Abstract
While the identification of biomarkers for Alzheimer’s disease (AD) is critical, emphasis must also be placed on defining the relationship between these and other indicators. To this end, we propose a network-based radial basis function-sparse partial least squares (RBF-sPLS) approach to analyze structural magnetic resonance imaging (sMRI) data of the brain. This intermediate phenotype for AD represents a more objective approach for exploring biomarkers in the blood and cerebrospinal fluid. The proposed method has two unique features for effective biomarker selection. The first is that applying RBF to sMRI data can reduce the dimensions without excluding information. The second is that the network analysis considers the relationship among the biomarkers, while applied to non-imaging data. As a result, the output can be interpreted as clusters of related biomarkers. In addition, it is possible to estimate the parameters between the sMRI data and biomarkers while simultaneously selecting the related brain regions and biomarkers. When applied to real data, this technique identified not only the hippocampus and traditional biomarkers, such as amyloid beta, as predictive of AD, but also numerous other regions and biomarkers.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is the most common form of dementia and is a global problem, especially in developed countries, where the aging population is growing rapidly. Several biomarkers that signal the risk for AD development, such as apolipoprotein E (apoE) ε4 allele1,2, before the appearance of dementia symptoms, could play a crucial role in implementing early treatment after a pathological diagnosis of AD based on amyloid beta (Aβ) in the brain tissue3.
One of the most popular imaging techniques is structural magnetic resonance imaging (sMRI), which is widely used to examine properties of brain regions. The regression method is one potential way to explore biomarkers, such as hippocampal volume, as a univariate outcome4; however, sMRI can also be used to evaluate morphological changes in gray matter density, directly at the voxel level, to classify features related to AD5,6,7,8,9,10,11. While cognitive and clinical symptoms are the traditional phenotypes (outcomes) for AD, sMRI data are useful as a quantitative intermediate phenotype, which can be evaluated more objectively than traditional phenotypes12,13.
MRI data consist of image intensities of millions of voxels in a three-dimensional (3D) array. Since one voxel corresponds to one variable in statistical terms, MRI data are high dimensional. Existing neuroimaging analysis methods require regions of interest (ROI) to be pre-specified in the brain, mainly because of the computational difficulties involved in using a vast number of voxels, while a voxel-based analysis can be used for a more precise analysis. A limitation of this voxel-wise neuroimaging system includes sample sizes that are generally small in comparison with the high dimensionality of the data, making it challenging to define correlations between the response (Y) and predictive (X) variables. In order to determine the associations among these complex multivariate data sets, partial least squares (PLS), canonical correlation, reduced rank regression, and independent component analyses have been used to detect morphological abnormalities from sMRI data, in association with non-imaging markers8,10,14,15.
To analyze sMRI data as an intermediate phenotype and, in parallel, to explore non-imaging biomarkers from over 100 candidates, we have defined multivariate variables for both X and Y in a PLS regression model. In our previous study, we proposed the RBF-sPLS approach, which involves application of the radial basis function (RBF) method to 3D sMRI data as a pre-processing step for the sparse partial least squares (sPLS) approach for investigating the relationship between clinical characteristics and brain morphology16. Wolz et al.17 have proposed the nonlinear dimension-reduction method for such studies, although with a limited number of clinical characteristics.
In the present study, we wanted to reduce the dimensionality of non-imaging data as a predictive variable matrix of sPLS in the prediction model of brain morphology. Since the RBF method has the benefit of maintaining the original positional information on brain MRI, even after dimension reduction, it was possible to interpret a region of the brain by re-mapping it onto a brain map from the set of variables extracted after dimension reduction. This was possible as the approach involved a linear (or explicit) formulation as well as a nonlinear relationship, rather than being based solely on a nonlinear function. Furthermore, in this study, we also set out to reduce the dimensions of non-imaging biomarkers as a pre-processing step for the PLS model while simultaneously retaining the information on the relations among variables. However, since the non-imaging biomarkers that we used did not include structural information, such as location information in the brain image, which would have been useful for pre-process dimension reduction, we applied the network-clustering method, which is useful for clarifying structure. Network-based approaches have emerged as powerful tools for studying complex disease models18.
To adapt the statistical method for this study, we 1) constructed a network for non-imaging biomarkers (CSF and blood biomarkers) and 2) selected components to simplify the interpretation of diagnostic information. First, the entire network was broken down into sub-networks based on graph theory. For the sPLS algorithm, the summarized values of sub-networks from non-imaging biomarkers were used as X, while radial basis functions were applied to brain imaging data and the resulting values were defined as Y. Although most previous studies using PLS analysis selected the first few components only, we here selected components predictive of disease. For this purpose, we specifically used the score of the PLS as the predictor and the disease status as the response in logistic regression analysis. Thus, the selected components can be interpreted as pairs of brain regions and biomarker networks associated with the AD stage.
The goal of this study was to investigate biomarkers associated with AD using MRI as well as non-imaging data, including the results of blood and CSF sample analyses. To this end, we examined two types of dimension-reduction methods as a pre-procedure for sPLS: the radial basis function method and network-based clustering method for X and Y, respectively. From the components extracted by the sPLS procedure, we selected certain components to define brain regions and non-imaging biomarkers associated with the incidence of AD, using logistic regression models.We show that the proposed method is particularly useful for achieving dimension reduction, based on typical real-life data.
Methods and Materials
Procedure of Network based RBF-sPLS
The network based RBF-sPLS consisted of three steps, as follows:
step 1: Dimension reduction procedure of X and Y, respectively
step 2: Sparse PLS
step 3: Selection of components relevant to AD.
All statistical analyses were performed in R version 3.2.5 by using the following analysis packages: for description of the network graph, [igraph (0.6.5–2)] and [gRapHD (0.2.3)]; for construction of network clustering, [linkcomm (1.0–11)]; for calculation of centralization of clusters, [sna (2.3–2)]; and for multiple imputations for missing data [random Forest (4.6–7)].
RBF: Pre-processing for MRI Data Analysis
We used the Statistical Parametric Mapping 8 software (SPM8, Wellcome Department of Cognitive Neurology, London, UK) and the VBM8 toolbox (http://dbm.neuro.uni-jena.de/vbm/download/), running under the MATLAB environment (MathWorks, Natick, MA), to pre-process MR brain images. Three-dimensional T1-weighted images of a subject were first segmented into gray matter, white matter, and CSF space, followed by anatomical normalization to a template image by using DARTEL19. The normalized image was finally smoothed with an 8 mm FWHM isotropic Gaussian filter.
Dimension Reduction based on Network Clustering for Non-Imaging Data Analysis
As one important facet of this study, we incorporated structural information of non-imaging data, based on network modeling, which might be helpful for interpreting results. This can also reduce the dimensionality of non-imaging data. We performed three steps to achieve this. First, with each non-imaging variable as a vertice (node) of the graph, the network whose edges represented statistical dependence through the decomposition of the joint probability density was estimated using graphical modeling. Second, the vertices of the estimated network were clustered based on similarity in the framework of graphical theory. This is referred to as network clustering, based on a recently developed overlapping clustering approach; Ahn et al.20 performed pioneering work on overlapping clustering, and Becker et al.21 developed a new algorithm with their proposed modularity. We used the R package [linkcomm] developed by Kalinka and Tomancak22 to implement the method proposed by Becker et al.21. Third, in order to reduce the dimensionality of the non-imaging data after the clustering procedure, we selected one representative node among each sub-cluster. If more than three nodes belonged to a cluster, we calculated centralization to select the highest information centrality node in each network. This was implemented in the R package [sna]. If only two nodes belonged to one cluster, we calculated the variance of each variable and selected the one with the larger variance as the representative variable of the network. The number of sub-clusters were noted p. Thus, we obtained the dimension-reduced n × p predictive matrix X for sPLS.
Sparse PLS
The PLS regression technique, which was introduced by Wold in 198623, searches for a set of components by using latent variables, and performs a simulation decomposition of X and Y, with the constraint that these components should explain as much of the covariance between X and Y as possible. This was followed by a linear regression step, in which the decomposition of X is used to predict Y.
In a previous study, we reported on the advantages of RBF by using a simulation method and its application in actual clinical datasets16. In this study, we also applied RBF for preprocessing brain data (Y0). We used the radial B-spline function \(\varphi (\cdot )\)24 to reduce the dimensions, which is represented as follows. For a given g ≥ 0,
where d ≥ 0. We used the distance between these knots to define g as \(g=\sqrt{3\times {g}_{0}^{2}}\), where \({g}_{0}\) is the distance between adjacent knots. Therefore, \({\boldsymbol{B}}=\varphi (d)\), \({{\boldsymbol{Y}}}_{0}={({{\boldsymbol{Y}}}_{01},\ldots ,{{\boldsymbol{Y}}}_{0n})}^{T}\) and the dependent variable matrix, Y, is constructed as
The n × q dimension reduction matrix Y was defined with the brain image data matrix as the response variable for the sPLS regression analysis.
We defined the response Y and the predictor X as follows: the MRI data-reduced dimension by RBF was Y, and the representative variables selected as the most central feature of each cluster of the non-imaging data, such as CSF and demographic data, were X. In PLS regression, the criterion used involved determining the Y-score vector, \({\boldsymbol{u}}={\boldsymbol{Yw}}\), and the X-score vector, \({\boldsymbol{t}}={\boldsymbol{Xv}}\). The weights w and v were estimated L(v, w) in two subjects to \(\Vert {\boldsymbol{v}}{\Vert }_{2}=\Vert {\boldsymbol{w}}{\Vert }_{2}=1\).
where \({\lambda }_{{\boldsymbol{X}}}\) and \({\lambda }_{{\boldsymbol{Y}}}\) are L1 penalization parameters for the weight vectors of matrices X and Y, respectively. The amplitudes of \({\lambda }_{{\boldsymbol{X}}}\) and \({\lambda }_{{\boldsymbol{Y}}}\) correspond to increases and decreases in the number of X and Y variables, which contribute to the regression. The algorithm for the computation of weight vectors v and w is as follows:
-
1)
Initialize v and w using, for instance, the first pair of singular vectors of the matrix \({{\boldsymbol{X}}}^{T}{\boldsymbol{Y}}\) and normalize \({\boldsymbol{v}}\leftarrow {\boldsymbol{v}}/\Vert {\boldsymbol{v}}{\Vert }_{2}\) and \({\boldsymbol{w}}\leftarrow {\boldsymbol{w}}/\Vert {\boldsymbol{w}}{\Vert }_{2}\).
-
2)
Until convergence of v and w with \({h}_{\lambda }(y)=sign(y){(|y|-\lambda )}_{+}\), where \({(a)}_{+}=max(0,a)\).
-
(a)
for fixed \({\boldsymbol{w}},\hat{{\boldsymbol{v}}}={h}_{{\lambda }_{{\boldsymbol{X}}}}({{\boldsymbol{X}}}^{T}{\boldsymbol{Y}}{\boldsymbol{w}})\) and then normalize \(\hat{{\boldsymbol{v}}}\) as in step 1;
-
(b)
for fixed v, \(\hat{{\boldsymbol{w}}}={h}_{{\lambda }_{{\boldsymbol{Y}}}}({{\boldsymbol{Y}}}^{T}{\boldsymbol{X}}{\boldsymbol{v}})\) and then normalize \(\hat{{\boldsymbol{w}}}\) as in step 1;
-
(c)
\({\boldsymbol{v}}=\hat{{\boldsymbol{v}}}\), \({\boldsymbol{w}}=\hat{{\boldsymbol{w}}}\).
-
(a)
-
3)
\({\boldsymbol{t}}={\boldsymbol{Xv}}\), \({\boldsymbol{\ell }}={{\boldsymbol{X}}}^{T}t/{t}^{T}t\), and \({\boldsymbol{m}}={{\boldsymbol{Y}}}^{T}t/{t}^{T}t\).
-
4)
The deflation step \({\boldsymbol{X}}\leftarrow {\boldsymbol{X}}-t{{\boldsymbol{\ell }}}^{T}\) and \({\boldsymbol{Y}}\leftarrow {\boldsymbol{Y}}-t{{\boldsymbol{m}}}^{T}\).
By repeating 1) to 4), k-th component scores and weights are obtained at the k-th repetition. The choice of penalization parameters \({\lambda }_{{\boldsymbol{X}}}\), \({\lambda }_{{\boldsymbol{Y}}}\) and number of components k are important in model construction. We fixed the number of components k = 10 and used a cross-validation technique with a prediction error sum of squares; PRESS jk using the following equation:
This was applied for the jth-dependent variable and the RBF-sPLS model with k components defined as follows. Let κ: {1, 2, …, n}→{1, 2, …, 5} be an indexing function that indicates the partition to which observation i is allocated to \(\kappa (i)\,\)th part of the data by the randomization. \({\hat{y}}_{(-\kappa (i))j}({\lambda }_{{\boldsymbol{X}}},{\lambda }_{{\boldsymbol{Y}}},k)\) is the predicted value for the jth dependent variable from the sPLS model with penalization parameters \({\lambda }_{{\boldsymbol{X}}}\) and \({\lambda }_{{\boldsymbol{Y}}}\) and the number of components k and estimated weight vectors from \(\kappa (i)\,\)th part of the data removed. That is, for any i subject, we predict that \({\hat{y}}_{(-\kappa (i))j}({\lambda }_{{\boldsymbol{X}}},{\lambda }_{{\boldsymbol{Y}}},k)={x}_{i}{\hat{b}}_{(-\kappa (i))j}({\lambda }_{{\boldsymbol{X}}},{\lambda }_{{\boldsymbol{Y}}},k)\), where \({\hat{b}}_{(-\kappa (i))j}({\lambda }_{{\boldsymbol{X}}},{\lambda }_{{\boldsymbol{Y}}},k)\) is the jth column of estimated regression coefficient matrix C from the RBF-sPLS model with penalization parameters \({\lambda }_{{\boldsymbol{X}}}\) and \({\lambda }_{{\boldsymbol{Y}}}\) and number of components k and \(\kappa (i)\,\)th part of the data removed. We selected the optimal set \(({\lambda }_{{\boldsymbol{X}}},{\lambda }_{{\boldsymbol{Y}}})\) based on the binary search with \(PRES{S}_{j}({\lambda }_{{\boldsymbol{X}}},{\lambda }_{{\boldsymbol{Y}}},k)\) as the objective function.
Component Selection
In normal PLS analyses, the number of components k is selected with the first k components using criteria such as the cross-validation error. These criteria, for example Q2 as proposed by Lê Cao25, can estimate k (the number of PLS components). In the present study, we opted for setting 10 components as k with reference to our experience and based on previous reports. If this analysis was applied, subcomponents of X and Y would be related within each component, but they would not be related to the outcome (i.e., they would be independent of whether AD is present). To address this, we applied a backward stepwise-selection based on the multivariable logistic regression model with the \(n\times k\) score matrix T of the PLS algorithm as predictors and the AD diagnosis at baseline as the response variable. The Bayesian information criterion (BIC) was used to select the optimal model corresponding to the selected components. Thus, the obtained components are the set of brain regions and networks of non-imaging biomarkers relevant to a diagnosis of AD.
Application to ADNI Data
Blood/CSF Biomarkers and Brain Imaging Data of ADNI
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data, but did not participate in analysis or writing of this report. The ADNI was launched in 2003 by a 5-year public-private partnership of several research institutions and private pharmaceutical companies.
This ADNI project was approved by the Institutional Review Boards of all the participating institutions. Informed written consent was obtained from all participants at each site. The ADNI database includes results from various types of imaging examinations and from clinical and neuropsychological assessments. Accordingly, meaningful results can likely be obtained by compiling this information to measure the progression of mild cognitive impairment (MCI) and early AD.
To analyze brain imaging data, we used T1-weighted MR images from the ADNI1 database. MRI acquisition had been performed according to the ADNI acquisition protocol26. CSF, proteomic, and blood-sample biomarkers obtained via peripheral veins and these, along with vital signs were defined as non-imaging biomarkers. First, we screened 398 non-imaging biomarkers in AD, MCI, and normal control (NC) individuals, as defined by ADNI criteria. In a number of cases, participants refused to give consent for the more invasive examinations, such as lumbar puncture; thus, CSF markers were not available for all, particularly in asymptomatic cases. We applied the “random forest” method27, which replaces missing values by using the R package [randomForest] and created a new data set (X0) by excluding cases with missing items in the examinations. The first step of the rfImpute method creates various subsets randomly using bootstrap methods, while the proximity matrix from these subsets is used to update the imputation of missing values.
Numerical Evaluation
We used an already well-studied dataset for the evaluation. In addition, we numerically evaluated the efficacy and stability of the result focusing on the network-based cluster analysis, which was newly applied in our analysis. Since this cluster analysis was used as the dimension-reduction method for the sPLS analysis, in order to focus on the effect of the clustering algorithm, we simplified the sPLS using a univariate response (the ratio of the hippocampal volume to the intra cranial volume). This corresponds to multiple regression analysis with representative variables of each cluster group in X extracted by each clustering as independent variables. This multiple regression analysis with L1 penalization and backward stepwise variable selection was repeated 1000 times using the resampling technique, and we calculated adjusted r-squared values and mean-squared error (MSE) from bootstrap samples that did not contain that observation for each regression model. The mean r squared values and MSE among repeats were compared among the three clustering algorithms: network clustering (our approach), hierarchical clustering28, and mixture distribution model29. These methods have been applied for some biomarker studies30,31.
Results
Pre-processing for MRI Data and Non-Imaging Biomarkers
In the present study, 534 baseline MRI scans at 1.5T were downloaded from the ADNI database for evaluation in March 2013; these consisted of 104 AD and 430 non-AD (NC + MCI) scans. The demographical baseline data of all subjects are presented in Table 1. First, we examined the dimension reduction procedure with respect to the response variable. Out of the 2,122,945 (121 × 145 × 121) voxels for each subject, all voxels representing grey matter were extracted, resulting in 839,089 voxels. The dimension of the basis function was q = 7,176, because the knots were equally distributed with a 4-voxel interval (h0 = 4; therefore, \(h=\sqrt{3\,\times \,{4}^{2}}=6.93\)). Thus, the \(n\times q\) brain data matrix Y was created.
A total of 398 non-imaging blood and CSF biomarkers, including the apoE ε4 allele, were screened as covariates (p0 = 398) and 209 clusters were constructed. The network for each cluster was constructed from extensively overlapping nodes. The largest network cluster grew to 12 vertices (variables) and the smallest one grew to two vertices. This network is illustrated in Fig. 1; nodes from different networks are represented by different colors. We created a new data set with 165 (the notation was p) representative variables as the center variable of each cluster, after the overlapping variables were combined into one. Thus, the \(n\times p\) biomarker matrix X was defined.
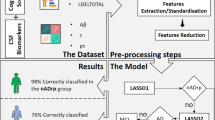
Schematic diagram of the study. We applied a network-based RBF-sPLS to the ADNI database. RBF was applied for dimension reduction of neuroimaging data, and network clustering was similarly applied for non-imaging biomarkers. Notations: k, number of components selected by sPLS; r, number of components selected by a multiple logistic regression analysis in their final forms.
Sparse Partial Least Squares and Component Selection
Next, we estimated the Q2 criterion25, which was defined by the residual and prediction error sum of squares in the PLS model and indicated the correlations between sub-components of X and Y. According to Q2 criterion, we found that the number of optimal PLS components was 2. However, these 2 components were not necessarily related to AD and could not to identify components relevant to AD, because the sPLS procedure did not utilize the AD diagnosis information. Thus, to select components relevant to AD from a larger number of components by means of logistic regression, we selected 10 sets of initial components as a complementary analysis to the PLS. We assessed how the results changed according to the number of initial components, namely a sensitivity analysis was conducted (Supplemental Fig. 1). For the sPLS algorithm, the optimal \({\lambda }_{{\boldsymbol{X}}}\) and \({\lambda }_{{\boldsymbol{Y}}}\) were chosen (\({\lambda }_{{\boldsymbol{X}}}=0.123\). \({\lambda }_{{\boldsymbol{Y}}}=0.012\)). Each component contained 547 to 1154 non-zero voxels and one to seven biomarkers. Of note, brain images were represented as “regions” comprised of neighboring voxels, and components of X were represented as a set of biomarkers constructed from the network. To select the components relevant to AD, we applied a logistic regression analysis using the score of each Y’s component (the notation was u) as explanation variables, and diagnosis (AD = 1, not AD = 0) as a response variable.
Three components were selected as relevant sets of brain regions and biomarkers for AD (Fig. 2a–c) by logistic regression, using BIC. The third of the first set of 10 components corresponded to both hippocampi, as well as to the apoE ε4 allele (indicated as #8 and #5 in Table 2 and Fig. 2a) and 14 other protein markers: phosphorylated tau 181P (#1), total tau (#2), apoE protein (#3), heart fatty acid-binding protein (#4), chitinase 3-like 1 (#6 and #7), gamma enolase (#9), pyruvate kinase M1/M2 (#10), peroxiredoxin-2 (#11 and #12), percent of neutrophils (#13), glial fibrillary acidic protein (#14), and body mass index (#15). These biomarkers were the representative variables for each cluster. At least one biomarker was present among these variables in each cluster (Table 2). In addition, two other brain regions and a set of non-imaging biomarkers linked with these brain regions were selected (Fig. 2b,c; Supplemental Table 2a,b, #16 to #58).
Three components selected by RBF-sPLS. (a) The most significant component (p value = 4.19 × 10−15) for Alzheimer’s disease, as identified by a logistic regression analysis, were derived from 10 pairs of brain regions and biomarkers extracted using RBF-sPLS. The red color on the brain images indicates significant atrophied areas, especially the hippocampi and left temporal lobe. The black circles on the network graph indicate representative variables of this component, and the gray circles indicate the nodes correlated with these representative variables. The numbers (#) on the network area match the numbers in Table 2. (b,c) The second and third most significant components (p value = 6.34 × 10−4 for b, p value = 1.41 × 10−4 for c) for Alzheimer’s disease are displayed analogous to (a). The numbers (#) on the network area match the numbers in Supplemental Table 1a,b.
The Efficacy of the Network-based RBF-sPLS Strategy
In order to show the benefits of our method, we compared three variations of clustering and dimension-reduction methods using a resampling technique, as a sensitivity analysis. The result of repeating 1,000 times in each clustering method, the median of the r-squared value was larger in our proposed method than in hierarchical clustering and mixture distribution model (Supplemental Fig. 2). These r-squared values indicated how well the representative variables, selected from each clustering method, fit the regression model as independent variables. The MSEs from bootstrap samples not containing that observation for each regression model, i.e., the distance between an estimator and the true underlying parameter, were sufficiently small and were less than those obtained by two other methods. Thus, more useful variables for the regression model were selected in our network clustering-based dimension-reduction method.
Discussion
The present study describes the application of the RBF-sPLS technique to performing a high-dimensional regression analysis, to assess correlations between brain MRI data and non-imaging biomarkers, from the ADNI database. Our proposed method, the network-based RBF-sPLS, applies two dimension-reduction methods: RBF and network clustering, as a pre-procedure for PLS. The advantage of these two dimension-reduction methods is that it allows the results from PLS to be more interpretable.
The strengths of the present study are as follows. First, we have newly applied the network-based RBF-sPLS technique for a high-dimensional regression analysis. We reduced the number of parameters without losing information about the correlation between non-imaging biomarkers using a network-clustering method, while preserving the advantages of RBF-sPLS that have been previously reported. In the present study, our proposed method showed results with high interpretability for AD. Although not assessed in detail, our method may be more stable in terms of a defined representative value for the predicting model than classical clustering methods, such as hierarchical clustering. Second, since we defined the network of biomarkers, the resulting components (i.e., the pairs of brain images and non-imaging biomarkers) can be interpreted not only based on the representative biomarkers used in the PLS model, but also by other biomarkers related to these representative biomarkers. Thus, this method would unearth a great deal of information for medical consideration. In addition, the traditional biomarkers, such as the apoE ε4 allele, tau, and several other proteins, were included as components significantly predicting AD. The network clustering suggests an indirect correlation between AD and these proteomic variables. Accordingly, through this strategy, it may be possible to identify the real markers for a disease that is initially diagnosed by a surrogate marker.
There are several limitations in the present study. First, not all of the participants’ baseline data from ADNI were used. This is primarily because samples from invasive examinations, such as lumbar puncture, were often not obtained. However, ADNI provides various different strategies for estimating missing data. A few methods for replacing missing values as block units have recently been proposed32,33. Therefore, in future studies, we will attempt to implement this technique to complete the database. Further, we focused on only the difference between AD and not-AD (MCI + NC), for the purpose of evaluating the approach. We did not attempt to distinguish between other combinations of AD stages (i.e. AD vs. MCI, MCI vs. NC). Our method could be extended to a multinomial response model, such as AD vs. MCI vs. NC according to a clinical interest, but this is the different scope of this paper. We will address this in future.
Second, this study employed a cross-sectional analysis. We relied on diagnoses that were based on tests of cognitive function at the time of screening, but a definitive diagnosis based on pathology was not available. Thus, we cannot exclude the possibility that the diagnoses used in this study might include misclassifications. The ADNI database also contains the following longitudinal data: ADNI1, ADNI2, and ADNIGO, which are valuable databases, because they contain long-term observations over the course of as much as 10 years. While we did not perform a longitudinal analysis, the RBF-sPLS technique could be applied effectively to such an analysis, thereby including a time-dependent analysis of the progression of AD, from onset or MCI to AD.
Third, we need to consider the method used to define the representative variables for each network and the method used to select the number of initial components for sPLS. After testing several methods for this study, the representative variables were defined using information centrality, and we extracted 10 sPLS components for logistic analysis. Because the purpose of present study was to investigate the effect of dimension reduction using a network model, the methods by which to select the representative variables and the number of sPLS components were out of the scope of the study. However, in future studies we plan to use the relationship described by the degrees between the network and the diagnosis, according to graph theory. Thereafter, we would like to investigate the robustness of our proposed method as only the minimal metric experiment was performed in the present study.
Conclusions
We here applied a network-based RBF-sPLS technique to identify several brain regions and non-imaging biomarkers that were related to AD, simultaneously.
References
de Leon, M. J. et al. Imaging and CSF studies in the preclinical diagnosis of Alzheimer’s disease. Ann N Y Acad Sci 1097, 114–145 (2007).
Shaw, L. M. et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol 65(4), 403–413 (2009).
Jack, C. R. Jr et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol 9(1), 119–128 (2010).
Tsuruya, K. et al. Clinical significance of fronto-temporal gray matter atrophy in executive dysfunction in patients with chronic kidney disease: The VCOHP study. Plos One 10(12), e0143706 (2015).
Gerardin, E. et al. Multidimensional classification of hippocampal shape features discriminates Alzheimer’s disease and mild cognitive impairment from normal aging. Neuroimage 47(4), 1476–1486 (2009).
Wolz, R. et al. Measurement of hippocampal atrophy using 4D graph-cut segmentation: application to ADNI. Neuroimage 52(1), 109–118 (2010).
Cuingnet, R. et al. Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage 56(2), 766–781 (2011).
Liu, M., Zhang, D. & Shen, D. Alzheimer’s Disease Neuroimaging Initiative. Ensemble sparse classification of Alzheimer’s disease. Neuroimage 60(2), 1106–1116 (2012).
Cho, Y., Seong, J. K., Jeong, Y. & Shin, S. Y. Alzheimer’s Disease Neuroimaging Initiative. Individual subject classification for Alzheimer’s disease based on incremental learning using a spatial frequency representation of cortical thickness data. Neuroimage 59(3), 2217–2230 (2012).
Wee, C. Y., Yap, P. T. & Shen, D. Alzheimer’s Disease Neuroimaging Initiative. Prediction of Alzheimer’s disease and mild cognitive impairment using cortical morphological patterns. Hum Brain Mapp 34(12), 3411–3425 (2013).
Eskildsen, S. F. et al. Prediction of Alzheimer’s disease in subjects with mild cognitive impairment from the ADNI cohort using patterns of cortical thinning. Neuroimage 65, 511–521 (2013).
Meyer-Lindenberg, A. & Weinberger, D. R. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci 7(10), 818–827 (2006).
Rasetti, R. & Weinberger, D. R. Intermediate phenotypes in psychiatric disorders. Curr Opin Genet Dev 21(3), 340–348 (2011).
Chu, C. et al. Does feature selection improve classification accuracy? Impact of sample size and feature selection on classification using anatomical magnetic resonance images. Neuroimage 60, 59–70 (2012).
Tong, T. et al. Multiple instance learning for classification of dementia in brain MRI. Med Image Anal 18(5), 808–818 (2014).
Yoshida, H., Kawaguchi, A. & Tsuruya, K. Radial basis function-sparse partial least squares for application to brain imaging data. Comput Math Methods Med 2013, 591032 (2013).
Wolz, R. et al. Nonlinear dimensionality reduction combining MR imaging with non-imaging information. Med Image Anal 16(4), 819–830 (2012).
Cho, D. Y., Kim, Y. A. & Przytycka, T. M. Network biology approach to complex diseases. Plos Comput Biol 8(12), e1002820 (2012).
Ashburner, J. & Friston, K. J. Voxel-based morphometry–the methods. Neuroimage 11(6 Pt 1), 805–821 (2000).
Ahn, Y. Y., Bagrow, J. P. & Lehmann, S. Link communities reveal multiscale complexity in networks. Nature 466(7307), 761–764 (2010).
Becker, E., Robisson, B., Chapple, C. E., Guénoche, A. & Brun, C. Multifunctional proteins revealed by overlapping clustering in protein interaction network. Bioinformatics 28(1), 84–90 (2011).
Kalinka, A. T. & Tomancak, P. linkcomm: an R package for the generation, visualization, and analysis of link communities in networks of arbitrary size and type. Bioinformatics 27(14), 2011–2012 (2011).
Wold, H. Estimation of principal components and related models by iterative least squares. In: Multivariate Analysis (Krishnaiah, P. R., ed.). New York, NY: Academic, 391–420 (1966)
Saranli, A. & Baykal, B. Complexity reduction in radial basis function (RBF) networks by using radial B-spline functions. Neurocomputing 18(1-3), 183–194 (1998).
Lê Cao, K. A., Rossouw, D., Christèle, R. G. & Besse, P. A sparse PLS: variable selection when integrating omics data. Stat Appl Gen Mol Biol 7(1), 35 (2008).
Jack, C. R. Jr et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 27(4), 685–691 (2008).
Breiman, L. Random forests. Machine Learning 45.1, 5–32 (2001).
Johnson, S. C. Hierarchical clustering schemes. Psychometrika 32(3), 241–254 (1967).
McLachlan, G. J., Basford, K. E. Mixture Models: Inference and Applications to Clustering. New York, NY: Marcel Dekker (1988).
Diez, I. et al. A novel brain partition highlights the modular skeleton shared by structure and function. Sci Rep. 5, 10532 (2015).
Spriensma, A. S., Hajos, T. R., de Boer, M. R., Heymans, M. W. & Twisk, J. W. A new approach to analyse longitudinal epidemiological data with an excess of zeros. BMC Med Res Methodol 13, 27 (2013).
Xiang, S. et al. Bi-level multi-source learning for heterogeneous block-wise missing data. Neuroimage 15, 192–206 (2014).
Thung, K. H., Wee, C. Y., Yap, P. T. & Shen, D. Alzheimer’s Disease Neuroimaging Initiative. Neurodegenerative disease diagnosis using incomplete multi-modality data via matrix shrinkage and completion. Neuroimage 1(91), 386–400 (2014).
Acknowledgements
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data, but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. To accomplish this study, we used the supercomputer of ACCMS, Kyoto University. This study was supported by a Grant-in-Aid for Scientific Research on Innovative Areas (Comprehensive Brain Science Network) from the Ministry of Education, Science, Sports, and Culture of Japan. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense Award Number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann−La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. We would like to thank Editage (www.editage.jp) for English language editing.
Author information
Authors and Affiliations
Contributions
Research idea and study design: H.Y., A.K.; data acquisition: H.Y., F.Y.; data analysis interpretation: H.Y., A.K., K.T.; statistical analysis: H.Y., A.K.; supervision or mentorship: A.K., K.T. Each author contributed important intellectual content during manuscript drafting or revision and accepts accountability for the overall work by ensuring that questions pertaining to the accuracy or integrity of any portion of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yoshida, H., Kawaguchi, A., Yamashita, F. et al. The utility of a network–based clustering method for dimension reduction of imaging and non-imaging biomarkers predictive of Alzheimer’s disease. Sci Rep 8, 2807 (2018). https://doi.org/10.1038/s41598-018-21118-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-21118-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.