Abstract
Segmentation of axon and myelin from microscopy images of the nervous system provides useful quantitative information about the tissue microstructure, such as axon density and myelin thickness. This could be used for instance to document cell morphometry across species, or to validate novel non-invasive quantitative magnetic resonance imaging techniques. Most currently-available segmentation algorithms are based on standard image processing and usually require multiple processing steps and/or parameter tuning by the user to adapt to different modalities. Moreover, only a few methods are publicly available. We introduce AxonDeepSeg, an open-source software that performs axon and myelin segmentation of microscopic images using deep learning. AxonDeepSeg features: (i) a convolutional neural network architecture; (ii) an easy training procedure to generate new models based on manually-labelled data and (iii) two ready-to-use models trained from scanning electron microscopy (SEM) and transmission electron microscopy (TEM). Results show high pixel-wise accuracy across various species: 85% on rat SEM, 81% on human SEM, 95% on mice TEM and 84% on macaque TEM. Segmentation of a full rat spinal cord slice is computed and morphological metrics are extracted and compared against the literature. AxonDeepSeg is freely available at https://github.com/neuropoly/axondeepseg.
Similar content being viewed by others
Introduction
Neuronal communication is ensured by the transmission of action potentials along white matter axons. For long distance communication, these axons, which are typically 1–10 µm in diameter, are surrounded by a myelin sheath whose main role is to facilitate the propagation of the electrical impulses along neuronal fibers and increase the transmission speed1,2. Pathologies such as neurodegenerative diseases (e.g., multiple sclerosis) or trauma are associated with myelin degeneration, which can ultimately lead to sensory and motor deficits (e.g., paraplegia)3,4. Being able to image axons and myelin sheaths at high resolution would help researchers understand the origins of demyelination and test therapeutic drugs5,6 and could also be used to validate novel magnetic resonance imaging biomarkers of myelin7. High resolution histology is typically done using electron microscopy following osmium staining to obtain myelin contrast. Then, axons and myelin can be analysed on the images to derive metrics such as axon density or myelin thickness. However, given that 1 mm2 of white matter can contain over 100,000 axons8, it is important to obtain a robust and reliable segmentation of individual axons and myelin as automatically as possible.
Several segmentation methods for axon and myelin have been proposed which are based on traditional image processing algorithms including thresholding and morphological operations9,10, axon shape-based morphological discrimination11, watershed12,13, region growing14, active contours without15,16 and with discriminant analysis16. However, a few limitations can be reported from the previous work: (i) traditional image-based methods are designed to work on specific imaging modalities and often fail if another contrast is used (e.g., optical image instead of electron microscopy); (ii) previous methods are not fully-automatic as they typically require either preprocessing, hand-selected features for axon discrimination and/or postprocessing; (iii) traditional image-based methods do not make full use of the contextual information of the image (i.e., multi-scale representation of axons, average shape of axons, etc.) and (iv) most of the previous methods are not publicly available (to our knowledge, only that from15,16 are).
In the last five years, deep learning methods have become the state of the art when it comes to computer vision tasks. Convolutional neural networks (CNNs) are particularly suited to image classification17,18,19,20 and semantic segmentation21. Cell segmentation is one of the popular application of CNNs22,23. The U-Net architecture introduced by Ronneberger and collaborators24 has inspired many medical segmentation applications, efficiently combining both context and localization of structures of interest. Segmentation of axons and myelin based on deep learning approaches offers significant advantages when compared with traditional image segmentation algorithms: (i) there is no need to hand-select relevant features because the network is able to learn the hidden structural and textural features by itself, (ii) this approach allows to segment both axons and myelin sheaths in two different labels with the same network, without the need of any explicit pre- or post-processing, (iii) the network can be trained for various imaging modalities without significantly changing its architecture and (iv) once trained, the model is relatively fast at the prediction step (only a few seconds) compared to more traditional image processing methods.
Few research groups have applied deep learning for axon and myelin segmentation. Naito and collaborators25 have implemented a two-step process that first performs clustering segmentation of myelinated nerve fibers in optical microscopic images, and then discriminates between true and false candidates by using a CNN classification network. This group did not exploit the CNN for the segmentation, but only for discrimination. The work from Mesbah and collaborators26 presented a deep encoder-decoder CNN that can segment both axon and myelin and claimed to achieve up to 82% pixel-wise accuracy. However, the network has been designed specifically for light microscopy images, the implementation is not publicly available and minimal regularization strategies have been employed in order to improve generalization.
We present AxonDeepSeg, a deep learning framework for robust and automatic segmentation of both axons and myelin sheaths in myelinated fibers. AxonDeepSeg features: (i) a CNN architecture for semantic segmentation of histological images; (ii) two ready-to-use models for the segmentation of scanning electron microscopy (SEM) and transmission electron microscopy (TEM) samples adapted to a variety of species and acquisition parameters; (iii) a well-documented training pipeline to generate models for new imaging modalities and (iv) free and open source code (https://github.com/neuropoly/axondeepseg).
Methods
Dataset
Microscopy images used in this study were acquired with two different imaging techniques: SEM and TEM. Different acquisition resolutions were used, in order to increase variability and obtain better generalization of the model, with isotropic pixel size resolution ranging from 0.05 to 0.18 µm (SEM) and 0.002 to 0.009 µm (TEM). SEM samples were stained with 2% osmium, embedded in epoxy, polished and imaged with the same SEM system (Jeol 7600F). TEM images were obtained from mice brain samples (splenium), as described in27. Additionally, a macaque sample of the corpus callosum was added to the test set. Preparation and imaging procedures are described in7. Table 1 lists the samples used for the experiments.
All methods were carried out in accordance with relevant guidelines and regulations. Experimental protocols involving rats were approved by the Montreal Heart Institute committee. Experimental protocols involving the human spinal cord were done at the anatomy laboratory of the University of Quebec at Trois-Rivieres. The spinal cord donor gave informed consent and procedures were approved by the local ethics committee (SCELERA-15-03-pr01). Similarly, TEM images shared by collaborators were obtained in accordance with the corresponding ethics committees (mice: Institutional Animal Care and Use Committee at the New York University School of Medicine, macaque: Montreal Neurological Institute Animal Care Committee).
Ground truth labelling
The ground truth labelling of SEM samples was created as follows: (i) Myelin sheaths were manually segmented (inner and outer contours) with GIMP (https://www.gimp.org/); (ii) Axon labels were obtained by filling the region enclosed by the inner border of the myelin sheaths; (iii) Small manual corrections were done on the axon and myelin masks (contour refinement, elimination of false positives) when necessary.
The ground truth labelling of TEM samples was created as follows: (i) Myelin was first segmented using intensity thresholding followed by manual correction, then the inner region was filled to generate axon labels. More details can be found about the generation of labels for the macaque7 and the mice27.
All ground truth labels were cross-checked by at least two researchers. The final ground truth consists of a single png image with values: background = 0, myelin = 127, axon = 255. Example SEM and TEM samples and corresponding ground truth labels are shown in Fig. 1. This figure also illustrates the large variability in terms of image features, especially for the SEM data (contrast, noise, sample preservation, etc.).
Overview of the data and ground truth labels for SEM (a) and TEM (b). Label masks contain 3 classes: axon (in blue in the figure), myelin (red) and background (black). All SEM and TEM samples shown here are cropped to 512 × 512 pixels. SEM patches have a pixel size of 0.1 µm, while TEM patches have a pixel size of 0.01 µm (see section “Pipeline overview”).
Pipeline overview
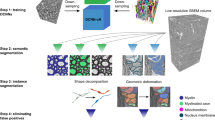
The pipeline of AxonDeepSeg is composed of four steps: data preparation, learning, evaluation and prediction. Figure 2 illustrates each step.
Overview of the AxonDeepSeg pipeline. During the data preparation step (a), microscopy samples and corresponding ground truth labels are resampled to have a common pixel size (0.1 µm for the SEM model, 0.01 µm for the TEM model), divided into 512 × 512 patches, and split into training/validation sets. The neural network is trained during the learning step (b) on the training/validation dataset. When the model is trained, performance is assessed on a test dataset (evaluation step (c)). For prediction (d), the new microscopy image to be segmented is first resampled to the working pixel size of the network, divided into 512 × 512 patches and analysed with the trained model. Segmented output patches are then stitched together and resampled back to the native pixel size.
In the data preparation step, raw microscopy images and corresponding axon/myelin labels are resampled to a common resolution space: 0.1 µm per pixel for SEM and 0.01 µm for TEM. These values are based on preliminary results and on the typical resolutions provided by each of these imaging systems. Resampled samples are divided into patches of 512 × 512 pixels due to memory constraints. This size was chosen to have around 15–75 axons per patch. Traditional pre-processing was applied patch-wise, including standardization and histogram equalization (not shown in Fig. 2 for clarity). For learning, the patches and corresponding labels were randomly split and then considered either for the training or for the validation sets (training/validation split of approximately 70/30%). For evaluation, full test images were randomly selected.
In the learning step, the training/validation dataset is fed into the network. Once the trained model is obtained, performance is evaluated on the test dataset (evaluation step). Finally, the trained model can be used for inference on new microscopy images (prediction step). The images are resampled to the pixel size of the model, divided into patches of 512 × 512 pixels, segmented, stitched to the native size, and resampled to the native resolution. Note that bilinear interpolation was used during the resampling steps.
Architecture of the network
The architecture is inspired by the original U-Net model24, combining a contracting path with traditional convolutions and then an expanding path with up-convolutions. Figure 3 illustrates the network architecture. The convolutional layers in the first block use 5 × 5 kernels, while the convolutional layers on remaining blocks use 3 × 3 kernels. The SEM network has 3 convolutional layers per block, while the TEM network has 2 convolutional layers per block. These decisions were based on preliminary optimizations (see section “Hyperparameter optimization”). In the contracting path, convolutions of stride 2 are computed after the last convolutional layer of each block to reduce the dimensionality of the features. Each strided convolution layer has a corresponding up-convolution layer in the expansion path in order to recover the localization information lost during the contraction path. Up-convolutions were computed by bilinear interpolation followed by a convolution. The merging of the context and localization information is done by concatenating the features from the contracting path with the corresponding ones in the expansion path. The number of features (channels) is doubled after each block, starting from 16, and then decreased at the same rate during the expansion path. All activation functions in the convolutional layers are rectified linear units (ReLU28). The last layer before the prediction is a softmax activation with 3 classes (axon, myelin and background). The SEM and TEM networks have a total of 1,953,219 and 1,552,387 trainable parameters, respectively.
Architecture of the convolutional neural networks designed for the segmentation of SEM and TEM images. For the SEM model, 3 convolutional layers are used at each block, while only 2 convolutional layers are used for the TEM model. Convolutional layers in dashed lines are removed for the TEM model. All activation functions used are rectified linear units (ReLU). Strided convolutions are used to downsample the features during the contraction path (left), while up-convolutions are used to recover the localization during the expansion path (right). Features of the contraction path are merged with features of the expansion path to combine localization and context (illustrated by the concatenation step). The pixel-wise classification is done by a 3-class softmax.
Data augmentation strategy
A data augmentation strategy was used on the input patches in order to reduce overfitting and improve generalization17,20,24. The strategy includes random shifting, rotation, rescaling, flipping, blurring and elastic deformation29. Table 2 summarizes the data augmentation strategy and the corresponding parameters.
Training procedure
For the training phase, we used a starting learning rate of 0.001 on which we applied a polynomial decay30 with a power of 0.9. The decay length was 200 epochs, after which the training stopped. We shuffled the samples list at the beginning of each epoch and used a batch size of 8 patches of 512 × 512 pixels. We have also implemented batch normalization31 before each activation. The momentum was exponentially decayed from 0.7 to 0.9. This was done to enable a quicker convergence at the beginning of the training by keeping a few samples for the batch normalization, while ensuring a stable training at the later epochs. A dropout32 rate of 0.25 is used in the convolutional layers to reduce the risk of overfitting and improve generalization. The network was trained with the Adam optimizer33. We minimized a spatially-weighted multi-class cross-entropy loss. The spatial weights ratios used to correct the class imbalance were respectively 1.1, 1.0 and 1.3 for background, myelin and axon. Those weights were chosen after hyperparameter optimization. The training phase took 86 minutes on an NVIDIA P100 GPU.
Inference procedure
During the inference step, we split the original images into patches of size 512 × 512 pixels. To overcome border issues (i.e. partial axons at edges not being properly identified as axons), the output segmentation mask is cropped around a smaller patch. Thus, patches overlap by d pixels to cover the entire image, as illustrated in Fig. 4. Based on preliminary optimizations, the default value d was set to 25.
Hyperparameter optimization
We used different grid searches in order to set the value of the hyperparameters with respect to the accuracy and error on the validation set. The following architecture parameters were optimized at the same time: number of layers, number of filters and convolutional kernel size. The starting learning rate and the batch normalization momentum were also optimized jointly using a grid search, as they both have an effect on the time the model takes to converge and the stability of the validation metrics (based on our experiments). We then jointly optimized the batch normalization momentum and the decay period of the momentum.
Evaluation method
For testing, the following metrics were computed: the Dice values (axon and myelin) and the pixel-wise accuracy to assess the quality of the segmentation, and the sensitivity and precision to assess the capability to detect true axonal fibers and avoid false axonal fibers.
Segmentation metrics
To assess the quality of the segmentation we used the Dice coefficient. For two binary images A and B, the Dice coefficient is defined as:
where A \(\cap \) B is the intersection between the two images (i.e. number of pixels that are true in both images), \(|A|\) is the number of pixels that are true in image A, and \(|B|\) is the number of pixels that are true in image B. The Dice coefficient is computed separately for axon and myelin segmentations, between the prediction and the ground truth masks.
Furthermore, the pixel-wise accuracy is evaluated in order to get a combined assessment of axon-myelin segmentation. The pixel-wise accuracy is computed as the ratio between correctly classified pixels (i.e. axon pixel classified as axon, myelin pixel classified as myelin, background pixel classified as background) and the total number of pixels in the test sample.
Detection metrics
To assess the performance of myelinated fiber detection, we computed the sensitivity and precision based on axon objects, using the positions of the centroids. Knowing the number of true positives (TP, axons present in both the prediction and the ground truth mask), false positives (FP, axons present in the prediction, but absent in the ground truth mask) and false negatives (FN, axons present in the ground truth mask, but absent in the prediction), we can compute the sensitivity (true positive rate) and the precision (positive predictive value) with the following equations:
Data availability
A part of the datasets generated during and/or analysed during the current study are available in the White Matter Microscopy Database repository (https://osf.io/yp4qg/). The remaining datasets are available from the corresponding author on reasonable request.
Results
Segmentation
Segmentation was evaluated on SEM (rat and human spinal cords) and TEM (mouse splenium and macaque corpus callosum) samples. Segmentation and ground truth masks for both axons and myelin sheaths are displayed on Fig. 5. Table 3 lists validation metrics computed on the segmentation outputs: axon Dice, myelin Dice, pixel-wise accuracy, sensitivity and precision. The SEM model trained on rat microscopy was able to achieve a pixel-wise accuracy between 85% and 88% on the rat test samples, while the pixel-wise accuracy on human test sample was 81%. The TEM model trained on mice microscopy achieved a pixel-wise accuracy of 95% on mice samples and a pixel-wise accuracy of 84% on macaque samples.
Example of segmentation results on SEM and TEM images on a variety of species. The corresponding ground truth segmentation is shown on the right. Overall, the agreement is good. A few discrepancies are noticeable, notably caused by ambiguous/untypical myelin structure (white arrows and white asterisks), inhomogeneous myelin thickness (yellow arrows) and untypical axon intensity (white squares). Some of these discrepancies could potentially be solved using post-processing methods.
To demonstrate the utility of AxonDeepSeg for large scale microscopy, segmentation of axon/myelin was performed on a full rat spinal cord SEM (cervical level). Processing time was 5 hours in a Mac laptop (2.9 GHz). Segmentation masks (axons in red, myelin sheaths in blue) are displayed on Fig. 6, along with a zoomed window of a small region for better visualization.
Full slice of rat spinal cord showing segmented axons (blue) and myelin sheaths (red). The zoomed panel illustrates the segmentation performance and sensitivity to fiber size: the left half of the panel contains smaller axons (mean diameter around 1.75 µm) while the right half contains larger axons (mean diameter around 2.5 µm).
Morphometrics extraction
As a proof-of-concept, morphometric statistics were extracted from a full spinal cord of rat using AxonSeg16. The segmented rat spinal cord shown in Fig. 6 was downsampled to 50 × 50 µm2 in order to generate maps of density (e.g., axon and myelin density). The following aggregate metrics were computed:
-
Axon diameter mean and standard deviation: arithmetic mean and standard deviation of the distribution of equivalent axon diameters (computed for each axon object as √(4*Area/π));
-
Axon density: number of axons per mm2;
-
Axon volume fraction (AVF): ratio between area of axons and total area of the region;
-
Myelin volume fraction (MVF): ratio between area of myelin and total area of the region;
-
G-ratio: ratio between axon diameter and myelinated fiber (axon + myelin) diameter, which can be estimated with the following formula7: √(1/(1 + MVF/AVF)).
A binary mask was used to only keep white matter pixels. Results are displayed in Fig. 7. Obtained metrics were compared with references of the white matter tracts of the rat spinal cord34,35,36. The distribution maps are in good agreement with known anatomy. In the corticospinal tract (tract #12 of the reference), we observe smaller axon diameters (around 1 µm), very high axon density (around 200,000 axons per mm2) and g-ratio values around 0.6. Larger axons are found close to the spinal cord periphery. See Discussion for more comparison with the literature.
Distribution maps of axon diameter mean and standard deviation, axon density, axon volume fraction, myelin volume fraction and g-ratio in a full rat spinal cord slice (cervical level). The SEM slice was segmented with AxonDeepSeg. The aggregate metrics of the white matter were generated by downsampling the axon/myelin segmentation masks to a 50 × 50 µm2 resolution. A schematic diagram of the main ascending and descending tracts of the white matter in the rat spinal cord based on the literature34,35,36 is provided as reference.
Discussion
This paper introduced AxonDeepSeg, a software framework to segment axon and myelin from microscopy data using deep learning. We showed that AxonDeepSeg can segment axon and myelin of SEM and TEM samples of various species with high accuracy. Moreover, AxonDeepSeg can serve as a tool to document nerve fiber morphometry, as demonstrated by the extraction of metrics from a full rat spinal cord slice.
Trained models
We propose a SEM model trained with a resolution of 0.1 µm per pixel, and a TEM model trained with a resolution of 0.01 µm per pixel. At inference, test image is resampled to meet the target resolution of the model. Other training set compositions were explored, with model trained on both SEM and TEM data in order to achieve better generalization. However, a few limitations arose: (i) SEM and TEM images exhibit very different resolution ranges, requiring large resampling factors to find a common resolution space; (ii) SEM and TEM modalities capture different microstructure/textures of the tissue (for instance, TEM microscopy can capture subcellular microstructure details of the axon); (iii) preliminary results of model simultaneously trained on SEM and TEM led to lower performance when compared to modality-specific models.
Performance metrics
In all test sets, sensitivity was high (>93%, see Table 3), indicating good capability to detect true positive axons. Lower performance metrics obtained in the human SEM sample are expected, as the human sample used exhibits different contrast/quality/noise properties when compared to the rat training set. Note that myelin sheaths of the macaque TEM sample are slightly underestimated when compared to the ground truth segmentation. In both models and all test samples, computed myelin Dice was lower than axon Dice. This could be explained by the fact that myelin objects have two interfaces: boundary ambiguity between myelin and axon, and boundary ambiguity between myelin and background. Therefore, the myelin Dice is affected by two types of myelin misclassifications: myelin pixel classified as axon or myelin pixel classified as background.
Overall, these results suggest that the trained SEM and TEM models are robust to a variety of species and contrast changes and can generalize well, given that the lowest pixel-wise accuracy observed was 81% (see Table 3). Similar work done on optical microscopy data26 have achieved a maximal pixel-wise accuracy of 82%. As pointed out in Fig. 5, most pixel misclassifications are due to ambiguous/untypical axon and/or myelin structure or intensity distribution. Note that these discrepancies could possibly be solved by implementing post-processing methods based on mathematical morphology or conditional random fields.
Morphometrics extraction
Morphological metrics were extracted from a full rat spinal cord slice at the cervical level (see Fig. 7). The metrics resulting from the segmentation are overall consistent with the known anatomy. The ventral spinothalamic tract (#3 in tract reference of Fig. 7) contains the largest axons34,36, while higher density and smaller axons are observed in the corticospinal tract (#12 in tract reference)35,36. Furthermore, the spinocerebellar tracts (#4 and #5 in tract reference) are mostly composed of large diameter fibers34. We also observe that axons in the cuneate fasciculus (#7 in tract reference) are larger than those found in the gracile fasciculus (#6 in tract reference), which is also in agreement with the literature37. G-ratio ranges between 0.5 and 0.75, which is in agreement with other rat microstructure studies38. Overall, concordance of metrics obtained with literature shows that AxonDeepSeg can serve as a tool to document distribution and size of myelinated fibers in microscopy samples.
Software
AxonDeepSeg is coded in Python and based on the TensorFlow deep learning framework. It can currently run on Linux and Mac OS X systems. Segmentation inference can be done on standard CPU computers at reasonable computational time. The code is available as open source in GitHub (https://github.com/neuropoly/axondeepseg) and an intuitive documentation is provided (https://neuropoly.github.io/axondeepseg/). A Binder link and a simple Jupyter notebook are available for getting started with AxonDeepSeg.
Future perspectives
The use of ensemble techniques, which consist of combining multiple neural network models, can potentially increase performance metrics. However, its drawback is that it increases computational time at inference. Another possible approach is to use transfer learning39 in order to obtain better generalization in new imaging modalities even when having a small training set. A partially trained model can be used as starting point for the training of another model of different modality. Note that AxonDeepSeg has been trained and tested on healthy tissues. It would be interesting to assess its performance on demyelinated microscopy samples, in which myelin sheaths might present smaller thickness and different morphology.
Even though current models are already performant, our long-term goal is to continuously improve these models by adding more training data from collaborators in order to improve generalization. Another objective is to build segmentation models for other modalities, such as optical microscopy and Coherent Anti-Stokes Raman spectroscopy (CARS). This vision is supported by the recent initiative of creating a White Matter Microscopy Database40, which provides to the community an open access microscopy data and associated labeled ground truth. We encourage people to share their data for fostering the development of performant segmentation methods.
References
Zoupi, L., Savvaki, M. & Karagogeos, D. Axons and myelinating glia: An intimate contact. IUBMB Life 63, 730–735 (2011).
Seidl, A. H. Regulation of conduction time along axons. Neuroscience 276, 126–134 (2014).
Lassmann, H. Mechanisms of white matter damage in multiple sclerosis. Glia 62, 1816–1830 (2014).
Papastefanaki, F. & Matsas, R. From demyelination to remyelination: the road toward therapies for spinal cord injury. Glia 63, 1101–1125 (2015).
Sachs, H. H., Bercury, K. K., Popescu, D. C., Narayanan, S. P. & Macklin, W. B. A new model of cuprizone-mediated demyelination/remyelination. ASN Neuro 6 (2014).
Pfeifenbring, S., Nessler, S., Wegner, C., Stadelmann, C. & Brück, W. Remyelination After Cuprizone-Induced Demyelination Is Accelerated in Juvenile Mice. J. Neuropathol. Exp. Neurol. 74, 756–766 (2015).
Stikov, N. et al. In vivo histology of the myelin g-ratio with magnetic resonance imaging. Neuroimage 118, 397–405 (2015).
Saliani, A. et al. Axon and Myelin Morphology in Animal and Human Spinal Cord. Front. Neuroanat. 11, 129 (2017).
Romero, E. et al. Automatic morphometry of nerve histological sections. J. Neurosci. Methods 97, 111–122 (2000).
Cuisenaire, O., Romero, E., Veraart, C. & Macq, B. M. M. Automatic segmentation and measurement of axons in microscopic images. In Medical Imaging1999: Image Processing 3661, 920–930 (International Society for Optics and Photonics, 1999).
More, H. L., Chen, J., Gibson, E., Donelan, J. M. & Beg, M. F. A semi-automated method for identifying and measuring myelinated nerve fibers in scanning electron microscope images. J. Neurosci. Methods 201, 149–158 (2011).
Liu, T., Jurrus, E., Seyedhosseini, M., Ellisman, M. & Tasdizen, T. Watershed Merge Tree Classification for Electron Microscopy Image Segmentation. Proc. IAPR Int. Conf. Pattern Recogn. 2012, 133–137 (2012).
Wang, Y.-Y., Sun, Y.-N., Lin, C.-C. K. & Ju, M.-S. Segmentation of nerve fibers using multi-level gradient watershed and fuzzy systems. Artif. Intell. Med. 54, 189–200 (2012).
Zhao, X., Pan, Z., Wu, J., Zhou, G. & Zeng, Y. Automatic identification and morphometry of optic nerve fibers in electron microscopy images. Comput. Med. Imaging Graph. 34, 179–184 (2010).
Bégin, S. et al. Automated method for the segmentation and morphometry of nerve fibers in large-scale CARS images of spinal cord tissue. Biomed. Opt. Express 5, 4145–4161 (2014).
Zaimi, A. et al. AxonSeg: Open Source Software for Axon and Myelin Segmentation and Morphometric Analysis. Front. Neuroinform. 10, 37 (2016).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet Classification with Deep Convolutional Neural Networks. in Advances in Neural Information Processing Systems 25 (eds Pereira, F., Burges, C. J. C., Bottou, L. & Weinberger, K. Q.) 1097–1105 (Curran Associates, Inc., 2012).
Ciresan, D. C., Meier, U., Gambardella, L. M. & Schmidhuber, J. Convolutional Neural Network Committees for Handwritten Character Classification. In 2011 International Conference on Document Analysis and Recognition 1135–1139 (2011).
Karpathy, A., Toderici, G., Shetty, S. & Leung, T. Large-scale video classification with convolutional neural networks. Proceedings of the (2014).
Simonyan, K. & Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv [cs.CV] (2014).
Long, J., Shelhamer, E. & Darrell, T. Fully convolutional networks for semantic segmentation. Proc. IEEE (2015).
Malon, C. D. & Cosatto, E. Classification of mitotic figures with convolutional neural networks and seeded blob features. J. Pathol. Inform. 4, 9 (2013).
Ciresan, D., Giusti, A., Gambardella, L. M. & Schmidhuber, J. Deep Neural Networks Segment Neuronal Membranes in Electron Microscopy Images. In Advances in Neural Information Processing Systems 25 (eds Pereira, F., Burges, C. J. C., Bottou, L. & Weinberger, K. Q.) 2843–2851 (Curran Associates, Inc., 2012).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 234–241 (Springer, Cham, 2015).
Naito, T. et al. Identification and segmentation of myelinated nerve fibers in a cross-sectional optical microscopic image using a deep learning model. J. Neurosci. Methods 291, 141–149 (2017).
Mesbah, R., McCane, B. & Mills, S. Deep convolutional encoder-decoder for myelin and axon segmentation. In 2016 International Conference on Image and Vision Computing New Zealand (IVCNZ) 1–6 (2016).
Jelescu, I. O. et al. In vivo quantification of demyelination and recovery using compartment-specific diffusion MRI metrics validated by electron microscopy. Neuroimage 132, 104–114 (2016).
He, K., Zhang, X., Ren, S. & Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. Proc. IEEE (2015).
Simard, P. Y., Steinkraus, D. & Platt, J. C. Best practices for convolutional neural networks applied to visual document analysis. ICDAR (2003).
Chen, L.-C., Papandreou, G., Schroff, F. & Adam, H. Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv [cs.CV] (2017).
Ioffe, S. & Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. in International Conference on Machine Learning 448–456 (2015).
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I. & Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014).
Kingma, D. P. & Ba, J. Adam: A Method for Stochastic Optimization. arXiv [cs.LG] (2014).
Kayalioglu, G. Chapter 10 - Projections from the Spinal Cord to the Brain. In The Spinal Cord 148–167 (Academic Press, 2009).
Watson, C. & Harvey, A. R. Chapter 11 - Projections from the Brain to the Spinal Cord. in The Spinal Cord 168–179 (Academic Press, 2009).
Schwartz, E. D. et al. Ex vivo evaluation of ADC values within spinal cord white matter tracts. AJNR Am. J. Neuroradiol. 26, 390–397 (2005).
Nunes, D., Cruz, T. L., Jespersen, S. N. & Shemesh, N. Mapping axonal density and average diameter using non-monotonic time-dependent gradient-echo MRI. arXiv [physics.med-ph] (2016).
Chomiak, T. & Hu, B. What is the optimal value of the g-ratio for myelinated fibers in the rat CNS? A theoretical approach. journals.plos.org (2009).
Oquab, M., Bottou, L., Laptev, I. Sivic -Proceedings of the IEEE, J. & Learning and transferring mid-level image representations using convolutional neural networks. cv-foundation.org (2014).
Cohen-Adad, J. et al. White Matter Microscopy Database., https://doi.org/10.17605/OSF.IO/YP4QG October 25 (2017).
Acknowledgements
The authors would like to thank Ariane Saliani and Tanguy Duval for helping with the acquisition of SEM data, Dr. Hugues Leblond for providing the human spinal cord sample, Nafisa Husein and Harris Nami for helping with the ground truth labelling of samples, Drs Adriana Romero Soriano and Yoshua Bengio (MILA - Montreal Institute for Learning Algorithms), and Dr. Robert Brown (McGill - Montreal Neurological Institute) for fruitful discussions on the network design, Drs. Nikola Stikov and Jennifer Campbell for sharing TEM data of macaque, and Dr. Els Fieremans for sharing TEM data of mice. The authors would also like to thank Compute Canada and Calcul Québec for access to computation units and the “NVIDIA Corporation” for offering a Tesla GPU. This study was funded by the Canada Research Chair in Quantitative Magnetic Resonance Imaging (JCA), the Canadian Institute of Health Research [CIHR FDN-143263], the Canada Foundation for Innovation [32454, 34824], the Fonds de Recherche du Québec - Santé [28826], the Fonds de Recherche du Québec - Nature et Technologies [2015-PR-182754], the Natural Sciences and Engineering Research Council of Canada [435897-2013], IVADO, TransMedTech and the Quebec BioImaging Network.
Author information
Authors and Affiliations
Contributions
A.Z., M.W. and J.C.A. wrote the paper. A.Z., M.W., V.H., P.L.A. and C.S.P. designed and developed the software. J.C.A. supervised the project and provided expert guidance. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zaimi, A., Wabartha, M., Herman, V. et al. AxonDeepSeg: automatic axon and myelin segmentation from microscopy data using convolutional neural networks. Sci Rep 8, 3816 (2018). https://doi.org/10.1038/s41598-018-22181-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-22181-4
This article is cited by
-
Machine learning enables non-Gaussian investigation of changes to peripheral nerves related to electrical stimulation
Scientific Reports (2024)
-
Convergence of machine learning with microfluidics and metamaterials to build smart materials
International Journal on Interactive Design and Manufacturing (IJIDeM) (2024)
-
DeepThink IoT: The Strength of Deep Learning in Internet of Things
Artificial Intelligence Review (2023)
-
Foretinib mitigates cutaneous nerve fiber loss in experimental diabetic neuropathy
Scientific Reports (2022)
-
Rapid, automated nerve histomorphometry through open-source artificial intelligence
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.