Abstract
Malaria is among the major threats to global health with the main burden of disease being in rural areas of developing countries where accurate diagnosis based on non-invasive samples is in high demand. We here present a novel molecular assay for detection of malaria parasites based on technology that may be adapted for low-resource settings. Moreover, we demonstrate the exploitation of this assay for detection of malaria in saliva. The setup relies on pump-free microfluidics enabled extraction combined with a DNA sensor substrate that is converted to a single-stranded DNA circle specifically by topoisomerase I expressed by the malaria causing Plasmodium parasite. Subsequent rolling circle amplification of the generated DNA circle in the presence of biotin conjugated deoxynucleotides resulted in long tandem repeat products that was visualized colorimetrically upon binding of horse radish peroxidase (HRP) and addition of 3,3′,5,5′-Tetramethylbenzidine that was converted to a blue colored product by HRP. The assay was directly quantitative, specific for Plasmodium parasites, and allowed detection of Plasmodium infection in a single drop of saliva from 35 out of 35 infected individuals tested. The results could be determined directly by the naked eye and documented by quantifying the color intensity using a standard paper scanner.
Similar content being viewed by others
Introduction
Malaria is a widespread tropical disease causing approximately half a million deaths worldwide. The major burden of the disease is in rural areas of developing countries with limited access to modern clinics with sophisticated equipment and often with limited electricity supply1. Reliable diagnosis under these conditions remain a challenge. The emerging DNA sensors or sensor systems based on DNA for highly sensitive detection of biomarkers present attractive potential solutions to this challenge. Such developments are spurred by the recent advances in chemical synthesis of modified DNA oligonucleotides. As a result of such advances a large number of DNA sensors or sensor systems for the detection of different biomarkers including small molecules2,3,4,5, proteins6,7, or enzyme activities7,8,9,10,11,12,13,14,15 have been reported. Some of the DNA based sensors have been integrated with complex nano-structures16,17,18,19 while others have been used as stand-alone units3,20. Common for most sensors is that they are composed of relatively simple structures, exemplified by single- or double-stranded DNA structures containing Förster resonance energy transfer (FRET) pairs or DNA sequences that alternate between different DNA conformations upon external stimuli3,20,21.
DNA is an excellent material for highly sensitive detection systems due to the ease by which it can be amplified by easily accessible DNA polymerases20,22,23. One example of the amplification methods utilized in relation to DNA sensor systems is the rolling circle amplification (RCA) reaction catalyzed by e.g. phi29 polymerase or other polymerases with high levels of processivity combined with high strand-displacement activity22,24,25. As implied by the name, RCA generates multiple copies of circular DNA and has traditionally been used for the detection of specific nucleotide sequences26,27,28 that facilitate circularization of specific DNA molecules. More recently RCA-based techniques were also demonstrated successful for detection of small molecules15,29,30,31,32 or signals generated from enzymatic DNA cleavage-ligation reactions12,33,34,35,36,37,38,39. RCA is an isothermal reaction that utilizes a circular DNA template to generate long tandem repeat DNA products. These can be detected at the single–molecule level by a variety of visualization techniques, including cryo-transmission electron microscopy40,41, atomic force microscopy,42,43 or fluorescence microscopy after incorporation of fluorescent labeled nucleotides or hybridization with fluorescently labeled probes15,34,36,44,45,46. Hence, DNA sensor systems combined with RCA offer the dual advantages of being directly quantitative and highly sensitive, since each target molecule that generates a DNA circle results in a detectable product. When detecting enzyme activities the sensitivity is even further increased by the fact that each enzyme per definition can generate many DNA circle products, which can each be detected at the single molecule level, without being consumed in the process.
Combined with a specialized microfluidics based extraction system RCA enhanced enzyme activity detection (in short REEAD) was previously exploited for highly sensitive detection of the malaria causing Plasmodium parasites34. This was achieved using a DNA sensor module that was circularized specifically by the DNA cleavage-ligation activity of the Plasmodium specific enzyme, topoisomerase I (pTopI). Subsequently, the generated DNA circles where amplified by RCA and the products hybridized with fluorescent labeled probes before they where visualized at the single molecule level in a fluorescence microscope resulting in a detection limit of 0.06 parasites/μL using only a single drop of blood34. This sensitivity of pTopI specific REEAD was superior to the already developed gold standard diagnostic methods47,48 and better than most of the reported PCR protocols49 These findings suggest that the REEAD technology has the potential for being further developed into a highly sensitive diagnostic test. On top of that, the assay allowed detection of all Plasmodium species causing human malaria34. However, although the core of the assay, i.e. the pTopI mediated DNA circle generation and the subsequent RCA, did not require any thermal cycling or other sophisticated equipment the pump-driven microfluidics based extraction and microscopic readout both represented high technological and equipment heavy methods. Hence, for practical use in rural areas of developing countries where malaria is predominant, the previously described assay34 setup would be hampered by the limitations imposed by extraction and readout. These steps relied on electrical pumps and a fluorescence microscope, respectively. Circumventing such obstacles, we here report the successful replacement of i) the pump driven microfluidics device with a pump-free system, and ii) the microscopic readout with a direct visible colorimetric readout based on horse radish peroxidase (HRP) activity. These developments represent two important steps towards a novel point-of-care diagnostic method.
Due to the lifecycle of the malaria causing Plasmodium parasites, which involves a red blood cell stage, blood is the preferred sample type for diagnosis of malaria using gold standard thick- or thin smear microscopy50, PCR49 or rapid diagnostic tests (RDTs)51,52. However, we and others have reported the presence of trace amounts of Plasmodium derived nucleotide sequences53,54,55,56 and proteins34,57,58 in saliva from infected individuals although it is unlikely that viable Plasmodium parasites may be present in this specimen. In line with the high sensitivity of the described pTopI based assay we here demonstrate the successful detection of active Plasmodium enzymes in the saliva from 35 out of 35 malaria positive individuals. The possibility of using a noninvasive sample type such as saliva as test material for the pTopI based assay holds great promise for the future, since the REEAD technology, once fully developed, may be used for diagnosis in areas with cultural reluctance of giving blood as well as for eradication programs, which may involve testing of large number of asymptomatic individuals.
Results
Detection of Plasmodium infection using Saliva as the Test Material
The principle of the pTopI specific REEAD assay, which measures the activity of the essential Plasmodium expressed pTopI enzyme, and the combination of this assay with droplet microfluidic assisted sample extraction (REEAD-on-a-chip) has previously been described for specific and highly sensitive detection of Plasmodium parasites in blood samples from malaria positive individuals34. Furthermore, testing of a few saliva samples indicated the presence of active pTopI also in this sample type34.
As illustrated in Fig. 1, the core of the REEAD assay is composed of a hairpin shaped DNA substrate, named S1 in the following (Table 1) that is converted to a closed DNA circle by the cleavage-ligation activity of pTopI. After hybridization to a specific primer the generated DNA circle functions as a template for a RCA reaction. This reaction generates a long tandem repeat product that can be visualized at the single molecule level by hybridization with fluorescently labeled probes with a nucleotide sequence complementary to a specific sequence on the RCA product. Subsequently, the individual RCA products, which each corresponds to a single cleavage-ligation reaction mediated by pTopI, can be visualized at the single molecule level under a fluorescence microscope. Consistent with the assay involving no thermal cycling it was demonstrated to be directly quantitative when measuring the number of signals obtained from serial diluted blood samples34.
Schematic depiction of the REEAD assay. (A) I) The DNA-substrate (S1) carrying a pTopI cleavage site (red arrow). II) pTopI (represented by a blue oval labeled pT) mediated cleavage leads to covalent attachment of the enzyme to the DNA-substrate. This covalent intermediate is denoted the cleavage complex. In a subsequent pTopI mediated ligation step, III), a covalently closed DNA circle is formed and pTopI is released from the DNA and therefore able to catalyze a new cleavage/ligation event. IV) The circular DNA product is hybridized to an immobilized RCA primer, which is elongated in a RCA reaction. V) Left panel: The tandem repeat RCA product is detected using fluorescent probes (green) and fluorescence microscopy. V) Right panel: A typical microscopic view obtained performing the REEAD assay.
The use of saliva as a test material for malaria diagnosis or detection of Plasmodium parasites in asymptomatic individuals will be highly valuable in areas with cultural reluctance of giving blood or for eradication programs. Therefore, in the current study we have addressed the potential of using pTopI specific REEAD for sensitive and specific detection of Plasmodium in saliva using a sample set of 52, including 23 frozen saliva samples from malaria positive individuals (Table 2, patient #1, #3–6, #8–25) and 29 frozen saliva samples from malaria negative individuals. The saliva samples collected from infected individuals were matched with corresponding blood samples taken from the same individuals at the same time. The blood samples were also tested by REEAD as well as by either RDT or, whenever possible, using standard thick smear microscopy, which allows estimation of the parasite concentrations. Information about the samples including their storage conditions and test results is summarized in Table 2. The negative samples were collected from uninfected individuals (including West Africans) residing in Denmark. Since it was relatively easy to obtain non-invasive samples it was possible to include more saliva (N = 29) than blood (N = 22) samples from uninfected individuals in the test. Matched saliva and blood samples from infected individuals were collected in Gabon. However, one of the blood samples was destroyed by coagulation and, therefore, 23 saliva and 22 blood samples from infected individuals were included in the test.
All samples were tested using REEAD and the results analyzed in a fluorescence microscope essentially as described previously34. An X-Y scatter plot of the individual test results is shown in Supplementary Figure S1. Only sporadic signals were observed in the 29 saliva or 22 blood samples from malaria negative individuals. In comparison, testing of saliva or blood from malaria positive individuals in all cases resulted in signals that were at least two standard deviations above the mean of the signals obtained from analyses of malaria negative samples. The discriminatory power of the assay was highlighted by the fact that the medians of the results obtained from the group of samples from uninfected individuals were significantly different (p value < 0.0001) from that of the positive samples when analyzed using a Mann-Whitney U test regardless whether saliva (see box plot in Fig. 2A) or blood (box plot shown in Fig. 2B) was used as the test material. This result clearly demonstrates the ability of the REEAD assay to detect the presence of Plasmodium in saliva from malaria patients and verifies the presence of active pTopI in the saliva from infected individuals. Plotting the results obtained from each saliva sample as a function of parasite concentrations (measured in the matching blood samples), however, demonstrated that the number of REEAD signals observed did not correlate to the parasitemia of the tested individuals (Figure S1). The REEAD assay was directly quantitative when analyzing dilutions of the same samples (see previous published results34 and Fig. 2C). Hence, the observed lack of correlation between the number of REEAD signals and parasitemia when analyzing different saliva (or the matching blood) samples (Figure S1) may best be explained by the samples being collected at different times and treated differently in the time interval between sampling and testing by REEAD. Due to the long distance between sample collection and place of analyses in the current study, collection time and the precise storage conditions from collection to testing were very difficult to control and concerns in relation to these issues remain a subject for future investigations.
Detection of pTopI in patient samples. (A) Boxplot representing the results of testing 29 saliva samples from uninfected individuals and 23 samples from Plasmodium infected individuals (Table 2, patient #1, #3–6, #8–25) using the REEAD assay depicted in Fig. 1. (B) Boxplot representing the results of testing 22 blood samples from uninfected individuals and 22 samples from Plasmodium infected individuals (Table 2, #1, #3–5, #8–25) using the REEAD assay depicted in Fig. 1. (C) Boxplot representing the results of testing saliva diluted in such a way that blood from the same individual diluted in the same way would have a Plasmodium parasite concentration of 0.2 parasite/µL (Pool 0.2, N = 7); or 2 parasites/µL (Pool 2, N = 10). As a control saliva from 12 uninfected individuals was tested (Pool 0). NB: a base 2 logarithmic scale is used for the y-axis. In all cases the results are shown as fold increase above the average number of signals per microscopic frame obtained from analysis of samples from uninfected individuals.
The detection limit of REEAD using blood as a test material was previously demonstrated to be as low as 0.06 parasites/µL34 simply by testing serial dilutions of blood with a known parasite concentration. Determination of the detection limit in saliva is more challenging since currently there is no standard method for quantification of parasite numbers in saliva samples. Therefore, only a rough estimation of the detection limit in saliva could be obtained. This was done by using the known parasite number in the matching blood samples as a reference for dilution of randomly picked saliva samples. Note, however, that this approach involved a number of uncertainties, not least due to the different quality of the available samples. In the experimental setup saliva samples from 7–10 individuals were each diluted with a factor equal to the dilution factor necessary to obtain a parasite concentration of 2 (referred to as pool 2 in the following) or 0.2 (referred to as pool 0.2 in the following) parasites/μL in the matching blood samples (termed blood equivalent parasite concentration (Table 2)) as described in materials and methods.
The results are shown as a box plot in Fig. 2C. Analyses of all samples (patients #1–3, #5, #7, #11, #14, #16, #18, #23; Table 2) (N = 10) in pool 2 resulted in a number of signals above the number of signals obtained when analyzing the uninfected control samples (Pool 0, N = 12) by at least 1.5 standard deviations. However, in pool 0.2 (patients #1, #2, #7, #14, #16, #18, #23; Table 2) (N = 7), one out of seven samples could not be distinguished from the pool 0 when analyzed by REEAD. A Kruskal-Wallis test shows that the three medians obtained when analyzing the three sample sets are not all equal (p-value = 2.6 × 10−5) and pairwise comparisons indicate that the medians of results obtained by analyzing pools 0 and 2, pools 0 and 0.2, and pools 0.2 and 2 are all statistically different with p-values of 7.577 × 10−5, 0.0009753 and 0.03179, respectively. These results suggest that Plasmodium infections may be detected in saliva samples even from patients with a relatively low parasite concentration of 2 parasites/μL in the blood stream.
REEAD enabled Plasmodium detection based on unfrozen saliva samples
For long term storage of field collected samples and to ensure experimental reproducibility, all saliva samples tested hitherto were frozen in aliquots prior to testing. For the potential future use of the REEAD assay as a diagnostic tool the method will most probably be used to test unfrozen saliva samples. Therefore, to investigate if freezing is a necessary step to achieve efficient extraction of pTopI from saliva and obtain positive REEAD results, fresh saliva samples were collected in Gabon and transported at 4 °C to Europe where they were tested by REEAD within 5 to 45 days post collection.
Immediately upon arrival of the samples (corresponding to five to six days post collection), saliva from seven malaria positive (#26–32, Table 2) and 19 uninfected individuals was analyzed using the REEAD assay. The box plot in Fig. 3A shows the average number of signals per image frame obtained from 16 images. To avoid noise arising from slide to slide variation all results were normalized to the results obtained using a sample from patient #12 (Table 2) as reference sample. The signals obtained from Plasmodium positive samples were in all cases above the mean of the signals arising from malaria negative samples with at least 2 standard deviations (data not shown), clearly demonstrating the ability of REEAD to detect malaria using unfrozen saliva as the test material. In concert with this finding, a Mann-Whitney U test shows that the median of the results obtained from the group of samples from uninfected individuals differ significantly from the median of the results obtained from the group of samples from Plasmodium infected individuals (P = 0.0001).
Detection of pTopI in unfrozen saliva from malaria patients: (A) Boxplot showing the results from the REEAD assay for testing unfrozen saliva from uninfected individuals (N = 19) or Plasmodium infected persons (N = 7) (#26–32, Table 2). (B) Bar chart showing the results of analyzing a saliva sample that had either been frozen upon arrival or kept at 4 °C for 11, 25, or 45 days. Due to limited amount of sample it was not possible to do repetitions of the analysis of the positive samples. The error bars on the bars representing the results of testing the positive samples therefore show the standard error of measurement for 16 randomly selected microscopic images. All results were normalized to the results obtained using a sample from patient #12 (Table 2) as reference sample.
Malaria is prevalent in areas with unstable electricity and, hence, without stable freezing capacity. In order to test the vulnerability of REEAD to storage of saliva samples outside a freezer, aliquots from patient #32 (see Table 2) were stored at 4 °C and tested by REEAD 11, 25, and 45 days after sample acquisition in comparison to 3 samples from malaria negative individuals stored under similar conditions. Moreover, aliquots of the sample from patient #32 (Table 2) were frozen five days after the time of collection and analyzed in comparison to the unfrozen aliquots. The results from the frozen aliquots were used for normalization to compensate for slide-to-slide variations.
As evident from the bar chart in Fig. 3B, the number of signals obtained when analyzing the unfrozen sample did not decrease relative to the starting point (five days post collection) for the first 25 days of storage, while after 45 days the number of signals declined with a factor ∼2. After 45 days of storage at 4 °C analyses of the unfrozen samples still resulted in approximately double amount of signals relative to the frozen aliquot of a sample taken from the same patient. These results demonstrate that freezing is not a necessary step for sample extraction and that unfrozen saliva may in fact be a more suited sample for REEAD analysis compared to the frozen saliva used for the analyses shown in Fig. 2.
Establishment of a colorimetric readout for REEAD based Plasmodium detection
The experiments described above were all performed using a fluorescence microscope to detect the REEAD signals. Such method is not suited for the low-resource areas where malaria is prevalent. To allow visualization of the RCA products resulting from pTopI activity, using a protocol that is more adaptable to the settings in low-income countries, a colorimetric readout was developed. In this setup, which is schematically depicted in Fig. 4A, the RCA reaction of the pTopI generated DNA circles is performed in solution in the presence of biotin conjugated deoxynucleotides, which are incorporated into the RCA products. The biotinylated RCA products are then bound to a silica filter and visualized through coupling to streptavidin-HRP followed by addition of the colorimetric HRP substrate, 3,3′,5,5′-Tetramethylbenzidine (TMB), directly to the silica membranes. A representative picture of the results obtained using this readout is shown in Fig. 4B. To the left are shown the results obtained when analyzing a negative sample containing no DNA circles (F1) and a positive control sample containing 0.1 pmol of premade DNA circles (F2), respectively. F3 and F4 show the results of analyzing two saliva samples from uninfected individuals. F5 and F6 are typical examples of the results obtained when analyzing saliva from malaria patients (#4 and #20, Table 2).
Detection of pTopI using a colorimetric readout. (A) The individual steps of the colorimetric readout are schematically depicted. (I) shows the RCA primer hybridized to the pTopI generated single stranded DNA circles. II) The primer is elongated in a RCA reaction performed in the presence of biotin labeled nucleotides (red asterisks). III) The biotinylated DNA generated is bound to a silica membrane, which is positioned in a column. IV) Silica bound biotinylated DNA is visualized by coupling to streptavidin conjugated HRP (black asterisks) followed by V) incubation with the colorimetric HRP substrate, TMB. (B) Representative pictures of the silica membranes after completion of the colorimetric readout using either a sample without DNA circles or spiked with premade DNA circles (F1 and F2 respectively) as well as after testing saliva samples from uninfected individuals (F3 and F4) or Plasmodium infected individuals (F5 and F6) (patients #4 and #20 respectively, see Table 2). (C) Box plot representing the result of using the above described colorimetric readout for testing 14 saliva samples from uninfected individuals and 16 samples from Plasmodium infected individuals. The results are shown as fold increase above the average signal obtained from the analysis of samples from uninfected individuals.
The colorimetric REEAD setup as exemplified in Fig. 4B, was tested using 16 frozen saliva samples from 12 different malaria patients (#1 (tested twice), #2, #4 (tested 3 times), #7, #11, #13, #16, #18, #20 (tested twice), #21–23 (tested once); see Table 2 for parasite concentrations) and 14 frozen samples from uninfected individuals. Note, that only some of the samples could be tested in repetition due to the limited sample volumes. The colorimetric signals generated when analyzing each of the samples were documented by scanning the silica filters using a paper scanner and quantified using the Image J software. The results are shown as a box plot in Fig. 4C. All malaria positive samples gave rise to signal intensities at least 2 standard deviations above the mean signal intensities obtained when analyzing malaria negative samples. Moreover, a Mann-Whitney U test shows that the medians of the results obtained from the samples taken from uninfected persons differ significantly from the median of the results obtained from analyzing samples from Plasmodium infected persons (P < 0.0001). Taken together these results strongly suggest that the malaria specific REEAD setup can be combined with a simple yet efficient colorimetric readout, which is a crucial step towards adapting the REEAD technology to low resource settings.
Development of a pump-free system for extraction of pTopI from saliva samples
In the tests described above, pTopI from patient samples were extracted using a so-called water-in-oil droplet microfluidics device. In this device (see Fig. 5A, top panel) the sample to be analyzed, the REEAD substrate (S1), and the hypotonic lysis buffer was loaded into individual channels of the microfluidic device. When these three aqueous streams merged they were broken up by an oil stream to form stable water-in-oil emulsions. Each aqueous drop served as a micro-reactor, which subsequently was pushed through a serpentine channel to ensure adequate mixing of reagents. As demonstrated previously the enhanced reaction kinetics observed in the picoliter droplets ensured efficient detection using even very small (a single drop) unprocessed samples for analysis. A drawback of the standard setup described above is that it involves pump driven control of the flow-rates of both the oil and the aqueous phases, which is unsuited for low resource settings.
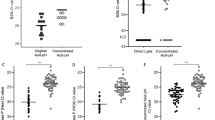
Detection of pTopI using a pump-free extraction method. (A) Schematic illustration of the microfluidic device used for pump-driven (top panel) and pump-free (lower panel) extraction of pTopI from saliva from Plasmodium infected individuals. In the case of pump-driven droplet microfluidics, patient sample, S1, and lysis buffer are fed into the microfluidic chip through the 3 inlets. By competition with oil (fed into the system through a 4th inlet) pL sized water in-oil-droplets are generated. The droplets are then led through a serpentine channel and collected. In the case of pump-free extraction, water-in-oil droplets are made by vortexing saliva, lysis buffer, S1, and oil. The droplets are then loaded into the microfluidic device using a handheld syringe and collected from the outlet after passage through a serpentine channel. (B) Extraction of saliva from uninfected or Plasmodium infected individuals was done using the pump-free microfluidics method depicted in A). The boxplot represents the results of testing two samples from uninfected persons and two samples from persons with malaria (patients #4 and #32, see Table 2). The results shown represent the results of three independent extractions of each sample. The results were normalized to the results obtained using a sample from patient #12 (Table 2) as reference sample (C) Left panel; representative image of the results obtained when combining pump-free microfluidics with the colorimetric read-out described in Fig. 4. The figure shows a filter after completion of the assay using a sample from an uninfected individual and a sample from a plasmodium infected individual (patient #33, see Table 2). Right panel; quantitative depiction of the result obtained when testing 6 samples using the pump-free extraction combined with colorimetric readout. The results are shown as fold-increase over the average of the readings obtained when testing negative samples. Hence, the negative samples give numbers varying around 1.
To circumvent the need of electrical pumps a pump-free extraction procedure was developed. This was achieved by generating the water-in-oil droplets by vortexing oil with a mixture of sample, substrate S1, and hypotonic lysis buffer. Though the droplets produced in this manner are heterogeneous, the confined droplets still offer the enhanced reaction kinetics necessary for effective extraction and substrate conversion34, when manually loaded in to a simplified microfluidic chip composed of a single inlet and outlet interrupted by a serpentine channel (Fig. 5A, lower panel). Droplets generated using this pump-free method were analyzed by REEAD following the same protocol as the droplets generated by the pump driven system and readout was based on microscopic visualization as described. Two saliva samples from uninfected individuals were analyzed together with two saliva samples from Plasmodium infected persons (Table 2, patients #4 and #32). The results of three independent extractions of each sample are shown in the box plot in Fig. 5B. As evident from the plot, analysis of the malaria positive samples resulted in signals above the mean of the signals obtained from the two malaria negative samples by at least 2 standard deviations. This result indicates that a handheld extraction method may replace the current pump driven method. Moreover, it was possible to combine the simplified hand driven extraction and mixing protocol with the above described colorimetric readout (Fig. 5C) holding promise for the future adaptation of the described method for a new malaria diagnostic tool based on saliva as the sample type and suitable for use in low-income countries. Note, that the analyses shown in Fig. 5C was only performed once due to low sample volume.
Conclusion
We here demonstrate the functionality of a REEAD protocol specific for the activity of the Plasmodium expressed enzyme pTopI for sensitive detection of malaria causing parasites using saliva from infected individuals as the test material. The documentation of saliva as a usable sample type for REEAD based detection of Plasmodium infections demonstrates the presence of active pTopI in this specimen. We found that although the REEAD method in principle is quantitative in nature the number of signals obtained when analyzing saliva samples from patients with different parasitemias did not correspond to the parasite concentrations in the blood. Since testing was done far from the sample collection in both space and time the lack of correlation between the number of REEAD signals and parasite load in tested individuals may reflect differences in storage conditions and time from collection to testing. Indeed, signals were observed to decline with storage of infected saliva over time even when the samples where kept at a steady temperature of 4 °C. Potential differences in storage conditions of the samples available for this study also impose a degree of uncertainty on the estimated detection limit (estimated to a blood equivalent parasite concentration of 2 parasites/μL) in frozen saliva. For a more precise determination of the detection limit and to address if the observed lack of correlation between REEAD signal and parasite load could be ascribed to sample quality, testing of larger number of freshly collected saliva samples close to the site of collection is being planned but is out of the scope of the current study. Indeed, such study will require the establishment of a test kit suitable for low resource settings. In the current manuscript, we present the development and testing of the individual components of such kit i.e. a pump free microfluidics based extraction of pTopI and a colorimetric readout allowing detection directly by the naked eye. Moreover, holding promise for future development of an all-in-one REEAD based kit for field testing of malaria in saliva specimen we demonstrate the feasibility of combining the pump free extraction with colorimetric readout for detection of malaria in laboratory settings. Even if future field tests should prove that saliva cannot allow quantitative testing of malaria, we believe that the use of non-invasive sample type for the detection of Plasmodium infections will continue to be appealing not least for eradication programs, which may involve tests of large number of asymptomatic individuals.
Materials and Methods
Oligonucleotides
Blood and saliva samples from malaria patients and uninfected individuals
Blood samples from individuals that are not infected with malaria were obtained from the blood bank at the Aarhus University Hospital, Skejby. Matching blood and saliva samples from patients diagnosed with malaria were obtained at the CERMEL, Albert Schweitzer Hospital, Lambaréné, Gabon. The patients were diagnosed using thick smear microscopy or by RDT’s (see Table 2). The study was conducted in accordance to the Declaration of Helsinki, and samples were collected and analyzed following local regulations and guidelines. Ethical clearance (#003/2014) for sampling in Gabon was obtained from the institutional ethics committee at the Centre de Recherches Médicales de Lambaréné. Informed consent was obtained from all study participants or their legal guardians in case of minors. Thick smear microscopy was performed by two independent readers. A third reading was performed if required (e. g. if first and second reading differed more than 50% of parasite count or if the same slide was read positive/negative by different readers). The number of counted parasites per µL blood were recorded. The final parasitemia was determined as the average of first and second reading (or, if third reading was performed: the average of the two closest results). In the case of testing by RDTs, the “Paracheck Pf” test was used.
Note that samples were diluted in PBS prior to microfluidics in order to avoid clotting of the microfluidic chip. This is not a prerequisite for the analyses but reduced the number of microfluidic chips necessary to conduct the analyses in this laboratory test.
Pump Driven Droplet Microfluidics
Unless otherwise stated in the result section pTopI was extracted from blood or saliva from malaria patients using pump driven droplet microfluidics. The droplet microfluidic devices (see Fig. 5A for design) were fabricated by conventional soft lithography techniques59, casting and curing the PDMS prepolymer on a SU-8 3025 (MicroChem) master of a channel height at around 25 μm. PDMS prepolymer (Sylgard 184) was prepared in a 10:1 (base:curing agent) ratio and cured at 65 °C for 1 hour. Prior to the experiments, the channels were wetted with oil/surfactant (Pico-Surf 1, 2% in HFE-7500, Dolomite Microfluidics) for at least 15 minutes. Two syringe pumps (Harvard Apparatus) were used to control the flow rates of oil/surfactant and reagents independently. The droplet volume and generation frequency were controlled by the flow rate ratio, determined by the competition between continuous phase and disperse phase34,60,61. Blood/saliva (diluted 1:4 in PBS), S1 substrate (Table 1 and12 − 167 nM final concentration in droplets) and lysis buffer (10 mM Tris pH 7.5; 5 mM EDTA; 0.2% Tween 20; 1 mM DTT; 1 mM PMSF) were subjected to droplet microfluidics essentially as described previously34. The procedure was completed by breaking the droplets by addition of 25%(v/v) 1 H,1 H,2 H,2H-Perfluoro-1-octanol (Sigma-Aldrich).
Pump-free Droplet Microfluidics
For pump-free droplet microfluidics (used in Fig. 5) a slightly modified microfluidic device design (see Fig. 5A) was used. This design consists of a serpentine channel flanked by a single inlet and a single outlet. Saliva was diluted 50 times in PBS and mixed with lysis buffer (10 mM Tris pH 7.5; 5 mM EDTA; 0.2% Tween 20; 1 mM DTT; 1 mM PMSF), the DNA substrate S1 (167 nM) (Table 1 and12), and Pico-Surf 1–2% in HFE-7500 in the same ratios as described previously34. Water-in-oil droplets were produced by vortexing for 1 minute. Using a hand-held syringe, the hereby generated droplets were subsequently fed into the single channel microfluidic chip. Following collection of the droplets from the outlet the droplets were broken by addition of 25% (v/v) 1 H,1 H,2 H,2H-Perfluoro-1-octanol.
pTopI assay
An amine coupled oligo (Amine-RCA primer, see Table 1) was immobilized on NHS-coated microscopy slides (CodeLink® HD Activated Slides) as described by the supplier (Surmodics).
Lysate supplemented with NaCl to a final concentration of 500 mM was added to the DNA conjugated slides to allow hybridization of pTopI generated DNA circles. Subsequent phi29 DNA polymerase enabled rolling circle amplification (RCA) and hybridization of a fluorescent detection probe (Table 1) to the RCA products was performed as described previously34. Fluorescence labeled RCA products were visualized in a fluorescence microscope (Olympus IX73) and quantified by counting the number of signals per microscopic image frame using the ImageJ software. The results were obtained by counting the number of pTopI specific signals on 16 randomly picked microscopic pictures (277,3 × 234 µm2) for each sample analyzed. Inherent to this readout is some slide to slide variation due to variations in coupling efficiency of the surface attached primer36. Therefore, to allow comparison across experiments/slides the results are shown as fold increase over the average of the number of signals achieved from the negative samples tested at the same time.
Generally, the data are presented as a box and whiskers plot with Tukey-style whiskers made using the GraphPad Prism software.
Colorimetric readout
The RCA primer was elongated by phi29 DNA polymerase (Life technologies) in the presence of 20 V% lysate (see Pump-driven droplet microfluidics), 10 µM RCA primer (Table 1) and 0.1 mM of each dNTP of which, 10% of the dCTP has been replaced by Biotin-16-Aminoallyl-2′-dCTP (TriLink Biotechnologies). Following primer elongation, the RCA product was diluted 10 fold in TE buffer (10 mM Tris-HCl, pH 7.5; 1 mM EDTA), and then 4 fold using CP buffer from E.Z.N.A. Cycle Pure Kit (OMEGA bio-tek). The diluted RCA products were bound to a DNA purification column (OMEGA bio-tek, Catalog number: D6492–02) and allowed to bind for 10 minutes on a shaker at room temperature. The entire reaction volume was then spun through the column (approximately 10 seconds in a table top centrifuge). Following binding of the RCA products, the column was washed with 200 µL PBS (pH 7.5) and blocked with 200 µL blocking buffer (5% skimmed milk in 100 mM Tris-HCl, pH 7.5) for 10 minutes. The column was then washed twice with 200 µL PBS, incubated with 200 µL 8 M Urea for 10 minutes, and washed with 200 µL PBS. 150 µL Streptavidin-HRP (Pierce Streptavidin Poly-HRP, Life Technologies) in blocking buffer (8 µL/mL blocking buffer) was then added to the column, which was allowed to incubate for 10 minutes. The column was then washed 3 times with 150 µL PBS supplemented with 0.2% Triton X-100. Finally, 150 µL TMB Enhanced HRP Membrane Substrate (ESPM) (Surmodics) was added to the columns. The reaction was allowed to run for 10 minutes after which the columns were spun at 10000 g for 10 seconds and the reaction stopped using 150 µL 1 M sulphuric acid. Finally, the columns were washed with 150 µL ddH2O.
The filters were removed from the columns and scanned using a HP Scanjet 5590 P paper scanner. The scanned image was split into the three RGB channels, and the red channel was analyzed using ImageJ. The “measure” function at standard settings was used to quantify the amount of reacted TMB. A standard value of 30.000, corresponding to an uncolored filter, was subtracted from all samples. The results are presented as fold increase over the average of the signal intensity achieved from the negative samples tested at the same time.
References
WHO. World Malaria Report 2016, Summary. (2016).
Halder, S. & Krishnan, Y. Design of ultrasensitive DNA-based fluorescent pH sensitive nanodevices. Nanoscale 7, 10008–10012, https://doi.org/10.1039/c5nr01158b (2015).
Baker, B. R. et al. An Electronic, Aptamer-Based Small-Molecule Sensor for the Rapid, Label-Free Detection of Cocaine in Adulterated Samples and Biological Fluids. (2006).
Liu, J. et al. Graphene Signal Amplification for Sensitive and Real-Time Fluorescence Anisotropy Detection of Small Molecules. (2013).
Zayats, M., Huang, Y., Gill, R., Ma, C.-A. & Willner, I. Label-Free and Reagentless Aptamer-Based Sensors for Small Molecules. https://doi.org/10.1021/ja0651456 (2006).
Bi, S., Li, L. & Zhang, S. Triggered polycatenated DNA scaffolds for DNA sensors and aptasensors by a combination of rolling circle amplification and DNAzyme amplification. Analytical chemistry 82, 9447–9454, https://doi.org/10.1021/ac1021198 (2010).
Zuccaro, L. et al. Real-Time Label-Free Direct Electronic Monitoring of Topoisomerase Enzyme Binding Kinetics on Graphene. ACS nano 9, 11166–11176, https://doi.org/10.1021/acsnano.5b05709 (2015).
Hardin, A. H. et al. Direct measurement of DNA bending by type IIA topoisomerases: implications for non-equilibrium topology simplification. Nucleic acids research 39, 5729–5743, https://doi.org/10.1093/nar/gkr109 (2011).
Jensen, P. W. et al. Real-time detection of TDP1 activity using a fluorophore-quencher coupled DNA-biosensor. Biosensors & bioelectronics 48C, 230–237, https://doi.org/10.1016/j.bios.2013.04.019 (2013).
Jepsen, M. L. et al. Specific detection of the cleavage activity of mycobacterial enzymes using a quantum dot based DNA nanosensor. Nanoscale 8, S1–S2, https://doi.org/10.1039/C5NR06326D (2016).
Kristoffersen, E. L. et al. Real-time investigation of human topoisomerase I reaction kinetics using an optical sensor: a fast method for drug screening and determination of active enzyme concentrations. Nanoscale 7, 9825–9834, https://doi.org/10.1039/C5NR01474C (2015).
Tesauro, C. et al. Specific detection of topoisomerase I from the malaria causing P. falciparum parasite using isothermal rolling circle amplification. Conference proceedings:… Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual Conference 2012, 2416–2419, https://doi.org/10.1109/EMBC.2012.6346451 (2012).
Andersen, F. F. et al. Multiplexed detection of site specific recombinase and DNA topoisomerase activities at the single molecule level. ACS nano 3, 4043–4054, https://doi.org/10.1021/nn9012912 (2009).
Jakobsen, A.-K. & Stougaard, M. Combining a Nanosensor and ELISA for Measurement of Tyrosyl-DNA Phosphodiesterase 1 (TDP1) Activity and Protein Amount in Cell and Tissue Extract. Nano LIFE 05, 1541001, https://doi.org/10.1142/S1793984415410019 (2015).
Stougaard, M., Juul, S., Andersen, F. F. & Knudsen, B. R. Strategies for highly sensitive biomarker detection by Rolling Circle Amplification of signals from nucleic acid composed sensors. Integrative Biology 3, 982, https://doi.org/10.1039/c1ib00049g (2011).
Banerjee, A. et al. Controlled release of encapsulated cargo from a DNA icosahedron using a chemical trigger. Angewandte Chemie - International Edition 52, 6854–6857, https://doi.org/10.1002/anie.201302759 (2013).
Goodman, R. P. et al. Reconfigurable, braced, three-dimensional DNA nanostructures. Nature Nanotechnology 3, 93–96, https://doi.org/10.1038/nnano.2008.3 (2008).
Juul, S. et al. Temperature-controlled encapsulation and release of an active enzyme in the cavity of a self-assembled DNA nanocage. ACS nano 7, 9724–9734, https://doi.org/10.1021/nn4030543 (2013).
Takenaka, T. et al. Photoresponsive DNA nanocapsule having an open/close system for capture and release of nanomaterials. Chemistry (Weinheim an der Bergstrasse, Germany) 20, 14951–14954, https://doi.org/10.1002/chem.201404757 (2014).
Fujita, H., Kataoka, Y., Tobita, S., Kuwahara, M. & Sugimoto, N. Novel One-Tube-One-Step Real-Time Methodology for Rapid Transcriptomic Biomarker Detection: Signal Amplification by Ternary Initiation Complexes. (2016).
Jacobsen, M. F. et al. Small molecule induced control in duplex and triplex DNA-directed chemical reactions. Org. Biomol. Chem. 8, 50–52, https://doi.org/10.1039/B919387A (2010).
Blanco, L. et al. Highly efficient DNA synthesis by the phage Φ29 DNA polymerase. Symmetrical mode of DNA replication. Journal of Biological Chemistry 264, 8935–8940 (1989).
Wei, H. et al. Production of dumbbell probe through hairpin cleavage-ligation and increasing RCA sensitivity and specificity by circle to circle amplification. Scientific Reports 6, 29229, https://doi.org/10.1038/srep29229 (2016).
Liu, D., Daubendiek, S. L., Zillman, M. A., Ryan, K. & Kool, E. T. Rolling Circle DNA Synthesis: Small Circular Oligonucleotides as Efficient Templates for DNA Polymerases. Journal of the American Chemical Society 118, 1587–1594, https://doi.org/10.1021/ja952786k (1996).
Fire, A. & Xu, S. Q. Rolling replication of short DNA circles. Proc Natl Acad Sci USA 92, 4641–4645 (1995).
Lohmann, J. S., Stougaard, M. & Koch, J. Detection of short repeated genomic sequences on metaphase chromosomes using padlock probes and target primed rolling circle DNA synthesis. BMC molecular biology 8, 103, https://doi.org/10.1186/1471-2199-8-103 (2007).
Stougaard, M., Lohmann, J. S., Zajac, M., Hamilton-Dutoit, S. & Koch, J. In situ detection of non-polyadenylated RNA molecules using Turtle Probes and target primed rolling circle PRINS. BMC biotechnology 7, 69, https://doi.org/10.1186/1472-6750-7-69 (2007).
Vagner, J., Steiniche, T. & Stougaard, M. In-situ hybridization-based quantification of hTR: a possible biomarker in malignant melanoma. Histopathology 66, 747–751, https://doi.org/10.1111/his.12501 (2015).
Cho, E. J., Yanq, L., Lew, M. & Ellinqton, A. D. Using a deoxyribozyme ligase and rolling circle amplification to detect a non-nucleic acid analyte, ATP. Journal of the American Chemical Society 127, 2022–2023, https://doi.org/10.1021/ja043490u (2005).
Ma, C., Wang, W., Yang, Q., Shi, C. & Cao, L. Cocaine detection via rolling circle amplification of short DNA strand separated by magnetic beads. Biosensors and Bioelectronics 26, 3309–3312, https://doi.org/10.1016/j.bios.2011.01.003 (2011).
Monsur Ali, M. & Li, Y. Colorimetric sensing by using allosteric-DNAzyme-coupled rolling circle amplification and a peptide nucleic acid-organic dye probe. Angewandte Chemie - International Edition 48, 3512–3515, https://doi.org/10.1002/anie.200805966 (2009).
Zhao, W., Ali, M. M., Brook, M. A. & Li, Y. Rolling circle amplification: applications in nanotechnology and biodetection with functional nucleic acids. Angewandte Chemie (International ed. in English) 47, 6330–6337, https://doi.org/10.1002/anie.200705982 (2008).
Juul, S. et al. Microfluidics-mediated isothermal detection of enzyme activity at the single molecule level. Conference proceedings:… Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual Conference 2011, 3258–3261, https://doi.org/10.1109/IEMBS.2011.6090885 (2011).
Juul, S. et al. Droplet microfluidics platform for highly sensitive and quantitative detection of malaria-causing Plasmodium parasites based on enzyme activity measurement. ACS nano 6, 10676–10683, https://doi.org/10.1021/nn3038594 (2012).
Roy, A. et al. Decreased camptothecin sensitivity of the stem-cell-like fraction of caco2 cells correlates with an altered phosphorylation pattern of topoisomerase I. PloS one 9, e99628, https://doi.org/10.1371/journal.pone.0099628 (2014).
Stougaard, M. et al. Single-molecule detection of human topoisomerase I cleavage-ligation activity. ACS nano 3, 223–233, https://doi.org/10.1021/nn800509b (2009).
Wang, J. et al. Novel DNA sensor system for highly sensitive and quantitative retrovirus detection using virus encoded integrase as a biomarker. Nanoscale 9, 440–448, https://doi.org/10.1039/c6nr07428f (2017).
Givskov, A. et al. Optimized Detection of Plasmodium falciparum Topoisomerase I Enzyme Activity in a Complex Biological Sample by the Use of Molecular Beacons. Sensors 16, 1916, https://doi.org/10.3390/s16111916 (2016).
Jakobsen, A.-K. et al. Correlation between topoisomerase I and tyrosyl-DNA phosphodiesterase 1 activities in non-small cell lung cancer tissue. Experimental and Molecular Pathology 99, 56–64, https://doi.org/10.1016/j.yexmp.2015.05.006 (2015).
Akhtar, S. et al. Real-space transmission electron microscopy investigations of attachment of functionalized magnetic nanoparticles to DNA-coils acting as a biosensor. The journal of physical chemistry. B 114, 13255–13262, https://doi.org/10.1021/jp105756b (2010).
Cheng, W., Yan, F., Ding, L., Ju, H. & Yin, Y. Cascade signal amplification strategy for subattomolar protein detection by rolling circle amplification and quantum dots tagging. Analytical Chemistry 82, 3337–3342, https://doi.org/10.1021/ac100144g (2010).
Cheglakov, Z., Weizmann, Y., Basnar, B. & Willner, I. Diagnosing viruses by the rolling circle amplified synthesis of DNAzymes. Organic & biomolecular chemistry 5, 223–225, https://doi.org/10.1039/b615450f (2007).
Mizuta, R., Mizuta, M. & Kitamura, D. Atomic force microscopy analysis of rolling circle amplification of plasmid DNA. Archives of histology and cytology 66, 175–181 (2003).
Blab, G. A., Schmidt, T. & Nilsson, M. Homogeneous Detection of Single Rolling Circle Replication Products. Analytical Chemistry 76, 495–498, https://doi.org/10.1021/ac034987 (2004).
Larsson, C. et al. In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes. Nature methods 1, 227–232, https://doi.org/10.1038/nmeth723 (2004).
Lizardi, P. M. et al. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature genetics 19, 225–232, https://doi.org/10.1038/898 (1998).
Joanny, F., Lohr, S. J., Engleitner, T., Lell, B. & Mordmuller, B. Limit of blank and limit of detection of Plasmodium falciparum thick blood smear microscopy in a routine setting in Central Africa. Malaria journal 13, 234, https://doi.org/10.1186/1475-2875-13-234 (2014).
In Guidelines for the Treatment of Malaria WHO Guidelines Approved by the Guidelines Review Committee (2015).
Berry, A., Benoit-Vical, F., Fabre, R., Cassaing, S. & Magnaval, J. F. PCR-based methods to the diagnosis of imported malaria. Parasite 15, 484–488, https://doi.org/10.1051/parasite/2008153484 (2008).
Wilson, M. L. Laboratory diagnosis of malaria: conventional and rapid diagnostic methods. Archives of pathology & laboratory medicine 137, 805–811, https://doi.org/10.5858/arpa.2011-0602-RA (2013).
Grobusch, M. P. et al. Comparison of three antigen detection tests for diagnosis and follow-up of falciparum malaria in travellers returning to Berlin, Germany. Parasitology research 89, 354–357, https://doi.org/10.1007/s00436-002-0764-7 (2003).
Ota-Sullivan, K. & Blecker-Shelly, D. L. Use of the rapid BinaxNOW malaria test in a 24-hour laboratory associated with accurate detection and decreased malaria testing turnaround times in a pediatric setting where malaria is not endemic. J Clin Microbiol 51, 1567–1569, https://doi.org/10.1128/JCM.00293-13 (2013).
Ghayour Najafabadi, Z. et al. Mitochondrial PCR-based malaria detection in saliva and urine of symptomatic patients. Transactions of the Royal Society of Tropical Medicine and Hygiene 108, 358–362, https://doi.org/10.1093/trstmh/tru061 (2014).
Putaporntip, C., Buppan, P. & Jongwutiwes, S. Improved performance with saliva and urine as alternativeDNA sources for malaria diagnosis by mitochondrial DNA-based PCR assays. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases 17, 1484–1491, https://doi.org/10.1111/j.1469-0691.2011.03507.x (2011).
Singh, R. et al. Comparison of three PCR-based assays for the non-invasive diagnosis of malaria: detection of Plasmodium parasites in blood and saliva. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology 33, 1631–1639, https://doi.org/10.1007/s10096-014-2121-z (2014).
Ghayour Najafabadi, Z. et al. Detection of Plasmodium vivax and Plasmodium falciparum DNA in human saliva and urine: loop-mediated isothermal amplification for malaria diagnosis. Acta tropica 136, 44–49, https://doi.org/10.1016/j.actatropica.2014.03.029 (2014).
Wilson, N. O., Adjei, A. A., Anderson, W., Baidoo, S. & Stiles, J. K. Detection of Plasmodium falciparum histidine-rich protein II in saliva of malaria patients. The American journal of tropical medicine and hygiene 78, 733–735 (2008).
Huang, H. et al. Proteomic identification of host and parasite biomarkers in saliva from patients with uncomplicated Plasmodium falciparum malaria. Malaria journal 11, 178, https://doi.org/10.1186/1475-2875-11-178 (2012).
Qin, D., Xia, Y. & Whitesides, G. M. Soft lithography for micro- and nanoscale patterning. Nature protocols 5, 491–502, https://doi.org/10.1038/nprot.2009.234 (2010).
Ho, Y. P., Grigsby, C. L., Zhao, F. & Leong, K. W. Tuning physical properties of nanocomplexes through microfluidics-assisted confinement. Nano letters 11, 2178–2182, https://doi.org/10.1021/nl200862n (2011).
Juul, S. et al. Detection of single enzymatic events in rare or single cells using microfluidics. ACS nano 5, 8305–8310, https://doi.org/10.1021/nn203012q (2011).
Acknowledgements
We are grateful to Noriko Hansen for skillful technical assistance. We thank Mr. Jones Tabi, Mrs. Sandra Hinge Petersen, and Dr. Mette Christiansen for help with providing samples from uninfected individuals in Denmark. BRK would like to acknowledge the support from Familien Erichsens mindefond, Aase & Ejnar Danielsen’s foundation, Aage & Johanne Louis-Hansens Foundation, Marie & M.B. Richters Foundation, Minister Erna Hamiltons Legat for Videnskab og Kunst, Civilingeniør Frode V. Nyegaard og hustrus Fond, Karen Elise Jensens Foundation, Arvid Nilssons Foundation, Ludvid og Franciska Andersens legat. MR acknowledges the Federal Ministry of Science, Research and Economy of Austria as part of the EDCTP-2 programme. This study is part of the EDCTP2 programme supported by the European Union. YPH would like to acknowledge the support from the Start-Up fund and Direct Research Grant provided by the Chinese University of Hong Kong.
Author information
Authors and Affiliations
Contributions
All authors contributed extensively to the work presented in this paper. B.R.K., Y.P.H., and M.S.H. conceived the study, designed or co-designed all experiments presented and wrote the major part of the manuscript assisted by S.F. M.R., F.L., R.M.Z. A.K., M.G., J.M., L.E., L.V., G.M.N. and E.P. organized and performed collection and screening of blood and saliva samples from febrile patients and identified plasmodium positive individuals. M.S.H., S.F., R.N. and A.K.W.S. performed and analyzed experiments. M.S., Y.P.H., and R.L. assisted with data analysis and experimental design.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hede, M.S., Fjelstrup, S., Lötsch, F. et al. Detection of the Malaria causing Plasmodium Parasite in Saliva from Infected Patients using Topoisomerase I Activity as a Biomarker. Sci Rep 8, 4122 (2018). https://doi.org/10.1038/s41598-018-22378-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-22378-7
This article is cited by
-
Liquid Biopsy for Promising Non-invasive Diagnostic Biomarkers in Parasitic Infections
Acta Parasitologica (2022)
-
Acceptance and perceived value of non-invasive malaria diagnostic tests in malaria-endemic countries
Malaria Journal (2021)
-
Fabrication of laser printed microfluidic paper-based analytical devices (LP-µPADs) for point-of-care applications
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.