Abstract
The clinical associations and correlations with other virulence factors such as cag pathogenicity island (PAI) of the Integrating Conjugative Elements Helicobacter pylori TFSS (ICEHptfs), a new type IV secretion system (TFSS) in H. pylori has not been described. Among 103 studied strains from Indonesia, almost all strains (99.0%) contained cag PAI with more than half (55.8%) were intact cag PAI. Patients infected with intact cag PAI strains showed significantly higher antral activity, inflammation and atrophy as well as corporal inflammation than those with non-intact cag PAI strains, confirming the virulence of cag PAI. Over half of strains (53.8%) contained ICEHptfs, predominantly consisted of ICEHptfs3-tfs4a (42.8%) and ICEHptfs3 (16.3%). Although patients infected with ICEHptfs-positive strains had lower H. pylori density, those with the complete ICEHptfs4b strains tended to have higher antral activity than the negative one. In combination, patients infected with combination of intact cag PAI-ICEHptfs-positive strains had more severe inflammation than those with non-intact cag PAI-ICEHptfs-negative, suggesting a possibility of a mutual correlation between these TFSS(s).
Similar content being viewed by others
Introduction
Helicobacter pylori is a human-specific bacterium, colonizing the stomach of approximately 50% of the modern human population1. Infection is associated with several gastro-duodenal pathologies, including chronic gastritis, peptic ulcers, and even gastric cancer in a subset of individuals, depending on the variation of bacterial virulence, host genetics and/or environmental factors2,3. H. pylori is the most genetically diverse pathogenic bacteria4,5, which might be associated with frequent horizontal gene transfer (HGT) and recombination within species as an adaptation process to the host over years of infection6. Therefore, its genome contains many putative genes, which are generally classified into three categories7. The first is phase-variable genes, defined as those with functional status that could change due to particular conditions. An example is the slipped-strand mispairing mechanism, which often shown in outer membrane proteins such as oipA and sabA, that can switch genes ‘on’ and ‘off’ very rapidly8,9. The second is genes with different structures/genotypes, such as the repeat region of CagA. The 3′ repeat region of CagA can differ between two genotypes, associated with a different risk of developing gastric cancer10,11. The last is strain-specific genes, defined if the gene only exists in a particular strain. The most studied genes in this category are cag pathogenicity island (PAI), which encodes a type IV secretion system (TFSS)7,12.
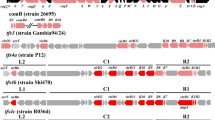
TFSS is a flexible secretion system found in both Gram-positive and -negative bacteria. In Gram-negative bacteria, it mediates secretion of various protein substrates, from monomeric proteins, multi-subunit protein toxins and nucleoprotein complexes13. Importantly, more than one TFSS could be found in one species of bacteria, including in the H. pylori. H. pylori has four types of TFSS with varied functionalities4,12,14,15. The first is cag PAI, which primarily injects CagA into host cells12. The second is the comB system which has a principal function of DNA uptake and natural transformation within H. pylori genome15. The most recently revealed TFSS is within Integrating Conjugative Elements (ICEs). In case of H. pylori, this is known as ICE H. pylori TFSS (ICEHptfs)16. ICEHptfs was initially named as plasticity regions, the regions within the H. pylori genome which have considerably lower guanine and cytosine content (~35%) compared with the rest of the genome (39%)17. The lower G + C content indicates that plasticity regions may be the result of HGT8,17. With the increasing number of H. pylori complete genomes deposited in the GenBank, plasticity regions are considered as conserved mobile elements, rather than a region with genomic plasticity, and are usually organized as a complete set of TFSS machinery. In addition, based on the acquisition of these elements through conjugative HGT, these elements are best described as ICEs. The TFSS within ICEHptfs(s) is called TFSS3 and TFSS4a/4b/4c4,16. Those differences between ICEHptfs3 and ICEHptfs4a/b/c were determined by the nucleotide diversity of the virB-virD orthologues genes, resulting very distinctive diversity between TFSS3 and TFSS4 in general16. In addition, the TFSS4 possesses sub-type based on nucleotide diversity in the virB2, virB3, virB4, topA, virB7 and virB8, discriminating TFSS4a and TFSS4b, and diversity in virB11, virD4 and virD2, distinguishing TFSS4a and TFSS4c16. The terminology of this region was inconsistent in several previous studies. A study conducted in 2009 reported a new TFSS termed as TFSS3, TFSS3a, and TFSS3b14 within a mobile element called transposon of plasticity zones (TnPZ) type 2 for TFSS3, TnPZ1 for TFSS3a and TnPZ1b for TFSS3b. However a year later the TFSS3b were termed as TFSS4 for the TFSS inside the mobile element plasticity zones 1 (PZ1) and TFSS3 for the TFSS inside the mobile element PZ3. Following with the newest terminology is ICEHptfs3 which containing TFSS3 and ICEHptfs4a/4b/4c which containing TFSS4a/4b/4c16. In order to make consistent terminology, in this study we used ICEHptfs3 and ICEHptfs4a/4b/4c for TFSS3 and TFSS4a/4b/4c, respectively16. Currently, the distribution and association of these new TFSSs to gastro-duodenal diseases are not fully described.
Indonesia is a country in South-East Asia, consisting of more than 13,600 islands and 400 ethnicities18. As described previously, H. pylori infected the ancestors of modern humans in Africa about 100,000 years ago (100 kya) and migrated with its host from Africa to Asian and American continents19,20. Therefore, ethnic diversity is associated with H. pylori infection as well as genome diversity, especially in Indonesia. We have described that ethnicity is a risk factor for H. pylori infection21. In addition, ethnicity is also a factor for the diversity of virulence genes in H. pylori. We have described that different ethnicity had a different genetic polymorphism on the several virulence genes in the nucleotides and amino acids level. For example, strains possessing pre-EPIYA motif of CagA isolated from Batak ethnic showed 6 bp deletion-type pre-EPIYA motif with East Asian-type CagA. The 6 bp deletion-type is unique type among East Asian-type CagA since almost all pre-EPIYA motif types of strains isolated from Japan and Vietnam was reported to have 39 bp deletion-type and 18 bp deletion-type, respectively22. Patients infected with this 6 bp deletion-type/East Asian-type CagA strains showed to have lower gastric mucosal histologic scores compared to those with Western-type CagA23, although it is well known that East Asian-type CagA had generally more virulent than Western-type CagA. In addition, the predominant type of CagA was also different in each ethnic group.
As the distribution and clinical association of ICEHptfs(s) have not been reported, it is interesting to investigate the distribution and clinical association of these regions as well as the correlation with other virulence genes in relation to the clinical outcome. Here, we reported the distribution of ICEHptfs in Indonesia using high throughput next-generation sequencing technology and revealed that strains from some geographic areas lack this genomic region, and the intactness of this region had an association with clinical outcome.
Results
Characteristic of patients and prevalence of ICEHptfs and cag PAI
We performed endoscopic examination on 1072 dyspeptic patients in 17 cities in Indonesia from August 2012 to August 2016, and a total of 103 H. pylori were isolated from patients (66 male and 37 female; mean age 49.2 ± 13 years; range 24–80 years), comprising 92 patients with gastritis, 10 with peptic ulcer disease (PUD) and 1 with gastric cancer. Among 103 isolates, 75 isolates were from our previous study with information of cagA genotypes24. Strains originated in Indonesia are shown in Supplementary Figure 1.
We evaluated the cag PAI and determined the functional status of each gene present (Table 1). We categorized these into i) Intact cag PAI, if all the genes were detected and there was no deletion, stop codon or frameshift in each gene; ii.) Non-intact cag PAI, if at least one of the genes were lacking or had stop codon and/or frameshift in the gene; and iii.) cag PAI-negative, if none of the cag PAI genes were detected. In total, cag PAI was detected in most isolates (99.1%), either intact or non-intact. Among the detected cag PAI strains, 57 strains possessed intact cag PAI (55.8%). The gastric cancer patient had intact cag PAI H. pylori. The cagA was detected in 101 strains (98%). Sequence analysis of the 27 new cagA-positive strains showed that 5 (18.5%) strains possessed Western-type CagA and 12 (44.4%) strains possessed East-Asian type CagA. In addition, we also confirmed a unique genotype of CagA (AB and B type) which mostly were isolated from Merauke city, Papua Island. Those B segment of CagA genotypes had very similar amino acids sequences with ABB type CagA from our previous report23 (Supplementary Figure 2). Therefore, we deemed it a subtype of the ABB type CagA. Taken together with our previous study24, the result was 60 (58.2%) strains possessed the East-Asian type CagA (AABD, AAD and ABD type), whereas 30 strains (29.1%) were Western-type CagA (ABC, ABCC and BC type) and 15 strains (14.5%) were ABB type CagA (ABB, AB and B) (Fig. 1A).
Distribution of CagA and CagL hypervariable motif (CagLHM) and the ICEHptfs. (A) The distribution of CagA genotype among Indonesian strains. More than half (58.2%) was East-Asian type CagA. (B) The proportion of ICEHptfs among CagA genotype. It showed the B, ABBD, AABD and ABCC type CagA did not possessed ICEHptfs. (C) The distribution of CagLHM among Indonesian cag PAI positive strains. We observed new CagLHM motif DKMGK. (D) The proportion of ICEHptfs observed in the CagLHM motif group showed all NKIGQ motif (n = 3) contained ICEHptfs elements. The new observed DKMGK motif strains showed the high prevalence (10/11, 90.9%) as high as the DKMGE motif strains (13/14, 92.8%), the progenitor motif of CagLHM.
ICEHptfs were detected in 56 of 103 (54.3%) strains. Among gastritis patients, 51 strains (55.4%) and 5 strains (50.0%) from PUD patients possessed ICEHptfs. Interestingly, the strain isolated from the gastric cancer patient did not contain ICEHptfs (Table 1). Sequence analysis showed there were no mutations leading to premature stop codons or frameshift mutation; thus, we concluded that all of the genes were functional. Among the ICEHptfs-positive strains the single ICEHptfs was observed as ICEHptfs3 (16.0%), ICEHptfs4a (14.4%) and ICEHptfs4b (10.7%). There was no strain with ICEHptfs4c. Aside from single ICEHptfs in the genome, strains possessing multiple ICEHptfs were also observed: ICEHptfs3-tfs4a (42.8%), ICEHptfs3-tfs4b (8.9%) and ICEHptfs3-4a/4b (7.2%) (Table 1).
The distribution of ICEHptfs and the ethnic groups
There was a significant association between ethnic group and prevalence of ICEHptfs (P = 0.031). Timor tribe strains had the highest prevalence of ICEHptfs (10/12, 83.3%) and the lowest prevalence was observed in Minahasanese strains (14.2%) (Table 2). There was also a significant association between ethnic groups and the type of ICEHptfs (P = 0.002). Batak tribes possessed predominantly ICEHptfs3-tfs4a (77.8%), whereas Chinese ethnicities possessed predominantly ICEHptfs3 (57.1%). As for the Timor ethnicity, the types of ICEHptfs were distributed evenly (Table 2).
Complete ICEHptfs were assessed as a cluster with complete TFSS machineries, composed of VirB2, VirB3, VirB4, VirB6, VirB7, VirB8, VirB9, VirB9, VirB10, VirB11, VirD2, VirD4, XerT and TopA. Among the positive ICEHptfs strains, 32 strains (57.1%) possessed incomplete ICEHptfs. The complete ICEHptfs were found in 24 strains: 19 (33.9%) with complete ICEHptfs3 and 5 (9.0%) with complete ICEHptfs4b. There was a significant association between the completeness of ICEHptfs and the ethnic groups (P = 0.03). Timor tribe strains showed the highest prevalence of complete ICEHptfs (80.0%), in which 5 strains (50.0%) possessed complete ICEHptfs3 and 3 strains (30.0%) possessed complete ICEHptfs4b (Table 2).
The ICEHptfs, CagA and CagL
Among totally 30 strains with Western-type CagA, we could find 18 strains (60.0%) containing ICEHptfs (Supplementary Table 2). However, we could not obtain any ICEHptfs elements from strains which possessed ABCC- and B-type CagA (Fig. 1B). Interestingly, the B-type CagA strains were isolated from Merauke city, Papua island, suggesting there is an association with the human population. Among the East Asian-type CagA, 32 strains (53.3%) possessed ICEHptfs. The ABBD type cagA strain did not contain any ICEHptfs elements. The ABB type cagA containing ICEHptfs were 4 of 11 strains (36.3%) and seemed to be equally distributed.
CagL Hypervariable Motif (CagLHM) had a close relationship with the geographical origin of H. pylori, as recently reported25. We evaluated the CagLHM and found 8 unique motifs. The predominant motifs were YEIGK, DEIGK and DKMGE (38.6%, 14.8% and 13.8%) (Fig. 1C). Interestingly we also found a novel motif DKMGK and this motif mostly was observed from H. pylori isolated from Samosir Island (Supplementary Table 1). This novel motif strains almost exclusively (91%) possessed ICEHptfs elements as exclusive as the DKMGE motif strains (92.8%) (Fig. 1D).
The cag PAI and histological findings
Comparison between histological findings and cag PAI intactness showed that patients infected with intact cag PAI had higher both corporal and antral inflammation than those with non-intact cag PAI (P = 0.011 and P < 0.001, respectively). Patients infected with intact cag PAI strains also showed higher activity and atrophy in the antrum than those with non-intact cag PAI strains (P < 0.001) (Fig. 2). Patients infected with intact cag PAI strains had significantly higher risk of antral activity, inflammation and atrophy and corporal inflammation and atrophy after adjusted with age and sex (Supplementary Table 3).
The ICEHptfs and histological findings
Histological examination showed the patients infected with strains possessing ICEHptfs elements (either complete or incomplete) had significantly lower antral H. pylori density than those without (P = 0.039) (Table 3). As for the comparison between complete and incomplete ICEHptfs(s), histological findings did not show any significant association; however, the patients infected with strains possessing complete ICEHptfs4b tended to have higher activity in the antrum than those possessing ICEHptfs-negative strains (P = 0.06) (Fig. 3).
Combination of the ICEHptfs, cag PAI and histological findings
We classified H. pylori strains according to both the cag PAI intactness and status of ICEHptfs, and examined the association between the combined classification and the histological scores. The patients infected with the strains possessing the combination of intact cag PAI-ICEHptfs-positive strains had significantly higher antral activity compared to those with non-intact cag PAI-ICEHptfs-negative as well as the non-intact cag PAI-ICEHptfs-positive strains (P = 0.002 and P = 0.002, respectively) (Table 4). However, patients infected with the intact cag PAI-ICEHptfs-negative strains did not show difference of antral activity compared to those with non-intact cag PAI-ICEHptfs-negative strains (P = 0.103), suggesting that intact cag PAI virulence for inducing acute inflammation in the antrum is dependent on the status of ICEHptfs. In addition, patients infected with intact cag PAI-ICEHptfs-positive strains showed significantly higher antral inflammation and atrophy compared to those with non-intact cag PAI-ICEHptfs-negative strains (P < 0.001 and P < 0.001). Corporal inflammation was also significantly higher in patients infected with intact cag PAI–ICEHptfs-positive strains than those with non-intact cag PAI-ICEHptfs-positive strains (P = 0.047). In addition, we also classified the strains based on the intactness of cag PAI and type of ICEHptfs, then evaluated association with the histological scores. Despite the number of the samples being small for strains possessing intact cag PAI-complete ICEHptfs4b (n = 3), we found that patients infected with these strains had higher antral activity and inflammation compared to those with non-intact cag PAI-incomplete ICEHptfs strains (P = 0.024 and P = 0.009, respectively) (Table 4).
Discussion
This is the first study to evaluate the pathogenic role of ICEHptfs in combination with cag PAI at a population level. We examined the prevalence of ICEHptfs and cag PAI using high throughput sequencing. The previous genomic comparison showed that the ICEHptfs has high prevalence (86.7%) in 45 strains worldwide16. We applied the same methods to determine the prevalence of ICEHptfs among Indonesian strains, which showed a lower prevalence of ICEHptfs (53.4%). In general, ICEs were transferred between genomes using conjugative HGT. This different prevalence might be due to observation only performed in one country compared to the worldwide observation. In addition, the distribution of ICEHptfs had a significant association with ethnic groups in Indonesia, suggesting the prevalence and type of ICEHptfs had an association with geographical origin. Some particular CagA genotypes strains did not possess any ICEHptfs, especially the strains isolated from Merauke city. All our strains isolated from Merauke city were assigned as hpSahul (data not shown) and other strains deposited in the GenBank belonging to hpSahul, PNG84A and ausaBRJ0526, also did not contain ICEHptfs, strongly supporting this association.
The cag PAI was transferred into H. pylori far prior human migrate from Africa 60 kya27 and interestingly, the cag PAI still can be observed in all the H. pylori populations after long period of human migration and shows the same evolution pattern as the house-keeping genes27. This suggests the importance of cag PAI towards the host colonization process. Our study showed almost all the Indonesian strains (98%) contained cag PAI, supporting its importance. In addition to the cag PAI, CagLHM may also help to discriminate geographical origin25. Our study showed that the predominant CagLHM in Indonesia were specifically observed in the East/Southeast Asia/Australasia groups, as previously reported25. We also found a new motif of CagLHM which showed as high prevalence of ICEHptfs as the DKMGE motif. DKMGE is believed to be the progenitor of the CagLHM motif25, and since the observed motifs only differed on the residue 62 (E62K), this observation suggests the new motifs were directly derived from the progenitor.
The cag PAI was originally designated as a TFSS, which mainly has a function to translocate CagA protein into host cell cytoplasm28,29. However, the virulence of this island dependent to the intactness of this island, therefore it may successfully inject the CagA protein12. Our previous study in Vietnam30 classified the intactness of cag PAI based on the existence of the gene using the PCR method. Our current results showed similarly that the intact cag PAI has more severe histological score than the non-intact cag PAI. However, the previous criterion evaluated the intactness of cag PAI only based on the presence or absence of the member genes and the resulting high prevalence of intact cag PAI, which may blur the association with histological scores. On the other hand, the evaluation of cag PAI sequences may give us a significant association with the histological scores. In addition, it may also discriminate the cluster of cag PAI genotype (East-Asian type and Western type cluster), of which the East-Asian type may bind stronger to the SHP-2 receptor11. Therefore, we recommend the criteria to evaluate intactness of cag PAI also considering the functional status of the genes, as a more reliable method to predict clinical outcome.
Although strains with ICEHptfs showed significantly lower H. pylori density, there was an association between a complete TFSS and the histological scores. It was reported that strains with a complete cluster of dupA, the VirB4 homologue of ICEHptfs4b, lead to a higher risk of developing duodenal ulcers than those with incomplete dupA clusters or dupA negative strains31. Our data also showed the same tendency, even with a lower density in the antrum, suggesting this region has a more significant association to the H. pylori virulence, resulting in a higher active inflammation rather than attachment to the gastric mucosa.
In addition, we combined the status and type of ICEHptfs with the cag PAI intactness. Our data showed patients infected with intact cag PAI-ICEHptfs-positive strains had higher antral activity than those with non-intact cag PAI-ICEHptfs-negative strains. However, patients infected with the intact cag PAI-ICEHptfs-negative strains did not show difference of antral activity compared to those with non-intact cag PAI-ICEHptfs-negative strains. These data suggest that the cag PAI and ICEHptfs were dependent each other to induce higher antral activity. The TFSS can be divided into three groups according to their function32. The first group is the conjugation system, translocating single-stranded DNA substrates to recipient cells in a contact-dependent manner, resulting in the adaptation of bacteria to environmental changes. The second group is the effector translocation system, delivering protein directly into eukaryotic cells. The third group is the DNA uptake mediators, which uptake or release DNA or protein substrates extracellularly, independently of contact with another cell33. Since there was an evidence that the ICEHptfs was a genetic mobile element which was transferred in the conjugation manner16,34, we assumed the function of ICEHptfs in the pathogenesis of H. pylori infection was belongs to the conjugation group33, suggesting the ICEHptfs might supporting the cag PAI to induce more severe clinical outcome.
Although we could not make strong conclusions due to a small sample size, particularly in the certain ICEHptfs groups, this study gives us new information about the distribution and clinical association of this relatively new TFSS in H. pylori. In addition, since there have not been many biological and structure evidences of this particular system, further study is needed to better understand the role of the TFSS in colonization by H. pylori.
Conclusion
In conclusion, our data showed a high prevalence of cag PAI in Indonesia, half of which were complete. Criteria determining intactness of cag PAI based on the gene functionality is more reliable to evaluate the influence of H. pylori on gastric mucosal status. The ICEHptfs strains tended to induce more active inflammation in the antrum even with a lower density of bacteria. In combination, it was shown that patients infected with intact cag PAI-ICEHptfs-positive strains had more severe inflammation than those with non-intact cag PAI-ICEHptfs-negative strains, suggesting possibility a mutual correlation between these TFSS(s).
Materials and Methods
Samples and DNA sequencing
We performed endoscopic examination on 1072 dyspeptic patients in 17 cities in Indonesia from August 2012 to August 2016. We excluded patients with partial/total gastrectomy, non-fasted patients and those with contraindication for upper endoscopy. Written informed consent was obtained from all patients and the study protocol was approved by the ethics committees of Dr. Soetomo Teaching Hospital (Surabaya, Indonesia), Dr. Cipto Mangunkusumo Teaching Hospital (Jakarta, Indonesia), Dr. Wahidin Sudirohusodo Teching Hospital (Makassar, Indonesia) and Oita University Faculty of Medicine (Yufu, Japan). We declare that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008 and 2013. We used antral gastric biopsy to isolate H. pylori as previously described24, resulting in 103 cultured isolates, including 75 isolates from our previous study24.
DNA extraction was performed using QIAamp DNA Mini Kit (QIAGEN, Valencia, CA, USA) following the manufacturer’s instructions. Whole genome sequencing was performed using a high throughput next generation sequencer; Illumina Hiseq. 2000 and Miseq as per the list in Supplementary Table 2. Briefly, high-quality genomic DNA was used, then was prepared using dual-indexed Nextera XT Illumina libraries and subjected to cluster generation and paired-end sequencing (2 × 300 bp) for Miseq and (2 × 150 bp) for Hiseq. We performed the quality control and de novo assembly prior the reference mapping to obtain the coverage and to select the result which may be used for further analysis using CLC Genomic Workbench v. 7.04, a commercial software (Qiagen Inc., Redwood, California, USA). The coverage we obtained was between 81–400 folds in each genome (Supplementary Table 1). The threshold for further analysis in this study, we use Q30 > 80% as recommended by Illumina and the average coverage more than 80 folds as had been described previously35.
Analysis of ICE and other virulence genes
Identification of the ICEHptfs-type was performed by using a reference mapping method. Short-read outputs were mapped to the corresponding reference sequences consisting of ICEHptfs3 (strain Gambia94/24), ICEHptfs4a (strain P12) ICEHptfs4b (strain G27) ICEHptfs4c (strain SouthAfrica7) using CLC Genomic Workbench v. 7.04, a commercial software (Qiagen Inc., Redwood, California, USA) as described previously16. The unmapped reads then also assembled by using de novo assembly by the CLC Genomic Workbench. The ICE genes were identified by BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) from the mapped reads. The cag PAI were identified using BLAST method and the query from strain 269958,27. The functional status of the each gene was evaluated by visual inspection using MEGA736.
Histological evaluation
All biopsy material for histological evaluation was fixed in 10% buffered formalin and embedded in paraffin. Serial sections were stained with hematoxylin and eosin as well as May-Giemsa stains. Gastric mucosa were evaluated based on the updated Sydney system37. Bacterial load was classified into four grades: 0, ‘normal’; 1, ‘mild’; 2, ‘moderate’; and 3, ‘marked’ according to the updated Sydney system37.
The degree of inflammation, neutrophil activity, atrophy and intestinal metaplasia were classified into four grades according to the updated Sydney system: 0, ‘normal’; 1, ‘mild’; 2, ‘moderate’; and 3, ‘marked’37. Immunohistochemistry for anti-H. pylori antibody was performed as previously described38.
Statistical analysis
Data were analyzed using IBM SPSS Statistics, version 22 (IBM Corp., USA). Discrete variables were tested using the chi-square test; continuous variables were tested using Mann-Whitney U test. An ordinal regression model was used to calculate risk for developing higher histological score. A two-tailed P value < 0.05 was considered statistically significant.
Availability of Nucleotide Sequences
The accession number for nucleotide sequences were deposited in DDBJ under accession number LC334483 – LC335589 and LC339076 – LC339479.
References
Hooi, J. K. Y. et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 153, 420–429, https://doi.org/10.1053/j.gastro.2017.04.022 (2017).
Salama, N. R., Hartung, M. L. & Muller, A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nature reviews. Microbiology 11, 385–399, https://doi.org/10.1038/nrmicro3016 (2013).
Cover, T. L. & Blaser, M. J. Helicobacter pylori in health and disease. Gastroenterology 136, 1863–1873, https://doi.org/10.1053/j.gastro.2009.01.073 (2009).
Fischer, W. et al. Strain-specific genes of Helicobacter pylori: genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic acids research 38, 6089–6101, https://doi.org/10.1093/nar/gkq378 (2010).
Fernandez-Gonzalez, E. & Backert, S. DNA transfer in the gastric pathogen Helicobacter pylori. Journal of gastroenterology 49, 594–604, https://doi.org/10.1007/s00535-014-0938-y (2014).
Suerbaum, S. et al. Free recombination within Helicobacter pylori. Proceedings of the National Academy of Sciences of the United States of America 95, 12619–12624 (1998).
Yamaoka, Y. Roles of the plasticity regions of Helicobacter pylori in gastroduodenal pathogenesis. Journal of medical microbiology 57, 545–553, https://doi.org/10.1099/jmm.0.2008/000570-0 (2008).
Tomb, J. F. et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388, 539–547, https://doi.org/10.1038/41483 (1997).
Fischer, W., Hofreuter, D. & Haas, R. in Helicobacter pylori: Physiology and Genetics (eds H. L. T. Mobley, G. L. Mendz, & S. L. Hazell) (ASM Press ASM Press., 2001).
Jones, K. R. et al. Polymorphism in the CagA EPIYA motif impacts development of gastric cancer. Journal of clinical microbiology 47, 959–968, https://doi.org/10.1128/JCM.02330-08 (2009).
Azuma, T. et al. Distinct diversity of the cag pathogenicity island among Helicobacter pylori strains in Japan. Journal of clinical microbiology 42, 2508–2517, https://doi.org/10.1128/JCM.42.6.2508-2517.2004 (2004).
Backert, S., Tegtmeyer, N. & Fischer, W. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future microbiology 10, 955–965, https://doi.org/10.2217/fmb.15.32 (2015).
Fronzes, R., Christie, P. J. & Waksman, G. The structural biology of type IV secretion systems. Nature reviews. Microbiology 7, 703–714, https://doi.org/10.1038/nrmicro2218 (2009).
Kersulyte, D. et al. Cluster of type IV secretion genes in Helicobacter pylori’s plasticity zone. Journal of bacteriology 185, 3764–3772 (2003).
Hofreuter, D., Odenbreit, S. & Haas, R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Molecular microbiology 41, 379–391 (2001).
Fischer, W. et al. A comprehensive analysis of Helicobacter pylori plasticity zones reveals that they are integrating conjugative elements with intermediate integration specificity. BMC genomics 15, 310, https://doi.org/10.1186/1471-2164-15-310 (2014).
Alm, R. A. & Trust, T. J. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. Journal of molecular medicine 77, 834–846 (1999).
Miftahussurur, M. et al. Identification of Helicobacter pylori infection in symptomatic patients in Surabaya, Indonesia, using five diagnostic tests. Epidemiology and infection 143, 986–996, https://doi.org/10.1017/S095026881400154X (2015).
Moodley, Y. et al. Age of the association between Helicobacter pylori and man. PLoS pathogens 8, e1002693, https://doi.org/10.1371/journal.ppat.1002693 (2012).
Linz, B. et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445, 915–918, https://doi.org/10.1038/nature05562 (2007).
Syam, A. F. et al. Risk Factors and Prevalence of Helicobacter pylori in Five Largest Islands of Indonesia: A Preliminary Study. PloS one 10, e0140186, https://doi.org/10.1371/journal.pone.0140186 (2015).
Uchida, T. et al. Analysis of virulence factors of Helicobacter pylori isolated from a Vietnamese population. BMC microbiology 9, 175, https://doi.org/10.1186/1471-2180-9-175 (2009).
Miftahussurur, M. et al. Helicobacter pylori virulence genes in the five largest islands of Indonesia. Gut pathogens 7, 26, https://doi.org/10.1186/s13099-015-0072-2 (2015).
Miftahussurur, M. et al. Surveillance of Helicobacter pylori Antibiotic Susceptibility in Indonesia: Different Resistance Types among Regions and with Novel Genetic Mutations. PloS one 11, e0166199, https://doi.org/10.1371/journal.pone.0166199 (2016).
Gorrell, R. J., Zwickel, N., Reynolds, J., Bulach, D. & Kwok, T. Helicobacter pylori CagL Hypervariable Motif: A Global Analysis of Geographical Diversity and Association With Gastric Cancer. The Journal of infectious diseases 213, 1927–1931, https://doi.org/10.1093/infdis/jiw060 (2016).
Montano, V. et al. Worldwide Population Structure, Long-Term Demography, and Local Adaptation of Helicobacter pylori. Genetics 200, 947–963, https://doi.org/10.1534/genetics.115.176404 (2015).
Olbermann, P. et al. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS genetics 6, e1001069, https://doi.org/10.1371/journal.pgen.1001069 (2010).
Tegtmeyer, N., Wessler, S. & Backert, S. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. The FEBS journal 278, 1190–1202, https://doi.org/10.1111/j.1742-4658.2011.08035.x (2011).
Fischer, W. Assembly and molecular mode of action of the Helicobacter pylori Cag type IV secretion apparatus. The FEBS journal 278, 1203–1212, https://doi.org/10.1111/j.1742-4658.2011.08036.x (2011).
Nguyen, L. T. et al. Clinical relevance of cagPAI intactness in Helicobacter pylori isolates from Vietnam. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology 29, 651–660, https://doi.org/10.1007/s10096-010-0909-z (2010).
Jung, S. W., Sugimoto, M., Shiota, S., Graham, D. Y. & Yamaoka, Y. The intact dupA cluster is a more reliable Helicobacter pylori virulence marker than dupA alone. Infection and immunity 80, 381–387, https://doi.org/10.1128/IAI.05472-11 (2012).
Alvarez-Martinez, C. E. & Christie, P. J. Biological diversity of prokaryotic type IV secretion systems. Microbiology and molecular biology reviews: MMBR 73, 775–808, https://doi.org/10.1128/MMBR.00023-09 (2009).
Trokter, M., Felisberto-Rodrigues, C., Christie, P. J. & Waksman, G. Recent advances in the structural and molecular biology of type IV secretion systems. Current opinion in structural biology 27, 16–23, https://doi.org/10.1016/j.sbi.2014.02.006 (2014).
Kersulyte, D. et al. Helicobacter Pylori’s plasticity zones are novel transposable elements. PloS one 4, e6859, https://doi.org/10.1371/journal.pone.0006859 (2009).
Ogawa, H. et al. Genetic variants of Helicobacter pylori type IV secretion system components CagL and CagI and their association with clinical outcomes. Gut pathogens 9, 21, https://doi.org/10.1186/s13099-017-0165-1 (2017).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular biology and evolution 33, 1870–1874, https://doi.org/10.1093/molbev/msw054 (2016).
Dixon, M. F., Genta, R. M., Yardley, J. H. & Correa, P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. The American journal of surgical pathology 20, 1161–1181 (1996).
Uchida, T. et al. Immunohistochemical diagnosis of the cagA-gene genotype of Helicobacter pylori with anti-East Asian CagA-specific antibody. Cancer science 98, 521–528, https://doi.org/10.1111/j.1349-7006.2007.00415.x (2007).
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (DK62813) and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (KAKENHI Grant Numbers 26640114, 15H02657, 16H05191 and 221S0002) (YY). Support was also provided by the Japan Society for the Promotion of Science (JSPS) Institutional Program for Young Researcher Overseas Visits (YY), the Strategic Funds for the Promotion of Science and Technology from the Japan Science and Technology Agency (JST) (YY). Support also were provided by Ministry of Research, Technology and Higher Education of Indonesia as part of grant for World Class Professor Program (no. 105/D2.3/KP/2017) (MIL and MM). PS and LAW are PhD students supported by the Japanese government (MEXT) scholarship program for 2013, and 2015, respectively.
Author information
Authors and Affiliations
Contributions
L.A.W., M.M. and Y.Y. design the study. P.S., L.A.W., T.U. and M.M. carried out the experiments. L.A.W., M.M., T.U., R.S. and Y.Y. carried out the analyzed the data. M.I.L., M.H., N.N., A.F.S., M.M., L.A.W. and Y.Y. carried out the sample acquisition and data collection. L.A.W., M.M., R.S. and Y.Y. writing and revising the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Waskito, L.A., Miftahussurur, M., Lusida, M.I. et al. Distribution and clinical associations of integrating conjugative elements and cag pathogenicity islands of Helicobacter pylori in Indonesia. Sci Rep 8, 6073 (2018). https://doi.org/10.1038/s41598-018-24406-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-24406-y
This article is cited by
-
Helicobacter pylori type 4 secretion systems as gastroduodenal disease markers
Scientific Reports (2021)
-
Variations in cag pathogenicity island genes of Helicobacter pylori from Latin American groups may influence neoplastic progression to gastric cancer
Scientific Reports (2020)
-
Characterization of a novel Helicobacter pylori East Asian-type CagA ELISA for detecting patients infected with various cagA genotypes
Medical Microbiology and Immunology (2020)
-
The role of integrating conjugative elements in Helicobacter pylori: a review
Journal of Biomedical Science (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.