Abstract
There is growing concern about the spreading of human microorganisms in relatively untouched ecosystems such as the Antarctic region. For this reason, three pinniped species (Leptonychotes weddellii, Mirounga leonina and Arctocephalus gazella) from the west coast of the Antartic Peninsula were analysed for the presence of Escherichia spp. with the recovery of 158 E. coli and three E. albertii isolates. From those, 23 harboured different eae variants (α1, β1, β2, ε1, θ1, κ, ο), including a bfpA-positive isolate (O49:H10-A-ST206, eae-k) classified as typical enteropathogenic E. coli. Noteworthy, 62 of the 158 E. coli isolates (39.2%) exhibited the ExPEC status and 27 (17.1%) belonged to sequence types (ST) frequently occurring among urinary/bacteremia ExPEC clones: ST12, ST73, ST95, ST131 and ST141. We found similarities >85% within the PFGE-macrorrestriction profiles of pinniped and human clinic O2:H6-B2-ST141 and O16:H5/O25b:H4-B2-ST131 isolates. The in silico analysis of ST131 Cplx genomes from the three pinnipeds (five O25:H4-ST131/PST43-fimH22-virotype D; one O16:H5-ST131/PST506-fimH41; one O25:H4-ST6252/PST9-fimH22-virotype D1) identified IncF and IncI1 plasmids and revealed high core-genome similarities between pinniped and human isolates (H22 and H41 subclones). This is the first study to demonstrate the worrisome presence of human-associated E. coli clonal groups, including ST131, in Antarctic pinnipeds.
Similar content being viewed by others
Introduction
The Antarctic regions are regarded as protected areas with limited anthropogenic influence. However, there is growing concern about the impact of scientific and tourism activities which increase the risk of spreading human microorganisms in such relatively untouched ecosystems1. Marine mammals are considered as bio-indicators of possible environmental changes2,3 but most studies focused on the effect of untreated sewage discharges next to research stations, with few data about the presence of human-associated microorganisms within these animals4.
Although it is well-known that Escherichia coli is a common member of the intestine microbiota of warm-blooded vertebrates, recovery of E. coli seems to be low in wild marine mammals living in pristine environments when compared to captive animals or those close to locations with high anthropogenic exposure5,6. Escherichia coli is also a major intestinal and extraintestinal pathogen by means of successful combinations of virulence factors that have classically defined specific pathotypes7. Most extraintestinal E. coli infections are caused by isolates from phylogenetic group B2 which includes clinically significant subgroups such as ST73, ST131 or ST958. ST131 has received special attention and much of its success is due to the spread of two fluoroquinolone-resistant subclones known as H30R/C1 and H30Rx/C2, frequently associated with human urinary tract infections and bacteriemia9,10,11. So far, the detection of ST131 in Antarctica has been twice reported from the isolation of water and/or marine sediments next to the research bases4,12.
The aims of the present study were i) to investigate the occurrence and characteristics of Escherichia spp. isolates in pinnipeds from the Antarctic regions ii) to assess the presence of high- risk clones in an ecological reserved region iii) to define possible sources of contamination and impact within this vulnerable ecosystem.
Results
O:H typing, phylogroups, pathotypes and ExPEC status
A total of 161 Escherichia spp. isolates recovered from Antarctic fur seals (80 isolates), Southern elephant seals (62 isolates) and Weddell seals (19 isolates) were phenotypically and molecularly characterized in this study. Of 63 different O:H antigen combinations, 10 accounted for 46.6% of the isolates: O11:H6 (five isolates), O73:HNM (five isolates), O15:H21 (six isolates), O51:H49 (six isolates), O25:H4 (seven isolates), O77:H45 (seven isolates), O120:H1 (seven isolates), ONT:H6 (eight isolates), ONT:H21 (10 isolates), O6:H1 (14 isolates). Phylotyping showed that the majority (84 isolates; 52.2%) belonged to phylogroup B2, followed by D (25; 15.5%), B1 (24; 14.9%), F (11; 6.8%), E (8; 5.0%) and A (6; 3.7%). Finally, three isolates (EC-25, EC-76 and EC-172b) that could not be assigned to any phylogroup were identified as Escherichia albertii (Supplementary Table S1).
The 161 isolates were analyzed by PCR for the presence of specific virulence factors (VF). None was positive for the vt1, vt2, ipaH, pcDV432, eltA, est or stb markers associated with verotoxigenic, enteroinvasive, enteroaggregative or enterotoxigenic pathotypes (Supplementary Table S1). However, 23 of the 161 (14.3%) isolates recovered from the three populations were carriers of the eae gene: Southern elephant seals (11 isolates, 17.7%), Antarctic fur seals (seven, 8.7%) and Weddell seals (five, 26.3%). These eae-positive isolates were identified as enteropathogenic E. coli (EPEC) (20 isolates) and E. albertii (three isolates), which belonged to 12 serotypes and harboured seven eae variants: α1 (six isolates), β1 (one isolate), β2 (two isolates), ε1 (three isolates), κ (seven isolates), θ1 (two isolates), and ο (two isolates). Additionally, the O49:H10 eae-κ isolate (EC-147) from an Antartic fur seal was bfpA-positive and therefore designed as typical enteropathogenic E. coli (tEPEC) (Table 1).
The PCR-screening of 20 extraintestinal VF showed that all 161 isolates carried one to 13 genes (Supplementary Table S1). Furthermore, 62 of the 158 (39.2%) E. coli isolates were positive for two or more of the five markers papC, sfa/focDE, afa/draBC, kpsM II and iutA which define ExPEC status13, with similar prevalence within the pinniped populations (26.3% of Weddell seals, 37.3% of Southern elephant seals and 43.8% of Antarctic fur seals). Of the 62 ExPEC isolates, more than 50% were positive for fimH (100%), sfa/focDE (66.1%), iutA (62.9%), iucD (58.1%), iroN (64.5%), kpsM II (82.3%), traT (71.0%), malX (93.5%) and usp (79.0%) (Supplementary Table S2). Interestingly, one of the 62 ExPEC (EC-88) also harboured the eae gene (Table 1). Significant differences were observed between the 62 ExPEC (VF range of 5–13) and the 96 non-ExPEC isolates (VF range of 1–9) regarding the phylogroup distribution (1.6% vs 24.0% of B1, 85.5% vs 32.3% of B2, 0% vs 26.0% of D, respectively) (P < 0.001 for all comparisons).
STs, virotypes, clonotypes and PFGE macrorrestriction profiles
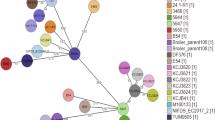
Eighty-five of the 161 isolates were further characterized by MLST and clonotyping including those identified as EPEC and/or ExPEC pathotypes and the three E. albertii. In total, 35 STs were established according to the Achtman scheme14, with 13 STs determined for first time in this study. Seven of those 13 new STs got numerical designations (ST4446, ST4448, ST4449, ST4607, ST4610, ST4611, ST6252) since they were performed before the requirement of WGS for new ST assignments, and the remaining six new allelic combinations were named here as STnew1 to STnew6. The Neighbor-Joining tree based on the concatenated sequences showed the phylogenetic relationship among the STs, consistently distributed in relation to their phylogroup and species (Fig. 1; Supplementary Table S3). The most prevalent STs were ST73 (14 isolates), ST1859 (eight isolates), ST131 (six isolates), ST589 (six isolates) and ST457 (five isolates).
Seven isolates belonged to the sequence type complex 131 (ST131 Cplx) and were further differentiated into three STs according to the Pasteur Institute scheme15 (PSTs): PST43 detected in five O25b:H4-ST131 isolates, PST9 in one O25b:H4-ST6252 and PST506 in one O16:H5-ST131 isolate. Besides, the virotype of these isolates was established based on the scheme of Dahbi et al.16 as virotype D1 (isolate EC-77), virotype D2 (isolates EC-23 and EC-24), virotype D-not typeable (EC-114, EC-115 and EC-160), while the virulence profile of the O16:H5 isolate (EC-152) did not match any of the 12 virotypes included in the scheme (Supplementary Table S1).
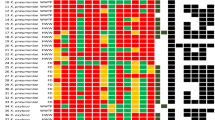
Clonotyping revealed 11 different clonotypes (CH) among the 23 eae-positive isolates, with five new fimH alleles in the analysis of the 489-nucleotide (nt) internal fragment designated in this study as fimH_A to E (Table 1; Supplementary Figure S1). The new allele fimH_A identified in two E. albertii (EC-25 and EC-76) showed 99.39% of identity with fimH424 with three nt changes and one aminoacidic change (threonine for isoleucine). The fimH_B allele identified in one E. coli isolate (EC-38) showed 99.80% of identity with fimH438, with one nt and one aminoacidic change (alanine for threonine). Interestingly, fimH_C, fimH_D and fimH_E were four, three and one nt variants of fimH41, respectively, but only fimH_E gave an aminoacidic change (asparagine for aspartic acid). Further 25 different clonotypes were determined among the ExPEC isolates, whose genetic relatedness and diversity were additionally analyzed by XbaI macrorestriction followed by pulsed-field electrophoresis (PFGE) (Fig. 2). In this comparison, an O16:H5-B2-ST131 isolate negative for the ExPEC status was also included. Thus, the 63 isolates grouped in the dendrogram in relation to their serotype/phylogroup/ST, with nine clusters of identity ≥ 85% (Clusters A to G) including the main clonal groups.
We compared the pinniped isolates of the main B2 clonal groups with E. coli clinical isolates (animal and human) of our collections. As a result, we found high similarities (>85%) (Fig. 3) between the PFGE macrorestriction profiles of O2:H6-B2-ST141 fimH5 (one Southern elephant seal and one human isolates), O25b:H4-B2-ST95 fimH27 (one Antarctic fur seal and one poultry isolates), O25b:H4-B2-ST131 fimH22 (two Southern elephant seal and four human isolates) and O16:H5-B2-ST131 fimH41 (one Southern elephant seal and one human isolates).
Resistance characterization
The antimicrobial susceptibility test using MIC panels including 19 antibiotics determined resistance values in six (3.7%) out of 161 isolates tested to ampicillin (two isolates), cefalotin (three isolates) and nalidixic acid (two isolates). Additionally, intermediate MIC values to ampicillin and cefalotin were detected in one and 14 isolates, respectively. By PCR, the 161 isolates were negative for blaSHV and blaCTX-M, and only two (EC-23 and EC-24) gave positive result for blaTEM whose sequence was identified as TEM-1.
Whole genome sequencing of ST131Cplx isolates
Whole genome sequencing was performed for six of the seven ST131 Cplx isolates obtained from the three pinniped species (EC-23, EC-24 and EC-77 from Southern elephant seals; EC-114 from a Weddell seal; EC-152 and EC-160 from Antarctic fur seals). The draft genomes of the six isolates yielded 63 to 327 contigs larger ≥ 200 bp, with assembly sizes ranging from 4.8 Mb (EC-152) to 5.5 Mb (EC-77) (average of 5.1 Mb). The in silico determination of O/H antigens and STs (7 gene scheme) predicted by EnteroBase (https://enterobase.warwick.ac.uk/) was in agreement with the experimental data, with the exception of adk allele EC-24 that could not be predicted. The resistome of the draft genome, analysed using ResFinder showed the presence of antimicrobial resistance determinants blaTEM-1C and tet(A) in EC-23, EC-24 and EC-77, while PlasmidFinder allowed the detection of plasmid replicons IncF (five isolates) and IncI1 (two isolates) (Table 2).
By genome comparison, we further investigated the relatedness between the six pinniped ST131 Cplx isolates and the four ST131 human clinic isolates with highly similar macrorrestriction profiles shown in Fig. 3, together with the ST131 reference genome EC958, all of them assembled in Enterobase (https://enterobase.warwick.ac.uk/). In the Minimum Spanning Tree of the core genome analysis based on the cgMLST scheme from Enterobase, the pinipped isolates EC-23 and EC-24 showed high genetic relatedness (58 and 147 differences, respectively) compared to human H1698 human isolate (all of subclone H22); likewise, the pinniped isolate EC-152 showed less than100 differences compared to human 60SA isolates (both of suclone H41) (Fig. 4). Five of the six WGS ST131 pinniped isolates carried contigs with IncF plasmid replicons of types F2:A-:B1 (predicted in EC-23 and EC-24) and F18:A-:B20 (predicted in EC-77, EC-114 and EC-160) (Table 2).
Discussion
In this study, we analysed a collection of 161 Escherichia spp. isolates obtained from three Antarctic pinniped species sampled in six locations with different human activity along the west coast of the Antarctic Peninsula. While none was positive for the verotoxigenic, enteroinvasive, enteroaggregative or enterotoxigenic virulence markers, eae-positive isolates were identified from 26.3% of the Weddell seals, 17.7% of Southern elephant seals and 8.7% of the Antarctic fur seals. The EPEC pathotype is characterised by the presence of the locus of the enterocyte effacement (LEE) containing the eae gene and do not produce verotoxins17. EPEC isolates can be further classified into typical (tEPEC) and atypical (aEPEC) EPEC on the basis of the presence (tEPEC) or absence (aEPEC) of the EPEC adherence factor (EAF) plasmid, which harbours the bfp operon encoding the bundle-forming pilus (BFP)17,18. The tEPEC is a major cause of diarrhoeal disease among children in developing countries, being humans the major natural reservoir for tEPEC which has been rarely reported from animals19,20,21, while aEPEC has been widely isolated from both humans and animals21,22. E. albertii is an emerging pathogen identified as a causative agent in human gastroenteritis. E. albertii frequently carries the eae gene causing it to be mistakenly identified as EPEC or Enterohemorragic E. coli (EHEC), and also typically possesses the II/III/V subtype group cdtB gene23.
The 23 eae-positive pinniped isolates (19 aEPEC, one tEPEC and three E. albertii) belonged to 12 serotypes and 11 STs, some of them previously reported in human diarrheagenic aEPEC (namely, serotypes O88:H25 and O51:H49; STs 28, 29, 328 and 589) but also in other sources24,25 (Table 1). O51:H49-B2-ST589 (eae-α1), the most prevalent clonal group within the pinniped EPEC isolates, was also reported in freshwater samples on the English Channel coast26, and the ST589 in aEPEC human diarrheagenic and animal isolates in China24. Of note is the wide host range of clonal group O88:H25-B1-ST328 eae-β1, including humans, lambs and even shellfish24,25,26, determined in the pinniped isolate EC-88 positive for the ExPEC status and for the F1C fimbriae focG gene associated to urinary strains. This heterogenic virulence capability was previously described by Dutta et al.27 in an ST328 aEPEC (eae-β) isolate from a child with acute diarrhea, which also harboured the elt gene in a conjugative plasmid (IncFII) together with the P fimbriae papG and papC genes. In our study, the 23 eae-positive isolates carried extraintestinal VF (range 5–9) conforming to a hybrid pathotype. Likewise, the three pinniped E. albertii isolates not only carried the reported eae and cdtB genes23, but also other associated with ExPEC: fimAv MT78 , iutA, iucD, traT and ibeA. Furthermore, one of the 23 eae-positive isolates (EC-147) was bfpA-positive (tEPEC) and belonged to the clonal group O49:H10-A-ST206 which is periodically detected in patients with diarrhea attending the hospital of our city (Supplementary Figure S2). Recently, Alonso et al.28 first reported this tEPEC clonal group from a wild boar, suggesting the acquisition from human sources. The surprising detection of this tEPEC from a marine mammal would need further investigation to explain its origin.
The ExPEC status was determined in 62 (39.2%) of the 158 E. coli recovered from the three pinniped species within different locations (16 of the 44 E. coli from rarely visited locations, four of the 17 from moderate visited locations, 42 of the 97 from highly visited locations), without significant difference in presence between rarely visited locations vs moderate plus highly visited (36.4% vs 40.3%) (P > 0.05).
ExPEC is a heterogeneous group, defined by isolation from infections outside the intestinal tract since no core set of virulence factors can unambiguously distinguish the ExPEC subgroups. Different researchers tried to elucidate this fact studying the phylogeny of the isolates, virulence traits and host factors29. In our study, the prevalence of certain extraintestinal VF was unexpectedly high and significantly associated to the 62 ExPEC isolates: papC, papG III, sfaS, focG, cnf1, hlyA, iutA, iucD, iroN, kpsM II-K5, neuC (K1), traT, ibeA, malX and usp (P < 0.001 for all comparisons). Many of those were previously correlated with urinary isolates together with the phylogenetic group B230,31. Here, the phylogroup B2 accounted for more than 50% of the Antarctic collection and 85.5% of the 62 ExPEC isolates, including 19 B2 clonal groups repeatedly implicated in human urinary tract infections and bacteremia worldwide8,32,33,34, such as ST73 (14 isolates O6:H1/O2:H1-B2-ST73), ST131 (five isolates O25b:H4-B2-ST131), ST12 (three isolates O4:H5-B2-ST12), ST141 (two isolates O2:H6-B2-ST141), ST95 (two isolates O25b:H4/O2:H7-B2-ST95), that additionally showed clonotypes found within clinical isolates35,36,37 (Fig. 2). Another minor clonal groups from the pinniped collection reported in clinic extraintestinal isolates were: ST363-B2 (CH38–5), ST648-F (CH4-58); ST328-B1 (CH23-31), ST457-F (CH88-145); ST640-B2 (CH147-5)37. Previous studies on commensal E. coli isolated from birds, non-human mammals and humans found that the prevalence of the phylogenetic groups was clearly different with predominance of D/B1, A/B1 and A/B2, respectively. Although the isolates from 323 mammals did not encompassed any from marine animals, the phylogroup B2 was clearly the less prevalent of this group38. Nevertheless, a recent study on 40 E. coli recovered from fruit bats, in areas with very low human population density of the Republic of Congo, reported a B2 prevalence of 46.2%, with detection of EPEC and ExPEC pathotypes. The authors stated that known human isolates might be present in wild animals as part of the naïve E. coli population more frequently as originally assumed39.
Power et al.4 also identified a high prevalence of phylogroup B2 (44.5%) among 353 Antarctic E. coli isolates with presence of STs 12, 73, 95, 127, 141 and 131; however, most were seawater and sediment isolates, and only ST95 was detected in isolates from three Weddell seals. The ST95 is associated with a highly pathogenic group of ExPEC belonging to different serotypes such as O1:H7/HNM, O18:H7, or O45:H7/HNM frequently implicated in neonatal meningitis, urinary tract infections and septicemia in humans, but also in avian pathology40,41. We found two ST95 isolates (O2:H7 and O25b:H4) in this study. It is noteworthy the detection of an O25b:H4-B2-ST95 (fimH27) isolate since this serotype is usually associated to B2-ST131, although it can also be found within other clonal groups such as O25b:H4-D-ST69 first described in raw sewage and river water samples in Barcelona42. Interestingly, we found high similarity (89.9%) between the macrorrestriction/virulence profiles of one pinniped and one chicken O25b:H4-B2-ST95 isolates (Fig. 3).
As far as we know, the ST131 had not been previously determined in isolates from Antarctic animals, even though CTX-M-15-producing ST131 isolates were recovered in 2011 from water samples close to research bases on the Antarctic Peninsula and King George Island12. ST131 is a recently emerged and globally disseminated multidrug-resistant clone associated with human urinary tract and bloodstream infections, but rarely reported among animal ExPEC43. Genomic studies on the ST131 clonal evolution determined that ST131 first acquired genes linked to an increased capacity to cause human infection, and then gained an arsenal of resistance genes to diver in different clades: clade A associated mostly with fimH41, clade B associated with fimH22 and precursor of clade C, which is associated with fimH30. Recently, two subclades were identified within clade C: C1 or H30R (associated with fluoroquinolone-resistance) and C2 or H30-Rx (associated with CTX-M-15). These two subclades are currently recognized as the E. coli lineages most likely to be responsible for human extraintestinal infections44, although wide longitudinal studies in Spain16 and England34 determined that all of the ST131 clades were present during the whole study period, which might indicate that the entire ST131 clonal group is successful and not just subclades C1 and C234. Here, we report the presence of ST131 Cplx in the three pinniped species from different locations, accounting for 4.3% of the 161 isolates. According to their fimH alleles, six isolates were of clade B (H22), one of clade A (H41) and none belonged to the C (H30) clade. The detection of O16:H5-ST131 subclone was especially unexpected since it is globally much less prevalent than O25b:H4-ST131, consistently accounting for approximately 1 to 5% of the E. coli clinical isolates overall16,45. All six fimH22 isolates showed virotype D in agreement with previous reports for this clade16,46.
Importantly, none of the sampled pinnipeds was positive for ESBL, and only six isolates exhibited resistance to ampicillin, cefalotin or nalidixic acid. Resistance within the classical ExPEC such as ST73 or ST95 has been unfrequently detected. However, emerging clonal groups such as ST131 or ST648 are known by their essential role in the global increase of antibiotic resistances34,47,48. Hernández et al.12 first reported ESBL-producing E. coli on the Antarctic region, specifically blaCTX-M-1 and blaCTX-M-15 genotypes (including ST131 isolates). Nevertheless, those clearly human-associated Enterobacteriaceae were not detected among 400 fresh penguin fecal droppings sampled near the bases which could indicate, according to the authors, infrequent interactions between penguins and humans or a less opportunistic diet compared with other feeders such as wild birds49.
Epidemic resistance plasmids belonging to IncF with divergent replicon types (such as FIA, FIB, and FII) have the ability to acquire resistance genes and then rapidly disseminate among Enterobacteriaceae50. Moreover, it has been postulated that the F-type plasmids shaped the evolution of the H30 subclone and specifically, an F1:A2:B20 plasmid would be strongly associated with the H30R/C1 clade, and an F2:A1:B- plasmid with the H30Rx/C2 clade51. Although the F2:A1 allele combination is dominating among H30Rx isolates, it also occurs among H22 and H41, and the F1:A2:B20 allele among H30Rx and H41 isolates. However, F18, F29, B1 and B10 were the most common replicon alleles among ancestral isolates according to the findings of Johnson et al.51. As a consequence, the IncF-type plasmids determined in the pinniped ST131 isolates might derived from ancestral lineages.
Conclusions
The present study provides evidence for Antarctic marine mammals carrying potentially pathogenic E. coli pathotypes which are frequently involved in enteropathogenic and extraintestinal human diseases. We report for first time the presence of ST131 Cplx recovered from three pinniped species sampled in different locations. The phylogenetic distribution, virulence profiles, STs and genomic traits would indicate possible ancestral sublineages of current clinic clonal groups. The challenge now is to answer how and when these isolates became part of the commensal microbiota of these marine mammals. A possible explanation might be geographic mobility of these animals, contacting areas of higher human activity. Mirounga leonine is widely distributed in Southern hemisphere, with specimens found in the Antarctic Peninsula and continent, including Southern Argentina. Leptonychotes weddellii have a more circumpolar distribution, making foraging trips up north with expanding ice pack during the winter, and with some wandering as far as New Zealand, South Australia and South America. Finally, Arctocephalus gazella usually wander in non-breeding season to Weddell Sea, Argentina coast, and some groups have been reported in Juan Fernandez Island (Chile)52,53. Surveillance studies are needed to know the evolution of those EPEC/ExPEC clonal groups within the normal pinniped microbiota as well as their possible transmission to other animals within the Antarctic niches.
Material and Methods
Escherichia spp. collection
A total of 193 faecal samples from three Antarctic pinniped populations were collected during the month of February of years 2007, 2010 and 2011 in six locations with different human activity, from low to highly visited, along the west coast of the Antarctic Peninsula: Deception Island, King George Island, Livingston Island (Hannah Point and Byers Peninsula), Ronge Island and Avian Island in a latitudinal gradient covering 5° of latitude (ranging from 62°15′S; 58°37′W-67°46′S; 68°43′W), distances greater than 600 km and differences in mean annual temperatures of up to 2 °C. The populations sampled included individuals with several years of age (adults or subadults from both sexes) of Weddell seals (Leptonychotes weddellii), Southern elephant seals (Mirounga leonina) and Antarctic fur seals (Arctocephalus gazella). Samples consisted of fresh faecal samples obtained directly from the ground close to animals or swab samples from the rectum of animals randomly selected, captured and physically restrained. All captured animals were tagged to ensure that no animal was sampled more than once. Faecal samples were preserved in FBP medium54 with 0.5% active charcoal (Sigma Ltd.) and frozen at −20 °C until analysis in the lab. For the analysis, each sample was thawed and placed in 5 ml of buffered peptone water broth (Oxoid). After incubation (37 °C for 18 h), an aliquot of 100 µl was plated onto MacConkey agar (Oxoid) and plates incubated (37 °C for 24 h). One suspected E. coli colony per sample was randomly selected and identified by classical biochemical methods (gram-staining, triple sugar iron, citrate, urea, indol). Confirmed colonies were stored at −80 °C until their subsequent characterization.
Finally, a collection of 161 Escherichia spp. isolates from 160 pinnipeds (80 Antarctic fur seals, 61 Sourthern elephant seals and 19 Weddell seals; 94 faeces and 66 rectal swabs) was analysed, including two different colonies from a rectal swab of a Sourthern elephant seal (Supplementary Table S1). Human and other animal isolates from the own LREC-USC collections were used for comparative purposes.
Typing, subtyping and phylogenetic grouping of the isolates
The isolates were characterized with regard to O:H antigens with all available O (O1–O181) and H (H1–H56) antisera55. Isolates that did not react with any antisera were designated as ONT or HNT (NT = nontypeable). Nonmotile isolates or cross-reactions were further tested by PCR using specific primers (Supplementary Table S4). The phylogenetic groups were determined by the quadruplex method of Clermont56 and those isolates that did not conform to any E. coli phylogroup were further investigated for the cryptic Escherichia lineages57 and E. albertii58.
All isolates were screened by PCR for the presence of the VF eae, vt1, vt2, ipaH, pcDV432, eltA, est or stb associated with the main intestinal pathotypes (enteropathogenic, verotoxigenic, enteroinvasive, enteroaggregative and enterotoxigenic) of E. coli. Likewise, specific extraintestinal VF were analyzed: fimH, fimAv MT78 , papC, (positive results tested for papG I, papG II and papG III), sfa/focDE (positive results tested for sfaS and focG), afa/draBC, cnf1, cdtB, sat, hlyA, iucD, iroN, kpsM II (establishing neuC-K1, K2 and K5 variants), kpsM III, cvaC, iss, traT, ibeA, malX, usp and tsh. E. coli isolates were designated presumptively as extraintestinal pathogenic E. coli (ExPEC status)13 if positive for two or more of five markers: papC, sfa/focDE, afa/draBC, kpsM II, and iutA (Supplementary Table S5).
Isolates identified as ExPEC and/or eae carriers were further typed for their fimH alleles (type 1 fimbrial adhesion) with the amplification and sequencing of a 489-nucleotide internal fragment as described elsewhere36, which were compared with those available in the Genomic Epidemiology repository https://bitbucket.org/genomicepidemiology/fimtyper_db/downloads/. The eae variant was established by amplification and sequencing of the variable region of the gene (Supplementary Table S5). Furthermore, ExPEC and eae-positive isolates were analyzed for their STs following the MLST schemes of Achtman14 (http://enterobase.warwick.ac.uk/species/ecoli/allele_st_search) and additionally, for those ST131, of the Pasteur Institute15 (http://bigsdb.pasteur.fr/ecoli/ecoli.html). To analyze the phylogenetic relationships among the isolates, a Neighbor-Joining tree was constructed by MEGA659 based on the concatenated sequences of the seven housekeeping genes for each isolate. Also, the virotype of the ST131 isolates was established based on the presence or absence of 13 specific VF according to the scheme described by Dahbi et al.16.
The XbaI-PFGE profiles of the ExPEC isolates were determined according to PulseNet protocol (http://www.pulsenetinternational.org/), imported into BioNumerics (Applied Maths, St-Martens-Latern Belgium) and clustered using the UPGMA algorithm and applying Dice coefficient and 1% tolerance.
Antimicrobial susceptibility testing and resistance genes
Antimicrobial susceptibility was tested using the commercial broth microdilution minimum inhibitory concentration (MIC) panels for gram negatives in a MicroScan WalkAway automated system (Siemens Healthcare Diagnostics, CA, USA) according to the manufacturer’s instructions. The antibiotics tested included ampicillin, cefazolin, cefalotin, cefuroxime, cefotaxime, ceftazidime, cefepime, amoxicillin-clavulanic acid, piperacillin-tazobactam, cefoxitin, imipenem, gentamicin, tobramycin, nalidixic acid, norfloxacin, ciprofloxacin, trimethoprim-sulfamethoxazole, nitrofurantoin and fosfomycin. The MICs were interpreted according to standard break points60. Additionally, detection of TEM, SHV and CTX-M genes was performed by PCR using specific primers (Supplementary Table S6).
Genome sequencing, assembly and analysis
DNA from ST131 Cplx isolates was extracted with the QIAamp 96 DNA Qiacube HT kit (Qiagen, Hilden, Germany) and libraries were prepared using the Nextera XT kit (Illumina). Pooled libraries were denatured following the Illumina protocol and 600 µl (approx. 20 pM) onto a MisSeq V2 -500 cycle cartridge (Illumina) and sequenced on a MiSeq to produces fastq files. Raw reads were uploaded and automatically assembled in Enterobase using SPAdes Genome Assembler v 3.5. with a contig threshold of minimum 200 nucleotides. Subsequently, the de novo assembled contigs were MLST (7 gene Achtman ST scheme, whole genome MLST and core genome MLST) and serotyped in silico using Enterobase typing tools (https://bitbucket.org/enterobase/enterobase-web/wiki/Home). A minimum spanning tree was constructed in Enterobase using the MSTree V2 algorithm61 and the cgMLST v1 scheme. This scheme consists of 2,513 genes that were present in over 98% of 3,457 genomes, which represented most of the diversity in Enterobase at the time (May 2016) https://bitbucket.org/enterobase/enterobase-web/wiki/Escherichia%20Statistics. Antimicrobial resistances, plasmid detection and typing were in silico determined by using ResFinder V2.162 and PlasmidFinder 1.3./pMLST 1.4.63, respectively.
Statistical analysis
Differences in the prevalence between groups were compared by a two-tailed Fisher’s exact test. P values < 0.05 were considered statistically significant.
Ethics statement
Permissions for captures and sampling of the Antartic pinnipeds were granted by the Spanish Polar Committee CPE- EIA-2006-2 and CPE-EIA-2008-9, complying with the Antarctic Treaty System.
Data availability
The whole genome sequenced samples are part of BioProject PRJEB19190 and correspond to BioSample IDs SAMEA92068168, SAMEA92071168, SAMEA92071918, SAMEA92086168, SAMEA92089168, SAMEA92092918, SAMEA92095168, SAMEA92096668, SAMEA92097418, SAMEA92099668.
References
Hernandez, J. & Gonzalez-Acuna, D. Anthropogenic antibiotic resistance genes mobilization to the polar regions. Infect. Ecol. Epidemiol. 6, 32112 (2016).
Rengifo-Herrera, C. et al. Detection of a novel genotype of Cryptosporidium in Antarctic pinnipeds. Vet. Parasitol. 191, 112–118 (2013).
Wallace, C. C., Yund, P. O., Ford, T. E., Matassa, K. A. & Bass, A. L. Increase in antimicrobial resistance in bacteria isolated from stranded marine mammals of the Northwest Atlantic. Ecohealth 10, 201–210 (2013).
Power, M. L. et al. Escherichia coli out in the cold: Dissemination of human-derived bacteria into the Antarctic microbiome. Environ. Pollut. 215, 58–65 (2016).
Delport, T. C., Harcourt, R. G., Beaumont, L. J., Webster, K. N. & Power, M. L. Molecular detection of antibiotic-resistance determinants in Escherichia coli isolated from the endangered Australian sea lion (Neophoca cinerea). J. Wildl. Dis. 51, 555–563 (2015).
Stoddard, R. A. et al. The effect of rehabilitation of northern elephant seals (Mirounga angustirostris) on antimicrobial resistance of commensal Escherichia coli. Vet. Microbiol. 133, 264–271 (2009).
Kaper, J. B., Nataro, J. P. & Mobley, H. L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140 (2004).
Clermont, O., Christenson, J. K., Daubie, A. S., Gordon, D. M. & Denamur, E. Development of an allele-specific PCR for Escherichia coli B2 sub-typing, a rapid and easy to perform substitute of multilocus sequence typing. J. Microbiol. Methods 101, 24–27 (2014).
Nicolas-Chanoine, M. H. et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61, 273–281 (2008).
Petty, N. K. et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. USA 111, 5694–5699 (2014).
Stoesser, N. et al. Evolutionary History of the Global Emergence of the Escherichia coli Epidemic Clone ST131. MBio 7, e02162 (2016).
Hernandez, J. et al. Human-associated extended-spectrum beta-lactamase in the Antarctic. Appl. Environ. Microbiol. 78, 2056–2058 (2012).
Johnson, J. R. et al. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47, 2161–2168 (2003).
Wirth, T. et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151 (2006).
Clermont, O. et al. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 11, 654–662 (2011).
Dahbi, G. et al. Molecular epidemiology and virulence of Escherichia coli O16:H5-ST131: Comparison with H30 and H30-Rx subclones of O25b:H4-ST131. Int. J. Med. Microbiol. 304, 1247–1257 (2014).
Trabulsi, L. R., Keller, R. & Tardelli Gomes, T. A. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8, 508–513 (2002).
Nataro, J. P. & Kaper, J. B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11, 142–201 (1998).
Ishii, S., Meyer, K. P. & Sadowsky, M. J. Relationship between phylogenetic groups, genotypic clusters, and virulence gene profiles of Escherichia coli strains from diverse human and animal sources. Appl. Environ. Microbiol. 73, 5703–5710 (2007).
Chandran, A. & Mazumder, A. Prevalence of diarrhea-associated virulence genes and genetic diversity in Escherichia coli isolates from fecal material of various animal hosts. Appl. Environ. Microbiol. 79, 7371–7380 (2013).
Krause, G., Zimmermann, S. & Beutin, L. Investigation of domestic animals and pets as a reservoir for intimin- (eae) gene positive Escherichia coli types. Vet. Microbiol. 106, 87–95 (2005).
Moura, R. A. et al. Clonal relationship among atypical enteropathogenic Escherichia coli strains isolated from different animal species and humans. Appl. Environ. Microbiol. 75, 7399–7408 (2009).
Ooka, T. et al. Clinical significance of Escherichia albertii. Emerg. Infect. Dis. 18, 488–492 (2012).
Xu, Y. et al. High prevalence of virulence genes in specific genotypes of atypical Enteropathogenic Escherichia coli. Front. Cell Infect. Microbiol. 7, 109 (2017).
Martins, F. H. et al. Lambs are an important source of atypical enteropathogenic Escherichia coli in southern Brazil. Vet. Microbiol. 196, 72–77 (2016).
Baliere, C., Rince, A., Delannoy, S., Fach, P. & Gourmelon, M. Molecular profiling of Shiga toxin-producing Escherichia coli and Enteropathogenic E. coli strains isolated from French coastal environments. Appl. Environ. Microbiol. 82, 3913–3927 (2016).
Dutta, S., Pazhani, G. P., Nataro, J. P. & Ramamurthy, T. Heterogenic virulence in a diarrheagenic Escherichia coli: evidence for an EPEC expressing heat-labile toxin of ETEC. Int. J. Med. Microbiol. 305, 47–54 (2015).
Alonso, C. A. et al. Occurrence and characterization of stx and/or eae-positive Escherichia coli isolated from wildlife, including a typical EPEC strain from a wild boar. Vet. Microbiol. 207, 69–73 (2017).
Kohler, C. D. & Dobrindt, U. What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol. 301, 642–647 (2011).
Safi, M. et al. Distribution of virulence associated traits among urine Escherichia coli isolates from patients in onco-hematology. J. Infect. Chemother. 22, 221–224 (2016).
Johnson, J. R. et al. Host Characteristics and Bacterial Traits Predict Experimental Virulence for Escherichia coli Bloodstream Isolates From Patients With Urosepsis. Open Forum Infect. Dis. 2 (2015).
Alhashash, F., Weston, V., Diggle, M. & McNally, A. Multidrug-resistant Escherichia coli bacteremia. Emerg. Infect. Dis. 19, 1699–1701 (2013).
Bengtsson, S., Naseer, U., Sundsfjord, A., Kahlmeter, G. & Sundqvist, M. Sequence types and plasmid carriage of uropathogenic Escherichia coli devoid of phenotypically detectable resistance. J. Antimicrob. Chemother. 67, 69–73 (2012).
Kallonen, T. et al. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res. 27, 1437–1449 (2017).
Tchesnokova, V. et al. Predictive diagnostics for Escherichia coli infections based on the clonal association of antimicrobial resistance and clinical outcome. J. Clin. Microbiol. 51, 2991–2999 (2013).
Weissman, S. J. et al. High-resolution two-locus clonal typing of extraintestinal pathogenic. Escherichia coli. Appl. Environ. Microbiol. 78, 1353–1360 (2012).
Matsumura, Y. et al. Population structure of Japanese extraintestinal pathogenic Escherichia coli and its relationship with antimicrobial resistance. J. Antimicrob. Chemother. 72, 1040–1049 (2017).
Escobar-Paramo, P. et al. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 8, 1975–1984 (2006).
Nowak, K. et al. Highly diverse and antimicrobial susceptible Escherichia coli display a naïve bacterial population in fruit bats from the Republic of Congo. Plos One 12, e0178146 (2017).
Mora, A. et al. Extraintestinal pathogenic Escherichia coli O1:K1:H7/NM from human and avian origin: detection of clonal groups B2 ST95 and D ST59 with different host distribution. BMC Microbiol. 9, 132 (2009).
Mora, A. et al. Poultry as reservoir for extraintestinal pathogenic Escherichia coli O45:K1:H7-B2-ST95 in humans. Vet. Microbiol. 167, 506–512 (2013).
Colomer-Lluch, M. et al. Detection of quinolone-resistant Escherichia coli isolates belonging to clonal groups O25b:H4-B2-ST131 and O25b:H4-D-ST69 in raw sewage and river water in Barcelona, Spain. J. Antimicrob. Chemother. 68, 758–65 (2013).
Nicolas-Chanoine, M. H., Bertrand, X. & Madec, J. Y. Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574 (2014).
Ben Zakour, N. L. et al. Sequential Acquisition of Virulence and Fluoroquinolone Resistance Has Shaped the Evolution of Escherichia coli ST131. MBio 7, e00347–00316 (2016).
Johnson, J. R. et al. Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131. J. Clin. Microbiol. 52, 1358–1365 (2014).
Mora, A. et al. Virulence patterns in a murine sepsis model of ST131 Escherichia coli clinical isolates belonging to serotypes O25b:H4 and O16:H5 are associated to specific virotypes. Plos One 9, e87025 (2014).
Horner, C. et al. Escherichia coli bacteraemia: 2 years of prospective regional surveillance (2010–12). J. Antimicrob. Chemother. 69, 91–100 (2014).
Johnson, J. R., Johnston, B. D. & Gordon, D. M. Rapid and specific detection of the Escherichia coli sequence type 648 Complex within phylogroup F. J. Clin. Microbiol. 55, 1116–1121 (2017).
Wang, J. et al. The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zool. Res. 38, 55–80 (2017).
Carattoli, A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238 (2009).
Johnson, T. J. et al. Separate F-type plasmids have shaped the evolution of the H30 subclone of Escherichia coli sequence type 131. mSphere 1 (2016).
Rengifo-Herrera, C. et al. Helminth parasites found in faecal samples of phocids from the Antarctic Peninsula. Polar Biol. 37, 685–695 (2014).
Shirihai, H. A complete guide to Antarctic wildlife: the birds and marine mammals of the Antarctic continent and the Southern Ocean. (Alula Press Oy, Degerby, 2002).
Gorman, R. & Adley, C. C. An evaluation of five preservation techniques and conventional freezing temperatures of -20 degrees C and -85 degrees C for long-term preservation of Campylobacter jejuni. Lett. Appl. Microbiol. 38, 306–310 (2004).
Guinée, P. A. M., Jansen, W. H., Wadström, T. & Sellwood, R. In Laboratory Diagnosis in Neonatal Calf and Pig Diarrhoea: Current Topics in Veterinary and Animal Science (eds Leeww, P.W. & Guinée, P.A.M.) 126–162 (Martinus Nijhoff Publisher, 1981).
Clermont, O., Christenson, J. K., Denamur, E. & Gordon, M. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Reports. 5, 58–65 (2013).
Clermont, O., Gordon, D. M., Brisse, S., Walk, S. T. & Denamur, E. Characterization of the cryptic Escherichia lineages: rapid identification and prevalence. Environ. Microbiol. 13, 2468–2477 (2011).
Smati, M. et al. Quantitative analysis of commensal Escherichia coli populations reveals host-specific enterotypes at the intra-species level. Microbiologyopen 4, 604–615 (2015).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
CLSI. Vol. 26 ed. CLSI supplement M100S (Clinical and Laboratory Standards Institute, Wayne, PA, 2016).
Zhou, Z., et al. 2017. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. bioRxiv 216788 (2017).
Zankari, E. et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644 (2012).
Carattoli, A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903 (2014).
Acknowledgements
The sampling was funded by the Spanish Ministry of Science and Innovation (CGL-2005-25073-E/ANT and CTM2008-00570) and the Sea World & Bush Gardens Conservation Fund. Work at USC-LREC was supported by projects AGL2013-47852-R from the Ministerio de Economía y Competitividad (MINECO, Spain) and Fondo Europeo de Desarrollo Regional (FEDER); AGL2016-79343-R from the Agencia Estatal de Investigación (AEI, Spain) and FEDER; PI16/01477 from Plan Estatal de I + D + I 2013-2016, Instituto de Salud Carlos III (ISCIII), Subdirección General de Evaluación y Fomento de la Investigación, and FEDER; CN2012/303 and ED431C 2017/57 from the Consellería de Cultura, Educación e Ordenación Universitaria, (Xunta de Galicia) and FEDER. We would like to thank the military personnel at the Spanish Antarctic Base “Gabriel de Castilla” for their help and assistance and the Marine Technology Unit (CSIC), the Spanish Navy’s Oceanographic Research Ship “Las Palmas” and the Unidad de Tecnología Marítima (UTM, CSIC) for logistics and transport. We also express our gratitude to J. Castro-Urda, F.T. García-Moreno, C. Rengifo-Herrera, S. Rojo-Montejo, I. Ferre, V. Navarro, M. Gómez-Bautista, T. Alvaro-Alvarez and J. Coello-Pérez for their invaluable help in the sample collection. A. Mora acknowledges the Ministerio de Educación, Cultura y Deporte (Spain) for the mobility grant for teachers and researchers from the Programa Estatal de Promoción del Talento y su Empleabilidad, Plan Estatal de Investigación Científica y Técnica y de Innovación 2013–2016.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: J.B., L.M.O., S.P., D.G., F.J.G., and A.M.; performed the experiments: F.J.G., M.P.A., S.P., C.L., S.V., G.D., J.M., J.M.S. and V.G.; analysed and interpreted the data: A.M., J.B., M.P.A., and L.M.O.; drafted the manuscript: A.M., F.J.G, M.J.S, L.M.O and J.B.; provided critical input and approved the final manuscript: A.M., F.J.G., M.P.A., S.P., L.M.O., D.G., C.L., S.V., G.D., J.M., M.J.S. and J.B.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mora, A., García-Peña, F.J., Alonso, M.P. et al. Impact of human-associated Escherichia coli clonal groups in Antarctic pinnipeds: presence of ST73, ST95, ST141 and ST131. Sci Rep 8, 4678 (2018). https://doi.org/10.1038/s41598-018-22943-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-22943-0
This article is cited by
-
Antimicrobial resistance in Antarctica: is it still a pristine environment?
Microbiome (2022)
-
The population genetics of pathogenic Escherichia coli
Nature Reviews Microbiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.