Abstract
Almost all cells require thiamin, vitamin B1 (B1), which is synthesized via the coupling of thiazole and pyrimidine precursors. Here we demonstrate that 5-(2-hydroxyethyl)-4-methyl-1,3-thiazole-2-carboxylic acid (cHET) is a useful in vivo B1 precursor for representatives of ubiquitous marine picoeukaryotic phytoplankton and Escherichia coli – drawing attention to cHET as a valuable exogenous micronutrient for microorganisms with ecological, industrial, and biomedical value. Comparative utilization experiments with the terrestrial plant Arabidopsis thaliana revealed that it can also use exogenous cHET, but notably, picoeukaryotic marine phytoplankton and E. coli were adapted to grow on low (picomolar) concentrations of exogenous cHET. Our results call for the modification of the conventional B1 biosynthesis model to incorporate cHET as a key precursor for B1 biosynthesis in two domains of life, and for consideration of cHET as a microbial micronutrient currency modulating marine primary productivity and community interactions in human gut-hosted microbiomes.
Similar content being viewed by others
Introduction
Thiamin (vitamin B1; called B1 herein), in the form of thiamin diphosphate, is an enzyme cofactor needed for energy generation and general metabolism in virtually all cells1. Despite the essentiality of B1, some populations in nature persist as B1 auxotrophs that cannot synthesize B1 de novo and so depend on exogenous B1 or related micronutrients to meet their B1 demands2,3,4. Cosmopolitan marine bacteria5, bloom-forming phytoplankton6, and cosmopolitan picoeukaryotic phytoplankton6,7, ubiquitous freshwater bacteria8, and about half of taxa inhabiting the human gut9 have been shown to be B1 auxotrophs – cumulatively highlighting the importance of exogenous B1 or related micronutrients to the operation of diverse ecosystems.
Aside from B1, precursors of B1 are also valuable exogenous micronutrients that some cells can use to meet their B1 demands2,3,4,10,11. B1 precursor use varies across prokaryotic and eukaryotic taxa2,3,4 and is thought to depend on the presence/absence of B1 biosynthesis and/or transporter genes in their respective genomes12. Prediction of B1 auxotrophy and/or precursor use based on gene repertoire recently helped reveal the importance of B1 precursors in sustaining environmentally significant and commercially valuable organisms. For example: (1) ubiquitous bacterioplankton, affiliated with the SAR11 clade, accounting for more than half of microbes in the oligotrophic surface ocean13, obligately require the pyrimidine precursor 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP) for growth5; (2) higher plants (Arabidopsis thaliana and Zea mays) salvage B1 from the thiazole precursor 4-methyl-5-thiazoleethanol (HET) via activity of ThiM14, a HET kinase previously described in model bacteria15; and (3) key cosmopolitan marine picoeukaryotic phytoplankton, which are significant contributors to oceanic primary production16,17 grow using an unidentified thiazole-related precursor(s), produced by de novo B1-synthesizing marine bacteria or phytoplankton, along with exogenous HMP18. For reference, a glossary of precursors and related enzymes referred to in this study is given in Table 1.
The picoeukaryotic phytoplankton (Ostreococcus, Micromonas spp.) do not grow on HET7, the only thiazole used in prior tests of B1 salvage from exogenous precursor(s)1,2,3,4. However, ThiM is required for Ostreococcus spp. to use the newly detected precursor(s) found in seawater and produced by de novo B1-synthesizing plankton - strongly suggesting that the compound(s) is thiazole-related18.
Intrigued that these marine picoeukaryotic phytoplankton potentially use a novel thiazole precursor, we noted with interest that carboxythiazole, 5-(2-hydroxyethyl)-4-methyl-1,3-thiazole-2-carboxylic acid (cHET), specifically phosphorylated cHET (cHET-P), is produced by bacterial thiazole synthase19, and that the thiazole synthase of plants and yeast similarly generates cHET-ADP (adenylation of the precursor is used rather than phosphorylation)20,21. Further, cHET-P is a functional substrate for bacterial thiamin monophosphate synthase (ThiE)22. This rigorous biochemical evidence points to cHET as a core component of de novo B1 biosynthesis; nonetheless, the vast majority of B1-related research and reviews to date make no mention of cHET (or phosphorylated or adenylated forms) and instead describe only synthesis and use of HET(−P)5,7,9,23,24,25. Given the apparent importance of cHET in B1 biosynthesis, and the ThiM (thiazole kinase) requirement for marine picoeukaryotic phytoplankton to use the newly detected thiazole precursor18, we hypothesized that cHET is a useful exogenous thiazole B1 precursor for phytoplankton.
Results
Marine picophytoplankton use exogenous cHET to meet their B1 demands
In experiments with vitamin-B1 limited Ostreococcus tauri RCC745, a ThiM-possessing marine picoeukaryotic phytoplankton unable to use HET18, low additions of cHET (plus the pyrimidine precursor HMP) promoted growth (Fig. 1), confirming our hypothesis and revealing that O. tauri is adapted to use minute amounts of cHET dissolved in seawater. The cHET stock used in these experiments contained no detectable B1 cross-contamination, but did contain trace (0.5%) amounts of HET contamination based on selected reaction monitoring mass spectrometry (LC-SRM) (Supplementary Table S1).
Ostreococcus tauri RCC745 grows on exogenous cHET (with HMP) in B1-deplete medium. Mean cell abundance data are for multiple days of the experiment (colored columns). (A) RCC745 grows when provided different concentrations of cHET (plus 1 nM HMP) or B1 (1 nM; as a positive control). The addition of cHET also facilitates use of low concentrations (pM) of HMP by RCC745 (Supplementary Fig. S2). (B) In contrast, a RCC745 ∆thiM mutant does not grow on supplied cHET (plus 1 nM HMP). Asterisks denote a significant difference (p < 0.05; n = 3; paired two-tailed t-test) relative to respective negative controls (−Con.).
Unlike wildtype O. tauri, a ∆thiM (lacking ThiM) mutant line did not grow on supplied cHET (Fig. 1B) indicating that cHET utilization requires ThiM, which agrees with prior results showing that O. tauri RCC745 requires ThiM in order to use the thiazole precursor produced by B1-synthesizing marine plankton18. Besides O. tauri, another cosmopolitan picoeukaryotic marine phytoplankton organism, Bathyococcus sp. RCC4222, also grew on supplied cHET (and HMP) under B1-limiting conditions (Supplementary Fig. S1), showing that use of exogenous cHET is a more general phenomenon in marine picoeukaryotic phytoplankton, particularly the Prasinophyceae.
ThiM prevalence in human microbiomes and cHET use by Escherichia coli
Diverse organisms, including freshwater algae, enteric bacteria, human pathogens, and terrestrial plants also possess ThiM11,14,15,18 and hence might similarly salvage exogenous cHET for use in B1 synthesis. Bioinformatic surveys revealed that metagenomes from the human microbiome contain ~10× higher relative abundance of ThiM sequences than marine and terrestrial metagenomes (Supplementary Table S2), prompting the hypothesis that ThiM-possessing human-associated bacteria use exogenous cHET, in the same way as picoeukaryotic marine phytoplankton (Fig. 1).
Human-associated enteric bacterium Escherichia coli K-12 as well as >400 other E. coli strains possess ThiM15 (Supplementary Table S3), making Escherichia coli a suitable model for testing our hypothesis. Experiments with an E. coli mutant lacking ThiG (∆thiG), the enzyme that synthesizes the thiazole precursor of B1 in de novo biosynthesis, showed the bacterium is also adapted to use low concentrations of exogenous cHET, specifically down to subpicomolar concentrations (Fig. 2). In contrast, ~1 million times more (>100 nM) HET was necessary to support comparable growth (Fig. 2), which confirms our hypothesis and is also consistent with prior experiments indicating that micromolar concentrations of HET are needed to sustain E. coli growth15. No trace cHET contamination was detected in HET stocks (Supplementary Table S1), suggesting that E. coli can synthesize B1 from HET, but only at relatively high extracellular concentrations of HET, which presumably enters the cell via low-affinity transporters and/or diffusion.
An E. coli ∆thiG mutant grows on B1-deplete M63 medium using exogenous cHET. All concentration values along the x-axes are in picomolar. (A) E. coli ∆thiG cells sustain growth using sub-picomolar concentrations of exogenous cHET or B1. (B) The E. coli ∆thiG mutant exhibited no notable growth upon supplied HET up to 105 pM, highly contrasting with responses to notably lower cHET additions. (C) Dramatically higher concentrations (≥106 pM) of exogenous HET are required to sustain growth of E. coli ∆thiG. Mean maximum yields for triplicate cultures are plotted along with their respective standard deviations. Asterisks denote significant differences (p < 0.05, n = 3; paired two-tailed t-test) versus the negative control (−Con.).
cHET use by the terrestrial plant Arabidopsis
Broadly contextualizing microbial use of cHET, we also tested whether ThiM-possessing terrestrial plants such as Arabidopsis14 can use exogenous cHET. Growth experiments with Arabidopsis, using the wild-type and a mutant (tz-1) unable to synthesize thiazole precursor, confirmed that Arabidopsis can use cHET because the mutant grew well when given high concentrations of cHET (Fig. 3). In contrast to picoeukaryotic phytoplankton and E. coli (Figs 1, 2), equivalent concentrations of HET also sustained growth of the Arabidopsis mutant (Fig. 3). This result highlights a key difference between plants and (aquatic) microbes, in that the latter are equipped to salvage B1 from very low concentrations of exogenous precursors (Figs 1, 2), likely as a result of adaption within an environment where precursor(s) are a community currency circulated between producers and consumers.
Thiazole-auxotrophic Arabidopsis thaliana plants can use cHET to sustain growth. Mutant tz-1 plants were grown with the indicated concentrations of HET or cHET. Pictures were taken 14 days after germination and are representative of at least 30 plants. A wild-type (WT) plant is shown for comparison.
Discussion
Collectively, our findings establish cHET as a valuable microbial B1-related currency and component of de novo B1 biosynthesis. Previously, HET was the only thiazole considered in research investigating B1 salvage1,2,3,4,7,9,14,25. Our results alter this paradigm as cHET is clearly useful for prokaryotic and eukaryotic microorganisms, and moreover is accessible at extremely low concentrations (Figs 1, 2). Specifically, acquisition of exogenous cHET sustains key primary producers in the ocean as well as important enterobacteria, presumably enabling them to bypass the energetic and/or elemental costs of de novo biosynthesis of cHET(−P). The ability of these cells, which are endemic to marine euphotic waters and the human body, to use very low exogenous cHET concentrations strongly suggests that cHET is bioavailable in nature and integral to interactions between B1 auxotrophs and B1-synthesizing microorganisms or hosts. However, direct evidence of this is lacking.
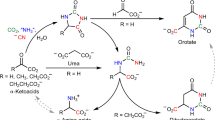
Our observation that diverse organisms grow on cHET (Figs 1–3, Supplementary Fig. S1), alongside prior biochemical evidence of cHET-P generation by bacteria and plants19,21, solidifies cHET-P as a fundamental and widely overlooked5,7,9,14,23,24,25 component of de novo B1 biosynthesis (Fig. 4). Establishing that cHET is central to de novo B1 biosynthesis pinpoints B1-prototrophic organisms as sources of the thiazole to co-occurring populations, i.e. key microorganisms that influence marine primary productivity (Fig. 1) or human health (e.g. Shiga-toxin producing E. coli (STEC)26 possessing ThiM, e.g. E. coli STEC_O31; Supplementary Table S3). Our findings also improve understanding of B1 biosynthesis in general - a vital process for life on Earth, and target for industrial and biomedical applications with human impact, e.g. efforts to increase crop nutritive value or resilience27,28 and to develop drugs targeting pathogenic microbes29.
An updated metabolic map including cHET in B1 salvage and de novo biosynthesis pathways. Orange arrows represent cHET utilization demonstrated here. Solid and dashed purple arrows represent usage and generation (via degradation) of HET reported previously1,2,4,30. Large black arrows denote core B1 biosynthesis processes. Cells acquire exogenous thiazole precursor via high (HA) or low affinity (LA) transport systems (with unknown sequence identity), or diffusion based on results shown here (Figs 1, 2, Supplementary Fig. S1) and in prior studies4,30. Shorthand compound names are in italics, while shorthand enzyme names are in bold. Thi80, TPK = thiamin pyrophosphate kinase; TMPsynthase = thiamin monophosphate synthase; P-tase = phosphatase; TMP = thiamin monophosphate; TDP = thiamin diphosphate; B1 = thiamin; −P = phosphate group; HMP = 4-amino-5-hydroxymethyl-2-methylpyrimidine; cHET = 5-(2-hydroxyethyl)-4-methyl-1,3-thiazole-2-carboxylic acid; HET = 4-methyl-5-thiazoleethanol.
For clarity, we propose that HET be regarded as a degradation-derived precursor as it comes from B1 degradation1,30, whereas cHET is generated via the de novo biosynthesis pathway19,20,21 (Fig. 4). Comparable descriptors also apply to pyrimidine B1 precursors 4-amino-5-aminomethyl-2-methylpyrimidine (AmMP; degradation product) and HMP (product of de novo biosynthesis)1,10,30.
In conclusion, prevailing views of thiazole precursor biosynthesis, use, and exchange require reassessment, as it is now evident that the role of B1 requirements in host-associated or aquatic microbiomes cannot be fully understood without consideration of the widely overlooked thiazole B1 precursor cHET(−P). Exchange of cHET and its influence upon interactions between taxa31, especially producers and consumers of cHET, deserves particular attention given that exogenous cHET acquisition is integral to the survival of microorganisms with environmental, industrial, and/or biomedical impacts, e.g. key marine microbial primary producers, E. coli, and other ThiM-possessing taxa7 (Figs 1, 2; Supplementary Table S2). Since model organisms readily utilize exogenous cHET, it should be possible for future research to decipher the intricacies of cHET flux and its influence upon cell interactions.
Methods
Chemicals
Thiamin hydrochloride and 4-methyl-5-thiazoleethanol (HET) (≥95% HPLC-determined purity) were purchased from Sigma Aldrich (St. Louis, MO, USA), and 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP) (>95% purity) was purchased from Enamine Ltd. (Kiev, Ukraine). 5-(2-Hydroxyethyl)-4-methyl-1,3-thiazole-2-carboxylic acid (cHET) (>98% HPLC-determined purity) was purchased from Finetech Industry Limited (Wuhan, China).
Marine phytoplankton growth experiments
Cultures of O. tauri RCC745 wild-type, the ∆thiM mutant line, and Bathycoccus sp. RCC4222 were maintained at 20 °C under 25 μE m−2 s−1 white light on artificial seawater (ASW) supplemented with trace metals and B1 and B12 vitamins as previously described18,32. Antibiotics (penicillin G 50 μg mL−1, streptomycin 200 μg ml−1) were added to prevent the growth of Marinobacter bacteria associated with O. tauri RCC745 cultures18. RCC745 cell growth and the absence of Marinobacter were determined on an Accuri C6 flow cytometer (Becton Dickinson) using SYBR Green Il staining. To start cHET bioassays, algal cells inoculated at 0.5 to 1 × 106 cells mL−1 were first grown for 7 days in B1-deplete ASW medium (ASW-B1). These B1-deprived cultures were used to inoculate ASW culture medium containing HMP (1 nM) and various concentrations of cHET at 0.5 to 1 × 106 cells mL−1 in triplicate. Similar experiments were performed to determine the requirements for B1 or HMP (in 1 nM cHET-supplemented ASW). Triplicate positive (1 nM B1) and negative (ASW-B1) controls were run in parallel. Microalgal cell abundances were determined on an Accuri C6 flow cytometer.
Escherichia coli growth experiments
E. coli JW5549 ΔthiG761::kan (Keio Collection) cells were cultured in LB medium. Cells were harvested via centrifugation (9 × 1000 g; 3 min), washed, and resuspended over three iterations in M63 minimal growth medium33 lacking B1 and only 1 mM glucose as a carbon source to minimize the potential for vitamin contamination from glucose stock. Washed/resuspended cells were added to triplicate sterile 4.5 mL polystyrene tubes containing M63 B1-free test medium (at a ratio of 0.5 µL cells: 1.5 mL medium) with various concentrations of HET or cHET, B1 (as a positive control), or without amendment (as a negative control). Static culture tubes were incubated at room temperature, in the dark. Cultures were thoroughly vortexed before sampling after one and two weeks of incubation. Optical density (590 nm) of cultures was measured using a FLUOstar Optima Plate Reader (Bmg-Labtech) and clear 96-well plates. Cell abundances were determined from fixed (2% formaldehyde) and frozen (−80 °C) culture samples via SYBR green I (Molecular Probes, Eugene, OR, USA) staining and flow cytometry34 using a FACS CANTO II (Beckton Dickenson, Heidelberg, Germany).
Plant growth experiments
Arabidopsis thaliana thiazole-auxotrophic mutant (tz-1; ABRC stock number CS3375)35 and wild type (Columbia-1; CS3176) seeds were surface sterilized and plated on ½ MS medium containing 0.6% (w/v) Phytagel, 1% (w/v) sucrose, and with or without various concentrations of HET or cHET. Plates were held in the dark at 4 °C for four days, then placed under fluorescent lights (130–150 µE m−1 s−1) on a 12:12 h light/dark cycle at 22 °C for 14 days.
Liquid chromatography-mass spectrometry
Sample preparation: Stock solutions (1 mg mL−1) and intermediate stock solutions (10 µg mL−1) of HET, cHET, HMP, and B1 were prepared in Milli Q water and stored at −20 °C in the dark. Working solutions were prepared as 0.02 µg µL−1 solutions in 5 mM ammonium formate, 0.1% formic acid and 10% methanol for LC-SRM analysis.
LC-SRM: Working stocks of all four compounds were prepared in 5 mM ammonium formate, 0.1% formic acid and 10% methanol. One microliter injections onto a 150 × 0.3 mm ID column (Acclaim PepMap RSLC, C-18, 2 µm, 100 Å), held at 50 degrees C and subject to an HPLC gradient of 2–6% B over 4 min, then 6 to 10% B over 1.5 min (A, 0.1% formic acid; B, 80% acetonitrile, 0.08% formic acid) at 7 µl per min. This was coupled to a Thermo Quantiva triple quadrupole mass spectrometer in selected reaction monitoring (SRM) mode, operating under the following conditions: Q1 and Q3 resolution 0.4 (FWHM), 50 msec dwell time, spray voltage 3500 (positive ion mode), sheath gas 6, aux gas 2, ion transfer tube 235 C, vaporizer temp 70 C°. SRM parameters for each compound are given in Table S4. Limits of quantitation and limits of detection were calculated as described previosuly36 using a standard curve created from repeat injections of 0, 10, 100, 200, 500 and 1000 fmol of each compound on column and are given in Table S3. Cross contamination was assessed and reported for triplicate injections of 1000 fmol of each compound on column.
Metagenomic queries
The E. coli ThiM amino acid sequence (Uniprot ID P76423) was searched against diverse metagenomic libraries using BLASTP (and default BLOSUM62 settings) via GenomeNet (http://www.genome.jp/tools/blast/; Kyoto University Bioinformatics Center). Queries of soil metagenomic libraries were performed using BLASTP searches via the Joint Genome Institute (JGI) Integrated Microbial Genome (IMG) portal37.
Data availability statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Change history
06 June 2018
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
References
Jurgenson, C. T., Begley, T. P. & Ealick, S. E. The structural and biochemical foundations of thiamin biosynthesis. Annu. Rev. Biochem. 78, 569–603 (2009).
Burkholder, P. R. in Symposium on marine microbiology (ed. Oppenheimer, C. H.) 133–150 (CC Thomas Springfield, 1963).
Lwoff, A. Some aspects of the problem of growth factors for protozoa. Annu Rev Microbiol 1, 101–114 (1947).
Droop, M. R. Requirement for thiamine among some marine and supra-littoral protista. J Marine Biol Assoc UK 37, 323–329 (1958).
Carini, P. et al. Discovery of a SAR11 growth requirement for thiamin’s pyrimidine precursor and its distribution in the Sargasso Sea. ISME J, https://doi.org/10.1038/ismej.2014.61 (2014).
Tang, Y. Z., Koch, F. & Gobler, C. J. Most harmful algal bloom species are vitamin B1 and B12 auxotrophs. Proc Natl Acad Sci 107, 20756–20761 (2010).
Paerl, R. W., Bertrand, E. M., Allen, A. E., Palenik, B. & Azam, F. Vitamin B1 ecophysiology of marine picoeukaryotic algae: Strain-specific differences and a new role for bacteria in vitamin cycling. Limnol Oceanogr 60, 215–228 (2015).
Garcia, S. L. et al. Auxotrophy and intrapopulation complementary in the ‘interactome’ of a cultivated freshwater model community. Mol. Ecol. 24, 4449–4459 (2015).
Magnúsdóttir, S., Ravcheev, D., de Crécy-Lagard, V. & Thiele, I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet 6, 148 (2015).
Jenkins, A. H., Schyns, G., Potot, S., Sun, G. & Begley, T. P. A new thiamin salvage pathway. Nat Chem Biol 3, 492–497 (2007).
Wrenger, C. et al. Vitamin B1 de novo synthesis in the human malaria parasite Plasmodium falciparum depends on external provision of 4-amino-5-hydroxymethyl-2-methylpyrimidine. Biol. Chem. 387, 41–51 (2006).
Rodionov, D. A., Vitreschak, A. G., Mironov, A. A. & Gelfand, M. S. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J Biol Chem 277, 48949–48959 (2002).
Morris, R. M. et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806–810 (2002).
Yazdani, M. et al. Identification of the thiamin salvage enzyme thiazole kinase in Arabidopsis and maize. Phytochem 94, 68–73 (2013).
Mizote, T. & Nakayama, H. The thiM locus and its relation to phosphorylation of hydroxyethylthiazole in Escherichia coli. Journal of Bacteriology 171, 3228–3232 (1989).
Li, W. K. W. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol Oceanogr 39, 169–175 (1994).
Worden, A. Z., Nolan, J. K. & Palenik, B. Assessing the dynamics and ecology of marine picophytoplankton: the importance of the eukaryotic component. Limnol Oceanogr 49, 168–179 (2004).
Paerl, R. W. et al. Use of plankton-derived vitamin B1 precursors, especially thiazole-related precursor, by key marine picoeukaryotic phytoplankton. ISME J. 11, 753–765 (2017).
Hazra, A., Chatterjee, A. & Begley, T. P. Biosynthesis of the thiamin thiazole in Bacillus subtilis: identification of the product of the thiazole synthase-catalyzed reaction. J. Am. Chem. Soc. 131, 3225–3229 (2009).
Chatterjee, A., Jurgenson, C. T., Schroeder, F. C., Ealick, S. E. & Begley, T. P. Thiamin biosynthesis in eukaryotes: characterization of the enzyme-bound product of thiazole synthase from Saccharomyces cerevisiae and its implications in thiazole biosynthesis. J. Am. Chem. Soc. 128, 7158–7159 (2006).
Godoi, P. H. C. et al. Structure of the thiazole biosynthetic enzyme THI1 from Arabidopsis thaliana. J Biol Chem 281, 30957–30966 (2006).
Hazra, A. B. et al. A missing enzyme in thiamin thiazole biosynthesis: Identification of TenI as a thiazole tautomerase. J. Am. Chem. Soc. 133, 9311–9319 (2011).
Sañudo-Wilhelmy, S. A., Gómez-Consarnau, L., Suffridge, C. & Webb, E. A. The role of B vitamins in marine biogeochemistry. Annu. Rev. Marine. Sci. 6, 339–367 (2014).
Helliwell, K. E. The roles of B vitamins in phytoplankton nutrition: new perspectives and prospects. New Phytol. 216, 62–68 (2017).
Goyer, A. Thiamine in plants: Aspects of its metabolism and functions. Phytochem 71, 1615–1624 (2010).
Luna-Gierke, R. E. et al. Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol. Infect. 142, 2270–2280 (2014).
Pourcel, L., Moulin, M. & Fitzpatrick, T. B. Examining strategies to facilitate vitamin B1 biofortification of plants by genetic engineering. Front Plant Sci 4, 160 (2013).
Hanson, A. D., Beaudoin, G. A., McCarty, D. R. & Gregory, J. F. Does Abiotic Stress Cause Functional B Vitamin Deficiency in Plants? Plant Physiol 172, 2082–2097 (2016).
Du, Q., Wang, H. & Xie, J. Thiamin (vitamin B1) biosynthesis and regulation: a rich source of antimicrobial drug targets? Int. J. Biol. Sci. 7, 41–52 (2011).
Dwivedi, B. K. & Arnold, R. G. Chemistry of thiamine degradation in food products and model systems: a review. J. Agric. Food Chem. 21, 54–60 (1973).
Steele, J. A. et al. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 5, 1414–1425 (2011).
Lozano, J.-C. et al. Efficient gene targeting and removal of foreign DNA by homologous recombination in the picoeukaryote Ostreococcus. Plant J 78, 1073–1083 (2014).
Elbing, K. & Brent, R. in Current Protocols in Molecular Biology https://doi.org/10.1002/0471142727.mb0101s59 (John Wiley & Sons, Inc., 2001).
Brussaard, C. P. D., Payet, J. P., Winter, C. & Weinbauer, M. G. in Manual of aquatic viral ecology ASLO 102–109 https://doi.org/10.4319/mave.2010.978-0-9845591-0-7.102 (American Society of Limnology and Oceanography, 2010).
Li, S. L. & Rédei, G. P. Thiamine mutants of the crucifer, Arabidopsis. Biochem. Genet. 3, 163–170 (1969).
MacDougall, D. & Crummett, W. B. Guidelines for data acquisition and data quality evaluation in environmental chemistry. Anal. Chem. 52, 2242–2249 (1980).
Chen, I.-M. A. et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45, D507–D516 (2017).
Acknowledgements
Thanks are extended to S. Svenningsen for providing E. coli JW5549 ΔthiG761::kan, C. Kachuk for technical assistance, and H. Paerl for comments on the manuscript. This research was supported by the BONUS Blueprint project that has received funding from BONUS, the joint Baltic Sea research and development programme (Art 185), funded jointly from the European Union’s Seventh Programme for research, technological development and demonstration, and the Danish Council for Strategic Research (LR). Additional funding was provided by the French Agence Nationale de la recherche (Photo-Phyto, ANR-14-CE02-0018) (FYB), and the projet d’investissement d’Avenir UCAJEDI ANR-15-IDEX-01 (MM), NSERC grant 950-230440 (EMB), and NSF award IOS-1444202 (ADH).
Author information
Authors and Affiliations
Contributions
R.W.P., F.-Y.B., E.M.B., A.D.H. conceived the research; R.W.P., P.S., M.M., E.M.B., E.R., F.S., T.D.N. conducted experiments; all authors contributed to the writing or editing of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Paerl, R.W., Bertrand, E.M., Rowland, E. et al. Carboxythiazole is a key microbial nutrient currency and critical component of thiamin biosynthesis. Sci Rep 8, 5940 (2018). https://doi.org/10.1038/s41598-018-24321-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-24321-2
This article is cited by
-
Genomic repertoire of Mameliella alba Ep20 associated with Symbiodinium from the endemic coral Mussismilia braziliensis
Symbiosis (2020)
-
A streamlined and predominantly diploid genome in the tiny marine green alga Chloropicon primus
Nature Communications (2019)
-
Modeling vitamin B1 transfer to consumers in the aquatic food web
Scientific Reports (2019)
-
Seasonal variation and species-specific concentrations of the essential vitamin B1 (thiamin) in zooplankton and seston
Marine Biology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.