS Family Study (SFS). SNPs that were located and previously genotyped in the TREM2 gene region were tested for association with TNF-α concentration. Among the five SNPs studied, the minor allele (T) of rs6918289 was significantly associated with increased TNF-α concentrations (P = 0.0015, Table 4).
Secondly, rs6918289 was genotyped in 393 adult and additional 277 children relatives from the SFS. Analysis was performed in the combined population of SFS (n total = 809) using three genetic models. The additive genetic model showed a positive association between polymorphism rs6918289, located in the TREM2 gene downstream region and TNF-α plasma levels (P = 0.0026, β = 0.13, Table 5) (Fig. 1). The recessive model showed stronger genetic association with TNF-α plasma levels (P = 0.0017, β = 0.498, Table 5).
Further association analysis was performed in an independent French population of European ancestry, consisting of 916 individuals. However, only a marginal association for the additive model was observed (P = 0.073, β = 0.042, Table 5). A significant association was observed for the recessive model (P = 0.023, β = 0.202, Table 5). Interestingly, the minor allele (T) of SNP rs6918289 was associated with elevated levels of TNF−α in the discovery and replication populations.
Meta-analysis of the SFS and the replication populations revealed similar results: A marginal association with TNF-α for the additive model results (P = 0.072) that becomes significant in the recessive model (P = 0.0003).
Genetic association of rs6918289 with IMT-F
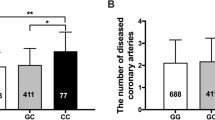
A sub-group of 350 individuals, consisting of adults and children from the SFS population, where IMT-F measurements were available, was used to identify association with SNP rs6918289. The additive genetic model showed significant association (P = 0.026, β = 0.02, Table 6). The association was also significant for the dominant model (P = 0.026, β = 0.024, Table 6). Thus, the minor allele (T), of SNP rs6918289 was associated with increased thickness of the femoral artery (Fig. 2).
Bioinformatics analyses
Polymorphism location
SNP rs6918289 is located on chromosome 6 in p21.1 region. Located at 41 134 089 bp, it is an intron variant of adenylate cyclase 10 pseudogene 1 (ADC10P1) (Sup. Figure 1). The polymorphism on the forward strand is G > T with a minor allele frequency of 0.07 for thymine in 1000 Genome project20,21.
Phylogenetic context
The guanine polymorphism of rs6918289 G > T is evolutionary and well-conserved in primates and mammals in general (Sup. Table 1). Further phylogenetic studies performed in 33 mammals and 46 vertebrates (Sup. Figure 2) showed that this polymorphism has a slower evolution rate than expected, having a PhyloP score of 0.061 and 0.056 respectively. This strengthens the idea that this variant is involved in important molecular mechanisms and that its preservation has been sustained throughout the natural selection processes.
Genomic context
SNP rs6918289 variant overlaps 2 transcripts and is located between two terminal exons of ADCY10P1: ADCY10P1-202 (3785 bp - between exon 17 and 18) and ADCY10P1-203 (4569 bp - between exon 18 and 19), both leading to transcripts that are not translated into proteins (Sup. Figure 2). Furthermore, according to the expressed sequence tags database (dbEST, https://www.ncbi.nlm.nih.gov/dbEST) available in Ensembl, the polymorphism rs6918289 is located within an intense transcriptionally active locus (Sup. Figure 3)20,21.
Discussion
The anti-inflammatory effects of TREM2 have been described in several studies. Indeed, knockdown or silencing of TREM2 gene results in increased levels of different pro-inflammatory molecules, among them TNF-α6,7,22. Despite the fact that several polymorphisms within the TREM2 gene have been related with numerous neurodegenerative and inflammatory diseases12,16,18,19, to date, no genetic determinants have been identified in the TREM2 locus (6p21.1) affecting TNF-α concentrations.
In this study, we evaluated the effects of SNPs located in the TREM2 gene region on TNF-α concentrations and IMT-F measurements. For 139 children from the SFS population, genotypes for five SNPs (rs7748777, rs6918289, rs7759295, rs9357347 and rs6915083) located in the TREM2 gene region were readily available. After conducting association analysis for the five aforementioned SNPs with TNF-α concentrations, we observed the minor allele (T) of SNP rs6918289, located in the downstream region of the TREM2 gene, to be associated with elevated levels of TNF-α (P = 0.0003). Importantly, further analysis in a larger sample of SFS population, as well as replication in an independent population and a meta-analysis of discovery and replication populations confirmed this novel association. Subsequently, we also performed association analysis of rs6918289 with intima-media thickness of the femoral artery in 350 individuals of the discovery population. Results suggest that the minor allele (T) of SNP rs6918289 is associated with increasing intima-media thickness of the femoral artery. Therefore, this is evidence that the minor T allele may be considered as a risk allele for inflammatory diseases and atherosclerosis.
The role of both studied phenotypes in the development of atherosclerosis and the prognosis of atherosclerotic patients is well documented. TNF-α is a key regulator of immune response and alterations of its levels lead to elevated inflammation and a subsequent deterioration of the outcome of patients with cardiovascular diseases23,24. Also, one of the early processes that lead to atherosclerosis is the arterial remodeling and one effective way that provides information about this process is measuring the intima-media thickness. Indeed, IMT is predictive of atherosclerosis in asymptomatic individuals25,26, and also provides information about the degree of atherosclerosis27,28 as well as predicting the future risk of suffering a myocardial infarction event29.
Chromosomal 6p21.1 genetic region, which is in proximity to the MHC and HLA regions, has been highly studied, and associated with autoimmune diseases3,4. Interestingly, previous GWAS studies have associated polymorphisms located within this genetic region with atherosclerotic stroke30. Although SNP rs6918289 is not in linkage disequilibrium with the variants associated with atherosclerotic stroke (rs556621 and rs556512), our results strengthen the idea that chromosomal 6p21.1 region could, indeed, be correlated with atherosclerosis risk. Our results are providing new insights on possible genetic regulations of pathological pathways that could lead to increased risk of atherosclerosis.
Despite the fact that this polymorphism contributes novel insights about its potential role in the development of cardiovascular diseases, the exact mechanisms by which this process is orchestrated is so far unknown and further studies are warranted. Indeed, there are no previous studies that associate this SNP or SNPs in linkage disequilibrium with TNF-α levels or IMT-F. In support of our findings, previous investigations suggest that TREM2 affects plasma TNF-α concentrations7,8,9,22. Also, our bioinformatic analyses indicate that rs6918289 is located in a transcriptional region of ADC10P1 gene that could impact TREM2 transcription levels31. Indeed, rs6918289 is referred by rSNPBASE (http://rsnp.psych.ac.cn/) as a post-transcriptional regulatory element and, in lymphocyte B cells, rs6918289 is associated with the PABPC1 protein. The PABPC1 protein binds to the 3′ poly(A) region of the mRNAs. Although the binding of this protein is necessary for the translation initiation, it is also required for poly(A) shortening, which is the first step in mRNA decay32. Thus, the minor allele (T) of SNP rs6918289 could affect the PABPC1 protein, which would at the same time affect the stability of the TREM2 mRNA, consequently contributing to elevated plasma TNF-α concentrations.
An alternative or synergistic mechanism is also conceivable. As shown by several studies, many RNAs bind to transcriptional repressor CTCF (11-zinc finger protein) to modulate its regulatory functions33. SNP rs6918289 is located between two CTCF genomic sequences that are “together” (Sup. Figures 2 and 3), which means they are forming a chromatin loop leading to a topologically associating domain (TAD) (Fig. 3).
TAD is a large, megabase-sized local chromatin interaction domain that forms a self-interacting genomic region34. The structural and organizational changes of such region affect gene expression and other cellular functions, such as re-organization of local interactions between enhancers and promoters. Moreover, DNA sequences within a TAD physically interact with each other more frequently than with sequences outside the TAD35. Thus, we hypothesize that CTCF, in association with the non-coding RNAs of ADCY10P1, could affect TREM2 expression levels. The minor allele (T) of SNP rs6918289 could promote the binding of CTCF and consequently trigger an insulation mechanism for TREM2 gene. These hypotheses should be tested in future works in order to conclude about the involved mechanisms.
Finally, the rs6918289 can be found in proximity to the TREML1 gene. In silico analysis showed significant correlation of variant with seven SNPs related to TREML1 gene (Sup. Table 2). Therefore, TREML1 could also be a possible mediator of the effect on plasma TNF-α concentration. TREML1 gene (Triggering Receptor Expressed On Myeloid Cells Like 1) encodes a protein, involved in platelet aggregation, inflammation, and cellular activation. Together with TREM 2 they are involved in common pathways in the setup of Alzheimer’s disease36.
Concerning the IMT-F results, we think that the effect of SNP rs6918289 could be indirect. Indeed, the remodeling of the artery is highly dependent on the inflammatory state, thus, increased TNF-α levels produced by the minor allele (T) of SNP rs6918289 could be responsible for the increased thickness of the femoral artery. Further studies will be necessary in order to clarify if this polymorphism or polymorphisms in linkage disequilibrium are capable of modulating the TREM2 protein levels and to discover which are the mechanisms behind this observation. However, important applications could be identified as these findings may be used for personalized treatments in patients with chronic inflammatory diseases37.
Limitation of our study is the unexplained mechanism of the rs6918289 effect on TNFα levels. Nevertheless, few pertinent hypotheses were proposed and future advances in genomic research with advanced bioinformatics tools will enable more precise explications of genetic interactions. Finally, genetic ancestry was not tested for populations used in this study; however strict inclusion criteria for origin were set aiming to collect homogeneous population, which are adequate for genetic association studies.
In summary, our study indicates variant rs6918289 located in the downstream region of the TREM2 gene as a candidate risk factor for inflammatory diseases because of its tight association with plasma TNF-α concentration. These findings support results of previous studies linking variants within chromosomal 6p21.1 loci to atherosclerosis and thus raising the awareness to consider genomic region 6p21.1 as candidate susceptibility loci for atherosclerosis.
Methods
Ethics statement
The samples are part of a human sample storage platform: BRC IGE-PCV number BB-0033–00051 located in Nancy, East France. All participants provided written informed consent. All populations involved in this study were recruited in accordance with the latest version of the Declaration of Helsinki for Ethical Principles for Medical Research Involving Human Subjects. All the protocols were approved by the local ethics committees for the protection of subjects for biomedical research: the Comité Consultatif de Protection des Persones dans la Recherche Biomédicale (CCPPRB).
Study participants
The discovery and replication populations are part of the Biological Resources Center ‘Interactions Gène-Environnement en Physiopathologie CardioVasculaire’ (BRC-IGE-PCV, number BB-0033–00051) in Nancy, France.
Individuals, comprising children and adults from the SFS38 were used as the discovery population. The SFS include more than 1,000 nuclear families, each composed of at least four individuals (two parents and two children). All families are of French origin (parents and grandparents of French origin and residence in the Lorraine region) and were recruited at the Centre for Preventive Medicine of Vandoeuvre-lés-Nancy (East France). Even though strict criteria of French (European) origin were set for the inclusion of individuals in both populations, genetic ancestry was not tested. All individuals were free of chronic diseases. Firstly, we used samples from 139 children of the SFS, where genotype information was readily available for the TREM2 gene region, to conduct association studies with plasma levels of TNF-α. Subsequently, 393 adult samples and 277 additional children samples from the corresponding families were included, reaching a total of 809 participants.
The replication cohort included 916 unrelated adults of French origin. Their inclusion criteria were the same as in the discovery cohort.
Blood samples and biological measurements
Blood samples were taken from individuals after an overnight fast (>8 hours). The plasma concentrations of TNF-α were measured by a commercially available enzyme-linked immunosorbent assay (ELISA) (R&D Systems, UK) according to manufacturer instructions. Body mass index (BMI) was calculated as weight divided by height squared (Kg/m²). IMT-F was measured in 350 SFS individuals (including children and adults) using B-mode ultrasound methods39. The right and left femoral arteries were examined with a 7.5 MHz probe, according to a protocol already described40,41. For each individual, two IMT-F measurements were obtained and right and left measurements were used to calculate the mean IMT-F (in mm). IMT-F data were not available in the replication population.
Genotyping
Genome-wide genotypes were readily available for all children samples from the SFS (n = 139). Genotyping was performed using Illumina® human CNV370-Duo array42. The Illumina® protocol for the BeadStation genotyping platform was used, followed by GenCall® software analysis to automatically collect, call genotypes, and designate confidence scores using the GenTrain clustering algorithm. The selection of the SNPs was done by first extracting from the genome-wide assay all the SNPs available in the region around TREM2 gene locus (50 kb upstream and downstream of the TREM2 gene). We employed PLINK software43 to conduct this analysis that resulted in the extraction and selection of five SNPs.
After performing initial association analysis in the child cohort of the SFS, polymorphism rs6918289 was de novo genotyped in the adult population (n = 393) and children population (n = 277) of SFS and the replication population (n = 916). Genotyping of rs6918289 in the replication population was conducted by Laboratory of the Government Chemist (LGC), using a PCR-based KASP assay44.
Statistical analysis
Normal distribution was tested by Kolmogorov-Smirnov test. If phenotypes did not conform to normal distribution, data were log transformed in order to reach normality. The Hardy-Weinberg Equilibrium (HWE) was tested using the chi-square test (P > 0.001). The SNP effects on the studied phenotypes were tested through linear regression adjusted for age, gender and BMI under three inheritance models (additive, dominant and recessive) and using the minor allele as the reference allele. Analyses were performed using the R package GWAF (Genome-Wide Association/Interaction Analysis and Rare Variant analysis with Family Data)45 taking into account familial resemblance. Alternatively, population characteristics were determined using SPSS statistical software version 20.0 (SPSS, Inc., Chicago, Illinois). Firstly, we tested the association of five SNPs located in the TREM2 gene region that were extracted from the GWAS available in children of the SFS (n = 139) with plasma TNF-α concentrations. Secondly, we tested the significance of SNP rs6918289 with TNF-α plasma levels in all available individuals of SFS (children and adults combined; n = 809) and in the replication population (n = 916). Meta-analysis of the discovery and replication cohorts was performed using the GWAMA software and a random-effect method. Thirdly, in 350 individuals of the SFS, rs6918289 was tested for association with IMT-F. Bonferroni correction was applied in order to adjust for multiple testing. The P-value threshold was set at P < 0.05/5 = 0.01 for the first analysis made in children, P < 0.05/1 = 0.05 for the analysis made in all individuals of the discovery population (children and adults) and P < 0.05/1 = 0.05 in the replication population and the meta-analysis.
Bioinformatics analysis
Location, genomic and phylogenetic context of rs6918289 were determined on the Human genome (GRCh38.p10) using Ensembl browser20. The putative regulatory role of rs6918289 was established using rSNPBASE21, and the PhyloP score was obtained using PhyloP software46.
Data availability
Extensive data is provided with this article and further information is available from the authors on request.
References
Committee, H. H. G. N. Gene names, https://www.genenames.org/ (2017).
Ford, J. W. & McVicar, D. W. TREM and TREM-like receptors in inflammation and disease. Curr Opin Immunol 21, 38–46, https://doi.org/10.1016/j.coi.2009.01.009 (2009).
Matzaraki, V., Kumar, V., Wijmenga, C. & Zhernakova, A. The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol 18, 76, https://doi.org/10.1186/s13059-017-1207-1 (2017).
Gough, S. C. & Simmonds, M. J. The HLA Region and Autoimmune Disease: Associations and Mechanisms of Action. Curr Genomics 8, 453–465, https://doi.org/10.2174/138920207783591690 (2007).
Lanier, L. L. DAP10- and DAP12-associated receptors in innate immunity. Immunol Rev 227, 150–160, https://doi.org/10.1111/j.1600-065X.2008.00720.x (2009).
Ito, H. & Hamerman, J. A. TREM-2, triggering receptor expressed on myeloid cell-2, negatively regulates TLR responses in dendritic cells. Eur J Immunol 42, 176–185, https://doi.org/10.1002/eji.201141679 (2012).
Turnbull, I. R. et al. Cutting edge: TREM-2 attenuates macrophage activation. J Immunol 177, 3520–3524 (2006).
Gao, X., Dong, Y., Liu, Z. & Niu, B. Silencing of triggering receptor expressed on myeloid cells-2 enhances the inflammatory responses of alveolar macrophages to lipopolysaccharide. Mol Med Rep 7, 921–926, https://doi.org/10.3892/mmr.2013.1268 (2013).
Liu, D. et al. Impact of TREM-2 gene silencing on inflammatory response of endotoxin-induced acute lung injury in mice. Mol Cell Biochem 394, 155–161, https://doi.org/10.1007/s11010-014-2091-6 (2014).
Raha, A. A. et al. Neuroprotective Effect of TREM-2 in Aging and Alzheimer’s Disease Model. Journal of Alzheimer’s disease: JAD 55, 199–217, https://doi.org/10.3233/jad-160663 (2017).
Rai, V., Rao, V. H., Shao, Z. & Agrawal, D. K. Dendritic Cells Expressing Triggering Receptor Expressed on Myeloid Cells-1 Correlate with Plaque Stability in Symptomatic and Asymptomatic Patients with Carotid Stenosis. PLoS One 11, e0154802, https://doi.org/10.1371/journal.pone.0154802 (2016).
Guerreiro, R. et al. TREM2 variants in Alzheimer’s disease. N Engl J Med 368, 117–127, https://doi.org/10.1056/NEJMoa1211851 (2013).
Jonsson, T. et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med 368, 107–116, https://doi.org/10.1056/NEJMoa1211103 (2013).
Guerreiro, R. J. et al. Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol 70, 78–84, https://doi.org/10.1001/jamaneurol.2013.579 (2013).
Cuyvers, E. et al. Investigating the role of rare heterozygous TREM2 variants in Alzheimer’s disease and frontotemporal dementia. Neurobiol Aging 35(726), e711–729, https://doi.org/10.1016/j.neurobiolaging.2013.09.009 (2014).
Benitez, B. A. & Cruchaga, C. & United States-Spain Parkinson’s Disease Research, G. TREM2 and neurodegenerative disease. N Engl J Med 369, 1567–1568, https://doi.org/10.1056/NEJMc1306509#SA4 (2013).
Rayaprolu, S. et al. TREM2 in neurodegeneration: evidence for association of the p.R47H variant with frontotemporal dementia and Parkinson’s disease. Mol Neurodegener 8, 19, https://doi.org/10.1186/1750-1326-8-19 (2013).
Correale, C. et al. Bacterial sensor triggering receptor expressed on myeloid cells-2 regulates the mucosal inflammatory response. Gastroenterology 144, 346–356 e343, https://doi.org/10.1053/j.gastro.2012.10.040 (2013).
Sieber, M. W. et al. Attenuated inflammatory response in triggering receptor expressed on myeloid cells 2 (TREM2) knock-out mice following stroke. PLoS One 8, e52982, https://doi.org/10.1371/journal.pone.0052982 (2013).
Aken, B. L. et al. Ensembl 2017. Nucleic Acids Res 45, D635–D642, https://doi.org/10.1093/nar/gkw1104 (2017).
Guo, L., Du, Y., Chang, S., Zhang, K. & Wang, J. rSNPBase: a database for curated regulatory SNPs. Nucleic Acids Res 42, D1033–1039, https://doi.org/10.1093/nar/gkt1167 (2014).
Takahashi, K., Prinz, M., Stagi, M., Chechneva, O. & Neumann, H. TREM2-transduced myeloid precursors mediate nervous tissue debris clearance and facilitate recovery in an animal model of multiple sclerosis. PLoS Med 4, e124, https://doi.org/10.1371/journal.pmed.0040124 (2007).
Ridker, P. M. et al. Elevation of tumor necrosis factor-alpha and increased risk of recurrent coronary events after myocardial infarction. Circulation 101, 2149–2153 (2000).
Koller-Strametz, J. et al. Circulating tumor necrosis factor-alpha levels in chronic heart failure: relation to its soluble receptor II, interleukin-6, and neurohumoral variables. J Heart Lung Transplant 17, 356–362 (1998).
Rosvall, M., Janzon, L., Berglund, G., Engstrom, G. & Hedblad, B. Incidence of stroke is related to carotid IMT even in the absence of plaque. Atherosclerosis 179, 325–331, https://doi.org/10.1016/j.atherosclerosis.2004.10.015 (2005).
Lorenz, M. W., von Kegler, S., Steinmetz, H., Markus, H. S. & Sitzer, M. Carotid intima-media thickening indicates a higher vascular risk across a wide age range: prospective data from the Carotid Atherosclerosis Progression Study (CAPS). Stroke 37, 87–92, https://doi.org/10.1161/01.STR.0000196964.24024.ea (2006).
Burke, G. L. et al. Arterial wall thickness is associated with prevalent cardiovascular disease in middle-aged adults. The Atherosclerosis Risk in Communities (ARIC) Study. Stroke 26, 386–391 (1995).
Nagai, Y. et al. Increased carotid artery intimal-medial thickness in asymptomatic older subjects with exercise-induced myocardial ischemia. Circulation 98, 1504–1509 (1998).
Salonen, J. T. & Salonen, R. Ultrasound B-mode imaging in observational studies of atherosclerotic progression. Circulation 87, II56–65 (1993).
Holliday, E. G. et al. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat Genet 44, 1147–1151, https://doi.org/10.1038/ng.2397 (2012).
Jones, B. L. & Swallow, D. M. The impact of cis-acting polymorphisms on the human phenotype. Hugo J 5, 13–23, https://doi.org/10.1007/s11568-011-9155-4 (2011).
Jalkanen, A. L., Coleman, S. J. & Wilusz, J. Determinants and implications of mRNA poly(A) tail size–does this protein make my tail look big? Semin Cell Dev Biol 34, 24–32, https://doi.org/10.1016/j.semcdb.2014.05.018 (2014).
Kung, J. T. et al. Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol Cell 57, 361–375, https://doi.org/10.1016/j.molcel.2014.12.006 (2015).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376, https://doi.org/10.1038/nature11082, https://www.nature.com/articles/nature11082#supplementary-information (2012).
Pombo, A. & Dillon, N. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 16, 245–257, https://doi.org/10.1038/nrm3965 (2015).
Carrasquillo, M. M. et al. A candidate regulatory variant at the TREM gene cluster associates with decreased Alzheimer’s disease risk and increased TREML1 and TREM2 brain gene expression. Alzheimer’s & dementia: the journal of the Alzheimer’s Association 13, 663–673, https://doi.org/10.1016/j.jalz.2016.10.005 (2017).
Jiang, T., Yu, J. T., Zhu, X. C. & Tan, L. TREM2 in Alzheimer’s disease. Mol Neurobiol 48, 180–185, https://doi.org/10.1007/s12035-013-8424-8 (2013).
Visvikis-Siest, S. & Siest, G. The STANISLAS Cohort: a 10-year follow-up of supposed healthy families. Gene-environment interactions, reference values and evaluation of biomarkers in prevention of cardiovascular diseases. Clinical chemistry and laboratory medicine 46, 733–747, https://doi.org/10.1515/cclm.2008.178 (2008).
Montauban van Swijndregt, A. D., De Lange, E. E., De Groot, E. & Ackerstaff, R. G. An in vivo evaluation of the reproducibility of intima-media thickness measurements of the carotid artery segments using B-mode ultrasound. Ultrasound Med Biol 25, 323–330 (1999).
Gariepy, J., Massonneau, M., Levenson, J., Heudes, D. & Simon, A. Evidence for in vivo carotid and femoral wall thickening in human hypertension. Groupe de Prevention Cardio-vasculaire en Medecine du Travail. Hypertension 22, 111–118 (1993).
Gariepy, J., Simon, A., Massonneau, M., Linhart, A. & Levenson, J. Wall thickening of carotid and femoral arteries in male subjects with isolated hypercholesterolemia. PCVMETRA Group. Prevention Cardio-Vasculaire en Medecine du Travail. Atherosclerosis 113, 141–151 (1995).
Bouatia-Naji, N. et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet 41, 89–94, https://doi.org/10.1038/ng.277 (2009).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575, https://doi.org/10.1086/519795 (2007).
He, C., Holme, J. & Anthony, J. SNP genotyping: the KASP assay. Methods Mol Biol 1145, 75–86, https://doi.org/10.1007/978-1-4939-0446-4_7 (2014).
Chen, M. H. & Yang, Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics 26, 580–581, https://doi.org/10.1093/bioinformatics/btp710 (2010).
Pollard, K. S., Hubisz, M. J., Rosenbloom, K. R. & Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20, 110–121, https://doi.org/10.1101/gr.097857.109 (2010).
Acknowledgements
This work was supported by the “Agence Nationale de la Recherche, programme d’Investissements d’avenir”, grant number ANR-15RHU-0004. The funder provided support in the form of salaries for author Vesna Gorenjak, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. This work was supported by the University of Lorraine with a “Contrat de travail de droit public”.
Author information
Authors and Affiliations
Contributions
Conceptualization: A.A.A. and S.V.S.; Formal analysis: A.A.A. and S.D.; Investigation: A.A.A., V.G., S.D., M.G.S., C.M.; Writing – original draft: A.A.A., V.G., S.D.; Writing – review & editing: A.A.A., V.G., S.D., S.V.S., M.G.S., D.R.V.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gorenjak, V., Aldasoro Arguinano, AA., Dadé, S. et al. The polymorphism rs6918289 located in the downstream region of the TREM2 gene is associated with TNF-α levels and IMT-F. Sci Rep 8, 7160 (2018). https://doi.org/10.1038/s41598-018-25553-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-25553-y
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.