Abstract
Identification of inter-individual variability for drug metabolism through cytochrome P450 2B6 (CYP2B6) enzyme is important for understanding the differences in clinical responses to malaria and HIV. This study evaluates the distribution of CYP2B6 alleles, haplotypes and inferred metabolic phenotypes among subjects with different ethnicity in Botswana. A total of 570 subjects were analyzed for CYP2B6 polymorphisms at position 516 G > T (rs3745274), 785 A > G (rs2279343) and 983 T > C (rs28399499). Samples were collected in three districts of Botswana where the population belongs to Bantu (Serowe/Palapye and Chobe) and San-related (Ghanzi) ethnicity. The three districts showed different haplotype composition according to the ethnic background but similar metabolic inferred phenotypes, with 59.12%, 34.56%, 2.10% and 4.21% of the subjects having, respectively, an extensive, intermediate, slow and rapid metabolic profile. The results hint at the possibility of a convergent adaptation of detoxifying metabolic phenotypes despite a different haplotype structure due to the different genetic background. The main implication is that, while there is substantial homogeneity of metabolic inferred phenotypes among the country, the response to drugs metabolized via CYP2B6 could be individually associated to an increased risk of treatment failure and toxicity. These are important facts since Botswana is facing malaria elimination and a very high HIV prevalence.
Similar content being viewed by others
Introduction
The human cytochrome P450 2B6 enzyme (CYP2B6) plays a pivotal role in the metabolism of different drugs used for malaria treatment (artemisinin derivatives such as artesunate, β-artemether and artemether) and for HIV life-long therapy (non-nucleoside reverse-transcriptase inhibitors such as efavirenz and nevirapine). CYP2B6 is a highly polymorphic enzyme that affects the therapeutic response including drug interactions in individuals1,2. Importantly, African populations show a high degree of variation in the CYP2B6 gene3. In Botswana, interventions towards the elimination of Plasmodium falciparum malaria have been intensified and HIV has been reported at a frequency of 18.5%4. So, a deeper knowledge of the genetic variability of CYP2B6 in the population of Botswana is necessary to improve the efficacy of ongoing fight against malaria and HIV.
Currently, the first line treatment for uncomplicated malaria in Botswana is a fixed-dose combination of artemether (AM) and lumefantrine (LU)5. After oral administration, AM is rapidly absorbed and metabolized to dihydroartemisinin (DHA) through demethylation, mainly via CYP2B6 enzymes6,7,8. DHA has a higher antimalarial activity than AM, and it is inactivated primarily by glucuronidation7,9.
Efavirenz (EFV) and nevirapine (NVP) are the most prescribed drugs in anti-HIV combination treatment in resource-limited countries10. There is a considerable literature on EFV and NVP pharmacogenetics and currently it has been proposed that personalizing the dosage of these anti-HIV drugs according to the CYP2B6 genotype of the patient will be of therapeutic benefit11.
The known single nucleotide polymorphisms (SNPs) within the CYP2B6 locus influencing AM and EFV/NVP plasma exposure are: 516 G > T (rs3745274), 785 A > G (rs2279343) and 983 T > C (rs28399499). The polymorphisms at position 516 and 983 confer a slow metabolic phenotype, leading to higher drug plasma exposure and increased toxicity risk. The variation at position 785 confers rapid drug metabolism, leading to lower drug plasma level and potentially poorer therapeutic outcome10. The above-mentioned SNPs, in combination or individually, may lead to the following CYP2B6 alleles: *4 (785 A > G only), *6 (516 G > T and 785 A > G), *9 (516 G > T only), *16 (785 A > G and 983 T > C), *18 (983 T > C only) (http://cypalleles.ki.se). The metabolic status for a given drug is defined as slow, intermediate, extensive or ultra-rapid as a result of the individual’s genetic make up12. Furthermore, in the anti-infectious therapy context, the therapeutic outcome based on the individual metabolic status for a given drug depends also on the length and frequency of therapy. For example, in the context of the short course uncomplicated malaria treatment, a slow AM metabolism induces a lower biotransformation rate to the most active metabolite DHA7 leading to a lower efficacy and possibly a longer tail during which malaria parasites are exposed to sub-inhibitory drug concentrations, inducing an increased permanence in the selective window for drug resistance13,14. Conversely, in the context of chronic EFV- or NVP-based antiretroviral combination therapy (ARTc), a slow metabolic profile reduces drug clearance leading to higher drug plasma exposure and consequent higher toxicity risk. This may cause poorer compliance by the patient with resultant emergence of viral resistance possibly related to suboptimal drug exposure10,15. Concerning the influence of rapid metabolism phenotypes on antimalarial and antiretroviral therapy, information is lacking10.
Another important aspect is that malaria treatment in sub-Saharan Africa may frequently overlap EFV- or NVP-based ARTc leading to possible drug-drug interactions. The co-administration of EFV or NVP and AM-LU was associated with low plasma concentration of AM and DHA, whereas LU concentration was reduced by EFV co-administration16,17. A recent study, looking at HIV-infected patients affected by malaria in Tanzania, has shown that the slow metaboliser genotype CYP2B6 *6/*6 leads to high EFV plasma concentration which are significantly correlated with low LU plasma concentrations and a high rate of recurrent parasitemia18, increasing the risk of the appearance of drug-resistant malaria parasites.
Finally, Botswana (together with other Southern African countries) is also home to individuals with unique southern African KhoeSan ancestry. The KhoeSan populations are the earliest known indigenous inhabitants of southern Africa and are distinguished by their unique phenotype(s), genetic divergence, click languages, and hunter-gatherer subsistence strategy compared to other African populations19,20,21. During evolution, hunter-gatherer practices exposed populations to several xenobiotics which sometimes imposed serious health and environmental risks, leading to the selection of specific mutations linked to an efficient detoxification. For example, significant differences in prevalence of acetylation phenotypes are found between hunter-gatherer and food-producing populations, both in sub-Saharan Africa and worldwide, and between agriculturalists and pastoralists in Central Asia22. In Africa, comparative studies on N-acetyl transferase (NAT2) haplotype frequencies and acetylation status inference revealed that the hunter-gatherer populations (San, but also Pygmies) are mainly composed of fast and intermediate acetylators23, in clear contrast with most agriculturalist populations (such as Bantu). A recent report showed differences in the allele frequency for CYP2C8*2 among San and Bantu-related communities in Botswana24. These general observations highlight the need for a detailed pharmacogenetic characterization of populations of pure or admixed KhoeSan ancestry in Botswana to accommodate individual/population genetic make-up and expedite optimum treatment strategies against important infections such as malaria and HIV infection.
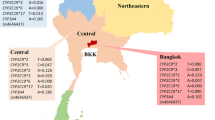
In this study we aim to: i) define the CYP2B6 haplotype status in Botswana via the three most important functional SNPs known to affect the enzyme activity; ii) examine the relationship, if any, of the haplotypes within ethnic groups; iii) apply the “metabolic score” in the study of inferred metabolic phenotypes among the populations. We selected three sites where the ethnic composition is different (San-related population in the Ghanzi health district and Bantu-related populations in the Chobe and Serowe/Palapye health districts, respectively) (see Fig. 1). This work is part of the ongoing screening activities for antimalarial drugs efficacy and safety study in Botswana unto malaria elimination by 2020, but it also provides useful information on antiretroviral drug metabolism in the population, making this research of public health relevance for the country.
Results
Out of 609 samples we obtained genotypic data for all the three polymorphisms from 570 samples (93.60%). The genotype and allele frequency for 516 G > T, 785 A > G and 983 T > C in the three selected districts are shown in Table 1 with statistical comparisons.
Hardy-Weinberg Equilibrium
Hardy-Weinberg equilibrium analysis showed that CYP2B6-516 displayed significant deviations from Hardy–Weinberg equilibrium in samples from Serowe/Palapye district due to a significant heterozygous defect (Wright’s F = 0.13), with the same sample having a significant CYP2B6-983 heterozygous excess (Wright’s F = −0.17). Genotypes in samples from Chobe and Ghanzi districts were in Hardy-Weinberg equilibrium. CYP2B6-785 genotypes were in equilibrium in all three districts analysed. Regarding the only two samples showing a rare CC genotype for 983 T > C polymorphism25,26, we sequenced them and confirmed the result.
Linkage Disequilibrium analysis
A significant linkage disequilibrium (LD) was found with Arlequin between all pairwise comparisons of the three polymorphic loci when samples were considered as a single population (n = 570). Chi-square test values ranged from 36.07 (P < 0.00001, 1 df) for CYP2B6-516 vs CYP2B6-983, to 443.55 (P < 0.00001, 1 df) for CYP2B6-516 vs CYP2B6-785, to 43.93 (P < 0.00001, 1 df) for CYP2B6-983 vs CYP2B6-785. When analysing the samples separately by district of origin, we found a significant LD in all districts for all pairwise comparisons between the three loci (Table 2). In general, LD between CYP2B6-516 and CYP2B6-785 was strongest, while LD between CYP2B6-983 and the other two loci was weakest but still highly significant.
Haplotype frequency estimation
Haplotype frequencies were estimated using Arlequin (Table 3). For the combined samples, the GAT haplotype was the most common and the GGC haplotype the rarest. Among the identified haplotypes, two not yet categorized were found, TGC and TAC, indicated as *6 + *18 and *9 + *18, respectively (Table 3). When districts were considered separately, GAT remained the dominant haplotype while GGC remained the rarest. However, differences in haplotype frequencies could be observed between districts. While there was no statistical difference in the population structure between the Serowe/Palapye and Chobe districts (Population pairwise FSTs test: P = 0.06 +/− 0.024, 110 permutations), there was a significant difference when both districts were separately compared to the Ghanzi district (Population pairwise FSTs test: P < 0.00001, 110 permutations for both comparisons). When the Serowe/Palapye and Chobe districts were combined and then compared to the Ghanzi district, the populations were still significantly different (Population pairwise FSTs test: P < 0.00001, 110 permutations). In particular the Ghanzi district displayed in particular a higher frequency (as estimated by ML) of the GAT (65.55%), TAT (10.98%) and TAC (1.53%) haplotypes and a lower frequency of the TGT (12.79%) and TGC (3.18%) haplotypes (Table 3). These data indicated a distinct population structure in terms of the CYP2B6 alleles in the Ghanzi district compared to both the Serowe/Palapye and Chobe districts.
Neutrality tests
For each sample, the most likely gametic phases (as determined by Arlequin) were selected to perform neutrality tests. The results of the Tajima’s D tests indicated positive values above 2 for the Serowe and Chobe populations with significant P-values and 1.26 for the Ghanzi samples with a non-significant P-value. The Chobe and Serowe combined population dataset (i.e. Bantu) also yielded a statistically significant D value above two (Table 4). The significantly positive Tajima’s D scores among the Bantu groups could either indicate the presence of balancing selection, migration or a sudden population contraction, or even indicate positive selection on pre-existing genetic variation27.
Metabolic score
The expected metabolic scores were analysed among the three populations (Table 5 and Fig. 2).
The Kolmogorov-Smirnov test did not find any statistically significant divergence from normal distribution for the metabolic scores in each of the three sub-samples (Table 5). The Bartlett test for homogeneity of variances showed that the three variances were not statistically different from each other (Bartlett’s chi square = 3.32, df = 2, P = 0.19). The chi-square statistic in the comparison of the absolute frequencies distribution of the metabolic scores among all districts was 9.43 with the associated P-value being 0.49, indicating that the distributions of metabolic scores in the three groups were comparable. The CYP2B6 inferred phenotypes associated with a reduced metabolism were found in 36.67% (n = 209/570) of the overall population studied, mainly intermediate (94.25%, n = 197/209) rather than poor metabolizer (5.75%, n = 12/209). Phenotypes associated with increased metabolism were found in 4.21% (n = 24/570) of the population studied, with only 2 individuals showing an inferred ultra-rapid metabolic phenotype.
Discussion
African populations represent the most genetically diverse populations in the world and this complicates the already inadequate treatment strategies developed for several communicable and non-communicable diseases. Our study in Botswana showed that haplotype composition is diverse between the Bantu-related communities from Serowe/Palapye and Chobe districts, and the San-related communities of the Ghanzi area. This probably reflects the different genetic background and evolutionary history of hunter-gatherers and food producing populations (farming and pastoralists) in Southern Africa28. We pooled together the haplotype data from Serowe/Palapye and Chobe districts considering them as a single population since there was no statistical difference in the population structure between the two districts. The frequency for CYP2B6 516 G > T is in line with few other studies from Botswana29,30. It should be pointed out that looking at the single SNP CYP2B6 516 G > T, Serowe/Palapye and Chobe districts do show statistical difference in genotypes distribution (see Table 1). However, haplotypes better predict the population structure than single SNPs31, and this observation is also true in CYP2B6 because of the known linkage among the sites2, as confirmed in this study. We identified two haplotypes not yet categorized into alleles that however were already described in other studies32,33.
A possible explanation of the difference observed for genotype distribution at 516 polymorphism between the two districts inhabited by Bantu could be due to the HWE of genotypes that was present only in Chobe district. The deviation from the HWE in Serowe/Palapye district was also found for the 983 polymorphism. Deviation from HWE could be due to lack of CC genotypes for CYP2B6-983, LD between SNPs at position 516 and 983. In addition, Serowe/Palapye has a wider sample size that increases the likelihood of deviation from the HWE when one of the genotypes (CC) is absent or very rare. Other studies in Africa found absence of 983-CC genotypes which prevented testing for HWE34,35. Another possible factor affecting both CYP2B6-516 comparisons and HWE in Serowe/Palapye district could be ethnic admixture since it is know that the Bantu-related population of Botswana carries a variable proportion of KhoeSan ancestry36,37.
An important result of our study is that the metabolic inferred phenotypes are similar among the three investigated sites. The main reason for this phenomenon could be due to the homeostatic effect of the mutations when taken together. Thus, despite clear and high levels of haplotype diversity among the sites, we observe an inferred phenotypic convergence, with the result that, globally, drug metabolism remains consistent and comparable among the populations studied. It is worth noting that convergent phenotypic evolution is a known phenomenon in biology38,39 and also relevant in human populations for several characters included skin pigmentation, lactose tolerance and immune responses40,41,42,43. The CYP2B6 enzyme plays an important primary role in bioactivating and detoxifying a certain number of procarcinogens and environmental agents44,45 as well as processing the arsenal of plant chemical defences introduced with diets46. The homogeneous phenotypes among the different sub-samples of this study are probably due to adaptation to the environmental and/or toxicological conditions of the Kalahari and surrounding areas. Based on the Tajima’s D neutrality test results, the two different patterns between Bantu and San could either indicate the presence of balancing selection or alternatively migration or a sudden population contraction in the Bantu population but not in the San. The results could also be possible evidence for positive selection on pre-existing genetic variation. A similar trend was already found by Fuselli et al.47 for the CYP2D6 gene and by Podgorná et al.22 for the NAT2 gene comparing hunter gatherers with food producing populations in Africa. There is thus some evidence that the current haplotypes might have evolved independently among the different ethnic groups and may thus represent a spectrum in terms of historical and potential adaptations to different ecological and toxicological niches. However, analysis of more genes and control groups will be required to exclude non-evolutionary events (e.g. migration into the Bantu populations) as the underlying cause of the observed results.
According to the metabolic score, expected metabolic phenotypes were obtained: 59.12% of the population study showed a CYP2B6 extensive metabolic phenotype, whereas 36.67% and 4.21% had a reduced and increased metabolic phenotype respectively (see Table 5). The poor and the ultra-rapid metabolic phenotype were observed in 2.1% (n = 12/570) and 0.35% (n = 2/570) of individuals respectively, whereas 34.56% (n = 197/570) showed an intermediate reduced metabolic phenotype, and 3.86% (n = 22/570) had an intermediate increased metabolic phenotype. Individuals with a CYP2B6 reduced metabolic phenotype may have an increased risk of malaria treatment failure when treated with artemisinin derivatives48, as well as an impaired outcome of EFV or NVP-based ARTc10,29. This effect on malaria therapy efficacy could constitute an important obstacle in the malaria elimination phase, suggesting the need to strengthen the surveillance of AM-LU drug efficacy in Botswana. It is important to note that Chobe is a seasonal malarious area bordering with Namibia, Zambia and Zimbabwe, whereas Serowe/Palapye is more prone to unstable epidemics based on weather patterns49,50. Differently, in Ghanzi, which does not usually receive high amounts of rainfall, malaria outbreaks do occur at times, as it is currently the case (http://www.moh.gov.bw/press%20release/MALARIA%20PRESS%20RELEASE.pdf).
Concerning individuals having an inferred ultra-rapid metabolic phenotype, they are rare and the effect of their metabolic pattern seems to affect mainly EFV or NVP-based ARTc by leading to sub-therapeutic drug exposure with a potential increase of risk of selection for viral resistance10. Further studies should focus on this fast metaboliser fraction of the population almost neglected in the scientific literature.
Finally, it is worth noticing that drug-drug interaction in malaria and HIV co-infections can lead to therapeutic consequences. For examples, EFV was shown to reduce AM AUC by 80%, tripling the dose of AM needed to compensate EFV-inductive effect51. In another study, EFV was shown to reduce LU bioavailability52. To ensure antimalarial treatment success in HIV/malaria co-infected patients on EFV-based ART, an increase of dosage or an extension of the duration of AM-LU treatment using the current dose was proposed from several authors17,53. Studying how this is affected by drug metabolism phenotype is another important topic that will deserve future attention.
Moreover, malaria infection stimulates HIV replication, causing transient elevation in viral load54 that can hamper the management of HIV infected patients, possibly amplifying HIV prevalence55 and this could be relevant for Botswana.
Our work has some limitations one being the absence of metabolic phenotypes, since this is a pure genetics study. Furthermore, we did not measure the extent of KhoeSan ancestry, instead we based our definition of San-related ethnicity on family names and sites. However, based on the results from our current study, we can conclude that although ethnically and genetically different, populations in Botswana display convergent evolution in their drug metabolism. The presence of significant numbers of slow and fast metabolisers can significantly impact the emergence and spread of drug resistance in malaria and HIV, either by exposing pathogens to sub-lethal drug doses or inducing non-compliance in patients, with similar consequences. The high frequency of co-infections and the negative interaction between anti-HIV and anti-malaria drugs further exacerbates the risk of resistance emerging. This warrants constant monitoring in the population to identify potential patients with abnormal drug metabolism and adapt treatments accordingly.
Methods
Sample collection
The survey was performed from March to May 2012 in Botswana in the broader context of a Malaria Indicator Survey56,57. A total of 609 unrelated children aged 2–12 years asymptomatic for malaria were enrolled from primary schools and child welfare clinics: 288 from the Serowe/Palapye district, 159 from the Chobe district and 162 from the Ghanzi (or Gantsi) district. These districts have peculiar ethnic compositions, as shown in previous studies24,58. The same protocol for enrollment was followed in all sites. Written informed consent for multiple genetic and epidemiological surveys was obtained from all the subjects’ parents/caregivers before enrollment in the study. This study was conducted in accordance with the guidelines of the Helsinki Declaration of 2000 revised 2013, with the approval from Human Research and Development Division of the Botswana Ministry of Health [PPME-13/18 V (380)] and the Institutional Review Board of the University of Pennsylvania [protocol number 820378]. Three ml of whole blood was collected into EDTA-containing tubes by qualified health care workers.
DNA extraction and PCR-RFLP conditions
DNA was extracted with Qiagen kits according to the manufacturer’s protocol. CYP2B6 516 G > T (rs3745274) detection was carried out using a PCR-RFLP technique according to the protocol of Lavandera et al.59 with minor modifications. For CYP2B6 983 T > C (rs28399499) detection we applied a touchdown PCR-RFLP assay developed in our laboratory34. Finally for the purpose of this study we adopted a new in-house protocol for the analysis of the CYP2B6 785 A > G (rs2279343) polymorphism. Briefly, we designed two primers that amplify a 223 bp fragment of the CYP2B6 gene (forward primer: 5′-AACCTGCAGGAAATCAATGC-3′; reverse primer: 5′-CCTTCTTCCCTCCCCATCTTC-3′). For PCR cycling, after 5 min of denaturation at 94 C, the PCR mixture was subjected to the following conditions: 30 s at 94 C, 30 s at 65 C and 30 s at 72 C for 35 cycles, with a final step of 10 min at 72 C. The PCR product was then incubated with NlaIV restriction enzyme that cuts the mutant allele (G) in three fragments of 144 bp, 62 bp and 17 bp; while the wild-type allele (A) is digested in two fragments of 161 bp and 62 bp. The digested fragments were visualized on a 4% agarose gel. Negative control comprised of PCR reaction without a DNA template and controls for human genotyping were utilized after sequencing the different genotypes.
Hardy-Weinberg Equilibrium evaluation
Evaluation of Hardy-Weinberg equilibrium was performed using the HWSIM software (freely available at http://krunch.med.yale.edu/hwsim/) and Monte-Carlo permutation test performed when genotypic classes had an expected cell size of less than five. Wright’s F statistics was applied to evaluate the expected level of heterozygosity.
Linkage Disequilibrium, haplotype frequency analysis and Neutrality test
Arlequin v3.560 was used to test for linkage disequilibrium between the three loci and for haplotype reconstruction. Several other studies were able to evaluate haplotype structure and population differentiation looking at functional SNPs in the same gene61,62,63,64. The input consisted of diploid genotypic DNA data with unknown gametic phase and assuming co-dominance. The expectation-maximization (EM) algorithm used to test for Linkage Disequilibrium (LD) was run for 20,000 permutations and 3 initial conditions, based on recommended criteria. For haplotype reconstruction the EM algorithm was run on the same input file used for the LD test at the haplotype level, with 50 starting points and 1,000 iterations. Other parameters were set to default. The Excoffier–Laval–Balding (ELB) algorithm (with default settings) was used to generate haplotype counts. Population haplotype frequencies were determined both by haplotype counts and estimated by Maximum Likelihood (ML) and were compared using pairwise FSTs calculated using default values.
After determining the gametic phase of the samples using the ELB algorithm in Arlequin, the DnaSP (v6) software65 was used to calculate and evaluate the Tajima’s D neutrality test using the total number of mutations.
Metabolic score
We elaborated and adopted a “metabolic score” to take into account the global effect of the three polymorphisms together, according to an extensive literature linking genetic polymorphisms to functional impact10,66,67,68. For the metabolic score we translated the genotype information into a measure of phenotype using an ‘activity score’ system already adopted for CYP2D647,69, for CYP2A670 and for CYP2C1971. The metabolic score adopted is based on the algebraic sum of the individual allele values, according to an additive model for CYP2B672,73. The scores were set conferring a −1 value for each slow metabolism allele and +1 for rapid metabolism allele, while an extensive metabolism allele was scored 0. Accordingly, we obtained: 516 GG = 0; 516 GT = −1; 516 TT = −2; 983 TT = 0; 983 TC = −1; 983 CC = −2; 785 AA = 0; 785 AG = +1; 785 GG = +2. Each composite genotype was attributed a final metabolic score by summing the single score for each SNP.
To test possible deviation from the normal distribution of the scores the Kolmogorov-Smirnov test was used and the statistic D values calculated for each distribution. A related P-value greater than 0.05 indicates normal distribution of data74. Bartlett’s test was performed to assess if the assumption of equal variances among the three populations was valid75. Comparisons among metabolic scores were calculated using the chi-square statistic.
All the statistical calculations were performed using Statistica 13.0 software (StatSoft).
References
Hedrich, W. D., Hassan, H. E. & Wang, H. Insights into CYP2B6-mediated drug-drug interactions. Acta Pharm. Sin B. 6, 413–425 (2016).
Turpeinen, M. & Zanger, U. M. Cytochrome P450 2B6: function, genetics, and clinical relevance. Drug Metabol. Drug Interact. 27, 185–197 (2012).
Čolić, A., Alessandrini, M. & Pepper, M. S. Pharmacogenetics of CYP2B6, CYP2A6 and UGT2B7 in HIV treatment in African populations: focus on efavirenz and nevirapine. Drug Metab. Rev. 47, 111–123 (2015).
Statistics Botswana. Botswana AIDS Impact Survey IV (BAIS IV). (Statistics Botswana, 2013).
Botswana Ministry of Health. Revised guidelines for the diagnosis and treatment of malaria in Botswana. (Botswana Ministry of Health, 2015).
Svensson, U. S. & Ashton, M. Identification of the human cytochrome P450 enzymes involved in the in vitro metabolism of artemisinin. Br. J. Clin. Pharmacol. 48, 528–535 (1999).
Djimdé, A. & Lefèvre, G. Understanding the pharmacokinetics of Coartem. Malar. J. 8, S4 (2009).
Honda, M. et al. Functional characterization of CYP2B6 allelic variants in demethylation of antimalarial artemether. Drug Metab. Dispos. 39, 1860–1865 (2011).
Aweeka, F. T. & German, P. I. Clinical pharmacology of artemisinin-based combination therapies. Clin. Pharmacokinet. 47, 91–102 (2008).
Russo, G. et al. Pharmacogenetics of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in resource-limited settings: Influence on antiretroviral therapy response and concomitant anti-tubercular, antimalarial and contraceptive treatments. Infect. Genet. Evol. 37, 192–207 (2016).
Mukonzo, J. K. et al. Pharmacogenetic-based efavirenz dose modification: suggestions for an African population and the different CYP2B6 genotypes. PLoS One 9, e86919 (2014).
Pirmohamed, M. Personalized pharmacogenomics: predicting efficacy and adverse drug reactions. Annu. Rev. Genomics Hum. Genet. 15, 349–370 (2014).
Paganotti, G. M. Reply to Cavaco et al. J. Infect. Dis. 207, 688–689 (2013).
Kay, K. & Hastings, I. M. Measuring windows of selection for anti-malarial drug treatments. Malar. J. 14, 292 (2015).
Ribaudo, H. J. et al. Pharmacogenetics of plasma efavirenz exposure after treatment discontinuation: an Adult AIDS Clinical Trials Group Study. Clin. Infect. Dis. 42, 401–407 (2006).
Kredo, T. et al. Interaction between artemether-lumefantrine and nevirapine-based antiretroviral therapy in HIV-1-infected patients. Antimicrob. Agents Chemother. 55, 5616–5623 (2011).
Byakika-Kibwika, P. et al. Significant pharmacokinetic interactions between artemether/lumefantrine and efavirenz or nevirapine in HIV-infected Ugandan adults. J. Antimicrob. Chemother. 67, 2213–2221 (2012).
Maganda, B. A., Minzi, O. M., Ngaimisi, E., Kamuhabwa, A. A. & Aklillu, E. CYP2B6*6 genotype and high efavirenz plasma concentration but not nevirapine are associated with low lumefantrine plasma exposure and poor treatment response in HIV-malaria-coinfected patients. Pharmacogenomics J. 16, 88–95 (2016).
Gronau, I., Hubisz, M. J., Gulko, B., Danko, C. G. & Siepel, A. Bayesian inference of ancient human demography from individual genome sequences. Nat. Genet. 43, 1031–1034 (2011).
Henn, B. M., Cavalli-Sforza, L. L. & Feldman, M. W. The great human expansion. Proc. Natl. Acad. Sci. USA 109, 17758–17764 (2012).
Uren, C. et al. Fine-Scale Human Population Structure in Southern Africa Reflects Ecogeographic Boundaries. Genetics 204, 303–314 (2016).
Podgorná, E. et al. Variation in NAT2 acetylation phenotypes is associated with differences in food-producing subsistence modes and ecoregions in Africa. BMC Evol. Biol. 15, 263 (2015).
Patin, E. et al. Sub-Saharan African coding sequence variation and haplotype diversity at the NAT2 gene. Hum. Mutat. 27, 720 (2006).
Motshoge, T. et al. Cytochrome P450 2C8*2 allele in Botswana: Human genetic diversity and public health implications. Acta Trop. 157, 54–58 (2016).
Wyen, C. et al. Impact of CYP2B6 983T > C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J. Antimicrob. Chemother. 61, 914–918 (2008).
Haas, D. W. et al. Associations between CYP2B6 polymorphisms and pharmacokinetics after a single dose of nevirapine or efavirenz in African Americans. J. Infect. Dis. 199, 872–880 (2009).
Przeworski, M., Coop, G. & Wall, J. D. The signature of positive selection on standing genetic variation. Evolution 59, 2312–2323 (2005).
Schuster, S. C. et al. Complete Khoisan and Bantu genomes from southern Africa. Nature 463, 943–947 (2010).
Gross, R. et al. Slow efavirenz metabolism genotype is common in Botswana. J. Acquir. Immune Defic. Syndr. 49, 336–337 (2008).
Vujkovic, M. et al. Brief Report: CYP2B6 516G > T Minor Allele Protective of Late Virologic Failure in Efavirenz-Treated HIV-Infected Patients in Botswana. Acquir. Immune Defic. Syndr. 75, 488–491 (2017).
Gattepaille, L. M. & Jakobsson, M. Combining markers into haplotypes can improve population structure inference. Genetics 190, 159–174 (2012).
Mehlotra, R. K., Bockarie, M. J. & Zimmerman, P. A. CYP2B6 983T > C polymorphism is prevalent in West Africa but absent in Papua New Guinea: implications for HIV/AIDS treatment. Br. J. Clin. Pharmacol. 64, 391–395 (2007).
Gengiah, T. N., Botha, J. H., Yende-Zuma, N., Naidoo, K. & Abdool Karim, S. S. Efavirenz dosing: influence of drug metabolizing enzyme polymorphisms and concurrent tuberculosis treatment. Antivir. Ther. 20, 297–306 (2015).
Paganotti, G. M. et al. CYP2B6 poor metaboliser alleles involved in efavirenz and nevirapine metabolism: CYP2B6*9 and CYP2B6*18 distribution in HIV-exposed subjects from Dschang, Western Cameroon. Infect. Genet. Evol. 35, 122–126 (2015).
Oluka, M. N., Okalebo, F. A., Guantai, A. N., McClelland, R. S. & Graham, S. M. Cytochrome P450 2B6 genetic variants are associated with plasma nevirapine levels and clinical response in HIV-1 infected Kenyan women: a prospective cohort study. AIDS Res. Ther. 12, 10 (2015).
Pickrell, J. K. et al. The genetic prehistory of southern Africa. Nat.Commun. 3, 1143 (2012).
Montinaro, F. et al. Complex Ancient Genetic Structure and Cultural Transitions in Southern African Populations. Genetics 205, 303–316 (2017).
Yeaman, S. & Whitlock, M. C. The genetic architecture of adaptation under migration-selection balance. Evolution 65, 1897–1911 (2011).
Ralph, P. L. & Coop, G. Convergent Evolution During Local Adaptation to Patchy Landscapes. PloS Genet. 11, e1005630 (2015).
Norton, H. L. et al. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol. Biol. Evol. 24, 710–722 (2007).
Tishkoff, S. A. et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39, 31–40 (2007).
Thomas, R. et al. Novel KIR3DL1 alleles and their expression levels on NK cells: convergent evolution of KIR3DL1 phenotype variation? J. Immunol. 180, 6743–6750 (2008).
Campbell, M. C. & Tishkoff, S. A. The evolution of human genetic and phenotypic variation in Africa. Curr. Biol. 20, R166–173 (2010).
Ekins, S. & Wrighton, S. A. The role of CYP2B6 in human xenobiotic metabolism. Drug Metab. Rev. 31, 719–754 (1999).
Hodgson, E. & Rose, R. L. The importance of cytochrome P450 2B6 in the human metabolism of environmental chemicals. Pharmacol. Ther. 113, 420–428 (2007).
Malenke, J. R., Magnanou, E., Thomas, K. & Dearing, M. D. Cytochrome P450 2B diversity and dietary novelty in the herbivorous, desert woodrat (Neotoma lepida). PLoS One 7, e41510 (2012).
Fuselli, S. et al. Evolution of detoxifying systems: the role of environment and population history in shaping genetic diversity at human CYP2D6 locus. Pharmacogenet. Genomics 20, 485–499 (2010).
Aung, A. K., Haas, D. W., Hulgan, T. & Phillips, E. J. Pharmacogenomics of antimicrobial agents. Pharmacogenomics 15, 1903–1930 (2014).
Mabaso, M. L., Kleinschmidt, I., Sharp, B. & Smith, T. El Niño Southern Oscillation (ENSO) and annual malaria incidence in Southern Africa. Trans. R. Soc. Trop. Med. Hyg. 101, 326–330 (2007).
MacLeod, D. A., Jones, A., Di Giuseppe, F., Caminade, C. & Morse, A. P. Demonstration of successful malaria forecasts for Botswana using an operational seasonal climate model. Environ. Res. Lett. 10, 044005 (2015).
Moss, D. M., Marzolini, C., Rajoli, R. K. & Siccardi, M. Applications of physiologically based pharmacokinetic modeling for the optimization of anti-infective therapies. Expert. Opin. Drug Metab. Toxicol. 11, 1203–1217 (2015).
Maganda, B. A., Ngaimisi, E., Kamuhabwa, A. A., Aklillu, E. & Minzi, O. M. The influence of nevirapine and efavirenz-based anti-retroviral therapy on the pharmacokinetics of lumefantrine and anti-malarial dose recommendation in HIV-malaria co-treatment. Malar. J. 14, 179 (2015).
Siccardi, M. et al. Use of a physiologically-based pharmacokinetic model to simulate artemether dose adjustment for overcoming the drug-drug interaction with efavirenz. In Silico Pharmacol. 1, 4 (2013).
Kublin, J. G. & Steketee, R. W. HIV infection and malaria–understanding the interactions. J. Infect. Dis. 193, 1–3 (2006).
Abu-Raddad, L. J., Patnaik, P. & Kublin, J. G. Dual infection with HIV and malaria fuels the spread of both diseases in sub-Saharan Africa. Science 314, 1603–1606 (2006).
Botswana Ministry of Health. Botswana malaria indicator survey 2012 report. (National Malaria Program, Ministry of Health, 2012).
Motshoge, T. et al. Molecular evidence of high rates of asymptomatic P. vivax infection and very low P. falciparum malaria in Botswana. BMC Infect. Dis. 16, 520 (2016).
Zetola, N. M. et al. Protocol for a population-based molecular epidemiology study of tuberculosis transmission in a high HIV-burden setting: the Botswana Kopanyo study. BMJ Open 6, e010046 (2016).
Lavandera, J. V., Parera, V. E., Rossetti, M. V., Batlle, A. & Buzaleh, A. M. Identification of CYP3A5 and CYP2B6 polymorphisms in porphyria cutanea tarda associated to human immunodeficiency virus. J. Clin. Exp. Dermatol. Res. S2, 006 (2011).
Excoffier, L. & Lischer, H. E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010).
Arenaz, I. et al. Haplotype structure and allele frequencies of CYP2B6 in Spaniards and Central Americans. Fundam. Clin. Pharmacol. 24, 247–253 (2010).
Carr, D. F., la Porte, C. J., Pirmohamed, M., Owen, A. & Cortes, C. P. Haplotype structure of CYP2B6 and association with plasma efavirenz concentrations in a Chilean HIV cohort. J. Antimicrob. Chemother. 65, 1889–1893 (2010).
Li, J. et al. Worldwide variation in human drug-metabolism enzyme genes CYP2B6 and UGT2B7: implications for HIV/AIDS treatment. Pharmacogenomics 13, 555–570 (2012).
Reay, R., Dandara, C., Viljoen, M. & Rheeders, M. CYP2B6 Haplotype Predicts Efavirenz Plasma Concentration in Black South African HIV-1-Infected Children: A Longitudinal Pediatric Pharmacogenomic Study. OMICS 21, 465–473 (2017).
Rozas, J. et al. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 34, 3299–3302 (2017).
Swart, M. et al. High predictive value of CYP2B6 SNPs for steady-state plasma efavirenz levels in South African HIV/AIDS patients. Pharmacogenet. Genomics 23, 415–427 (2013).
Zanger, U. M. & Schwab, M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138, 103–141 (2013).
Alessandrini, M. & Pepper, M. S. Priority pharmacogenetics for the African continent: focus on CYP450. Pharmacogenomics 15, 385–400 (2014).
Gaedigk, A. et al. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin. Pharmacol. Ther. 83, 234–242 (2008).
Kumondai, M. et al. Genetic Polymorphisms of CYP2A6 in a Case-Control Study on Bladder Cancer in Japanese Smokers. Biol. Pharm. Bull. 39, 84–89 (2016).
Dodgen, T. M. et al. Evaluation of predictive CYP2C19 genotyping assays relative to measured phenotype in a South African cohort. Pharmacogenomics 16, 1343–54 (2015).
Rotger, M. et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet. Genomics 15, 1–5 (2005).
Haas, D. W. et al. Associations between CYP2B6 polymorphisms and pharmacokinetics after a single dose of nevirapine or efavirenz in African americans. J. Infect. Dis. 199, 872–880 (2009).
Facchinetti, S. A. Procedure to Find Exact Critical Values of Kolmogorov-SmirnovTest. Ital. J. Appl. Stat. 21, 337–359 (2009).
Bartlett, M. S. Properties of sufficiency and statistical tests. Proc. R. Stat. Soc. Series A 160, 268–282 (1937).
Acknowledgements
We thank all the children in this study, and their parents and teachers, for their understanding and assistance. Furthermore, we thank all the staff for technical assistance in the blood collection and DNA extraction. We also thank Dr Gaseitsiwe S. from Botswana-Harvard Partnership (Gaborone, Botswana) for sequencing of the control genotypes included the two CC genotypes for 983 T > C polymorphism. This study was supported by Botswana Ministry of Health, the Penn Center for AIDS Research (CFAR), an NIH-funded program [grant # P30 AI 045008] and partly supported through the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative [grant # DEL-15-006]. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [grant # 107752/Z/15/Z] and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust or the UK government.
Author information
Authors and Affiliations
Contributions
G.M.P., I.K.Q. and E.P. conceived the study; L.T., T.M., P.R., N.M., C.W.M. and G.B.D.D. conducted the genetic analysis; G.M.P., A.M. and G.R. analysed the results; G.M.P., L.T. and G.R. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tawe, L., Motshoge, T., Ramatlho, P. et al. Human cytochrome P450 2B6 genetic variability in Botswana: a case of haplotype diversity and convergent phenotypes. Sci Rep 8, 4912 (2018). https://doi.org/10.1038/s41598-018-23350-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-23350-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.