Abstract
Early pregnancy is a critical time for successful reproduction; up to half of human pregnancies fail before the development of the definitive chorioallantoic placenta. Unlike the situation in eutherian mammals, marsupial pregnancy is characterised by a long pre-implantation period prior to the development of the short-lived placenta, making them ideal models for study of the uterine environment promoting embryonic survival pre-implantation. Here we present a transcriptomic study of pre-implantation marsupial pregnancy, and identify differentially expressed genes in the Sminthopsis crassicaudata uterus involved in metabolism and biosynthesis, transport, immunity, tissue remodelling, and uterine receptivity. Interestingly, almost one quarter of the top 50 genes that are differentially upregulated in early pregnancy are putatively involved in histotrophy, highlighting the importance of nutrient transport to the conceptus prior to the development of the placenta. This work furthers our understanding of the mechanisms underlying survival of pre-implantation embryos in the earliest live bearing ancestors of mammals.
Similar content being viewed by others
Introduction
While eutherian mammals primarily nourish their embryos via a placenta, a key feature of marsupial reproduction is a very short period of placentation during a short gestation, followed by an extended investment in lactation1. In eutherians, the embryo becomes closely apposed to the uterine epithelium, before implanting into the uterine tissue very early in pregnancy to form the placenta e.g.2,3,4,5. In contrast, marsupial implantation and placentation do not occur until at least two thirds of the way through pregnancy, making marsupials ideal models for studying the uterine environment required for survival of the mammalian early embryo. In marsupials, the embryo remains unattached within the uterine lumen for most of pregnancy, and is reliant on uterine secretions for nutrient supply4,6. The conceptus is coated in several layers, including a tough outer shell coat secreted by the epithelial cells and endometrial glands of the utero-tubal junction and cranial part of the uterus7,8. The shell coat persists until implantation, and is permeable to gases and other small molecules of up to 40 kDa in size, permitting histotrophic nutrition9. The shell coat may also prevent maternal immune attack of the embryo8.
At implantation, the embryo hatches from the shell coat, enabling placentation through direct contact between the trophoblast and the receptive maternal uterine epithelium3,10. Placentation in marsupials has been well-studied from morphological e.g.5,11,12, physiological e.g.13,14 and genetic e.g.15,16,17 perspectives. In contrast, pre-implantation marsupial pregnancy has received much less attention, particularly from genetic studies, which have focused on the immunological changes in the uterus15,18. Understanding the complete physiology of pre-implantation marsupial pregnancy is important, because this period represents the majority of gestation, when the embryo is growing and undergoing early organogenesis19. The physiology of this period of mammalian pregnancy is an important area of medical research e.g.20, due to the high rate of human pregnancy failure [~40–50% of human pregnancies are lost before 20 weeks, 75% of which have been attributed to implantation failure21]. Failure to implant is also a major impediment to assisted reproductive technologies such as IVF21. As successful establishment of pregnancy requires both a healthy conceptus and a receptive uterus, information about both the maternal and the embryonic components during mammalian pregnancy is required to fully understand implantation22.
In this study, we describe the uterine transcriptome of the model marsupial Sminthopsis crassicaudata (fat-tailed dunnart) in the period of pre-implantation uterine receptivity. The fat-tailed dunnart has a very brief (13.5 day) pregnancy23. Prior to implantation, which occurs around day 10 of pregnancy, the conceptus lies closely apposed to maternal tissues within folds of the uterine epithelium8,24,25. Subsequently, a yolk sac placenta forms, which erodes part of the maternal epithelium but does not breach maternal capillaries i.e. endotheliochorial placentation3. As the pre-implantation shelled embryo spends twice as long in the uterus as the period of placental attachment, modifications of the uterine environment for efficient gas, nutrient and waste transport must occur during the pre-implantation phase early in pregnancy. The ultrastructural modifications to cell-cell adhesion in the early pregnant S. crassicaudata uterus are possibly related to these functional requirements12,26,27. Here, we describe the uterine pre-implantation transcriptome in S. crassicaudata and identify the broad genetic underpinnings of maternal maintenance of the early marsupial conceptus during pregnancy. We focus on identifying the genes underpinning nutrient transport, which we hypothesise are critical in nourishing the developing embryo prior to the formation of the placenta.
Results
Transcriptome sequencing and annotation
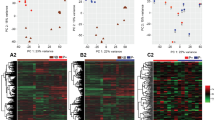
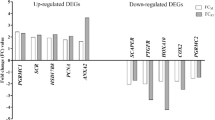
Our transcriptome sequencing recovered ~29–35 million paired reads from each of 3 pregnant (days 6–8 of pregnancy) and 3 non-pregnant dunnart uteri. After normalisation, 50.7 million reads were assembled into 234,671 transcripts from 136,066 ‘genes’ using Trinity28. The longest was 25,519 bp, the shortest 201 bp and the mean length 1,371.3 bp. We assessed the assembly completeness using BUSCO29 and recovered 90% complete or partial alignments of 3950 mammalian orthologs. All sequence data have been uploaded to GenBank (BioProject ID PRJNA399240). We used Kallisto30 to estimate abundance and DESeq231 to call differential expression. In total, 1,871 transcripts were differentially expressed between pregnant and non-pregnant animals (FDR-adjusted P < 0.001). Approximately 43% of these differentially regulated transcripts were annotated by Trinotate v3.0.228; on the basis of similarity to known genes in the PFam (v31.0) and SwissProt (release 2017_2) databases. Pearson correlation and Principal Component analyses of gene expression data across all samples show that gene expression is more highly correlated within sample groups than between them (Supplementary Figure 1). The 50 most significantly up- and down-regulated genes were identified for further analysis (Tables 1 and 2).
Gene ontology analysis
We conducted analyses of gene ontology for differentially expressed S. crassicaudata genes and identified broad functional categories on which to focus our analysis. These analyses are ideal for examining system-level gene expression changes in non-model species32. GO functional annotation of transcripts upregulated in pregnant compared with non-pregnant uteri identified 102 GO terms (Supplementary Table 1). In particular, there was significant enrichment for genes involved in metabolism, biosynthesis, lipid metabolism, transport and cellular structures (Supplementary Figure 2). There were 269 significantly enriched Gene Ontology categories for genes that are downregulated during pregnancy (Supplementary Table 2). There was enrichment for genes involved in development, transport, cell signalling, morphogenesis, metabolism and cellular structures membrane (Supplementary Figure 3). KEGG pathway analysis of pregnancy-upregulated genes showed significant enrichment of 13 pathways involved in metabolism, biosynthesis, lysosome, peroxisome, protein processing and export, signalling, one of which (metabolic pathways) survived Benjamini-Hochberg correction (Table 3). In contrast, KEGG pathway analysis of downregulated genes during pregnancy showed significant enrichment of 11 pathways involved in axon function, cell cycle, signalling, cancer, cell adhesion, metabolism, and receptor interaction, none of which survived Benjamini-Hochberg correction (Table 4).
Comparison between Monodelphis domestica and Sminthopsis crassicaudata
Ninety-seven percent of differentially expressed Monodelphis domestica (grey short-tailed opossum) genes18 between non-pregnant and pre-implantation uterus were shared in the S. crassicaudata uterine transcriptome. 20% of the top 50 annotated M. domestica pregnancy upregulated genes were upregulated in S. crassicaudata pregnancy, and 14% of the top 50 annotated M. domestica pregnancy downregulated genes were downregulated in S. crassicaudata pregnancy (Supplementary Tables 3 and 4). Of the M. domestica genes upregulated in pregnancy, 10% were upregulated in dunnart pregnancy; of the M. domestica genes downregulated in pregnancy, 13% were downregulated in dunnart pregnancy. Less than one percent of the differentially regulated opossum genes were differentially regulated in the opposite direction in dunnart (Fig. 1).
Gene ontology clustering analysis using DAVID33 indicated an overrepresentation of shared genes between dunnart and opossum that were upregulated during pregnancy, which are involved in a variety of functions, including membrane function, metabolism and biosynthesis, transport and lysosome function, cellular remodelling, motility, apoptosis and cell adhesion, and immunity (Supplementary Table 5). The same clustering analysis indicated an overrepresentation of shared genes downregulated during pregnancy that are involved in morphogenesis and development, transport, cellular motility, protein localization, focal adhesion, cytoskeletal function (laminin and focal adhesion function), and immune roles (Supplementary Table 6). KEGG pathway analysis of shared pregnancy-upregulated genes showed significant enrichment of 16 pathways involved in metabolism, protein processing and export, secretion, and lysosome function, three of which (metabolic pathways, protein export, protein processing in endoplasmic reticulum) survived Benjamini-Hochberg correction (Supplementary Table 7). In contrast, KEGG pathway analysis of downregulated genes during pregnancy showed significant enrichment of 11 pathways involved in axon function, cancer, signalling, metabolism, and receptor interaction, one of which (axon guidance) survived Benjamini-Hochberg correction (Supplementary Table 8).
Discussion
Our transcriptomic analysis of dunnart uterus reveals differential expression of a range of genes putatively involved in the processes of early pregnancy, prior to implantation of the unshelled conceptus into the lining of the uterus. GO and pathway analyses indicate that there is significant differential regulation of groups of genes involved in metabolism and biosynthesis, and almost one third of the top 50 upregulated genes in pregnancy have these roles (Table 1), an unsurprising result that highlights the importance of these processes in the metabolically active uterus during pregnancy. Our results also point to a role for differential regulation of genes encoding nutrient transporters, cytoskeletal molecules, and immune factors in the uterus to support histotrophy, immunological protection and tissue remodelling required for early development of the embryo. Similar functions have been identified using transcriptomic studies of species representing independent origins of viviparity, indicating that these processes are critical to maintaining pregnancy across taxa15,32,34,35.
Nutrient provisioning to the unimplanted embryo
In marsupials and eutherian mammals, the initial pre-attachment embryonic development is supported by histrotrophes secreted by uterine glands36. Following embryonic attachment, nutrient supply typically shifts to haemotrophy (i.e. secretion of material from the maternal blood circulation4). Haemotrophic nutrient transfer either occurs through direct embryonic contact with maternal blood, or through diffusion or active transport of haemotrophes from maternal blood, followed by secretion by the uterine epithelium into the uterine lumen37. In marsupials, the shift from histotrophic to haemotrophic nutrient transfer typically occurs following rupture of the embryonic shell coat38. In S. crassicaudata, this shift is accompanied by structural changes to the uterus. Early in S. crassicaudata pregnancy (the period at which our pregnant transcriptome samples were collected), uterine stromal glands are abundant and actively secreting12,24. As pregnancy progresses, gland abundance decreases and glandular secretion is replaced by secretory activity in the luminal epithelium12. We identified a number of genes putatively responsible for nutrient transport to the early conceptus:
Histotrophy
Almost one quarter of the top 50 upregulated genes in early S. crassicaudata pregnancy have putative transport-associated function, suggesting that nutrient transport underpins histotrophy in supporting the conceptus pre-implantation (Table 1), even before haemotrophic nutrient transport via the placenta. A number of secretion-related genes upregulated in early pregnancy may be associated with glandular secretion of histotrophe (e.g. AP4S1, HYOU1, SRPRA) (Table 5). Early pregnancy involves significant upregulation of nutrient transporter genes, including APOL6, involved in cholesterol transport39, PLA2G10, involved in hydrolysis of fatty acids during pregnancy40, and a suite of solute carrier proteins (SLCs) involved in transport of nucleoside sugars, ions and anions, glucose, fatty acids, calcium and zinc (Table 5). Upregulation of solute carrier proteins also occurs during pregnancy in the uterus of the viviparous skink Chalcides ocellatus35,41 and the post-implantation uterus of the marsupial M. domestica15. Similarly, cathepsin L (CTSL), upregulated during pregnancy in C. ocellatus35 and pigs42,43, is also significantly upregulated during pregnancy in S. crassicaudata (Table 5). Cathepsins are involved in remodelling of the uterine epithelium, which may enable transport of gases, macromolecules and micronutrients for embryonic development43. These molecules are also components of secreted uterine fluid in horses, pigs, sheep and cattle, along with phospholipases44. Additionally, cathepsins are present in the mouse and human yolk sac during early pregnancy, where they may degrade proteins to free amino acids for uptake by the fetus20, and we suggest that CTSL may play a similar role during early pregnancy in the dunnart uterus.
Macromolecule catabolism
Lysosomal activity is also one of the most significantly upregulated KEGG pathways during pregnancy in S. crassicaudata (Table 3). This result indicates that breakdown of macromolecules into small subunits for uterine secretion41,45 occurs during the period of receptivity in dunnarts. Such catabolism is probably required during histotrophic nutrition to provide molecules small enough for uptake through the permeable shell coat of the conceptus. Lysosomes and lysosomal-associated genes are also upregulated during pregnancy in the uterine epithelium of both pigs46 and viviparous skinks during pregnancy35,41,45, and lysosome-associated genes are abundant in the human yolk sac20. Increased lysosomal activity is consistent with an increased protein content of luminal fluid in the marsupial uterus pre-implantation24,47. Lysosomal activity is also congruent with morphological observations of dark electron-dense vesicles in uterine glandular epithelial cells, which become electron-lucent pre-implantation in S. crassicaudata12,26. This morphological pattern also occurs during pregnancy in viviparous skinks45 and pigs48. The lysosomal genes upregulated in pre-implantation S. crassicaudata uterus suggests that similar genetic mechanisms mediate nutrient breakdown for histotrophy in diverse viviparous groups.
Adenogenesis
Interestingly, both cadherins and the Wnt signaling pathway, involved in mammalian uterine adenogenesis (gland development, which is essential for histotrophy49), are down-regulated in the pregnant S. crassicaudata uterus (Tables 4, 6). This finding suggests a cessation of gland development in the uterine stroma as pregnancy progresses, which is consistent with a morphological decrease in gland density in the uterine stroma of S. crassicaudata during the period of uterine receptivity12. Hence, the shift from histotrophic nutrient transfer may begin prior to implantation to allow a rapid shift to haemotrophic nutrient provisioning upon implantation.
Steroid biosynthesis
The steroid biosynthesis pathway is also significantly enriched in the list of upregulated genes during pregnancy (Table 3). CYP27A1 (sterol 27-hydroxylase P450) is involved in the conversion of cholesterol to its primary metabolite 27-hydroxycholesterol, after which 27-hydroxycholesterol is converted to bile salt precursors by HSD3B7 (3-beta-hydroxysteroid dehydrogenase-7); the conversion of the 5-beta-reduction of bile acid intermediates and steroid hormones carrying a delta (4)-3-one structure is effected by AKR1D1 (aldo-keto reductase family 1 member D1)50. All four of these genes are significantly upregulated during pregnancy, especially AKR1DA and HSD3B7, which are in the top 50 differentially expressed annotated genes (Table 5). While deficiencies in this pathway cause adrenal dysfunction and bile acid reduction51, the reasons for their upregulation here is less clear. 27-hydroxycholesterol is a selective modulator of the estrogen receptors52, and bile acid intermediates are also nutrient signalling molecules53; both functions may be important in the pre-implantation uterus. Linked with this pathway is the upregulation of steroid biosynthesis pathways (Table 5). The production of 7-dehydrocholestrol is followed by a sequence of gene expressions culminating in the expression of 17-beta hydroxysteroid 7 (HSD17B7), which is involved in the conversion of steroid precursors to androgens51. The upregulation of these pathways may be linked to steroid recruitment mechanisms, but may also be important in other functions during pregnancy, including the transport and utilisation of fatty acids and electrolytes in the pre-attachment phase.
Immunity
The top five most significantly enriched GO categories in pregnancy downregulated genes are related to immune function (Supplementary Table 2), and 18% of the top 50 downregulated genes during pregnancy have putative immune function (Table 2). Many of these downregulated genes are immunoglobulins that make up subunits of antibodies (Table 6), which may simply reflect a lower relative number of B cells in pregnant uterine tissue. Other genes involved in maternal-fetal tolerance are also downregulated, including IL3454. This result reflects an important role of the uterus in immunosuppression to prevent maternal rejection of the semi-foreign embryo, even before the invasion of the embryo into the uterine epithelium. The dunnart embryonic shell membrane disintegrates prior to implantation, which in combination with remodelling may place maternal and embryonic tissues in close association3,10. The apposition of maternal and fetal tissues has likely driven the evolution of adaptations to ‘hide’ the embryo from the mother’s immune system, despite a lack of tissue invasion at that point in pregnancy. A similar downregulation of some immune genes occurs in the uteri of other vertebrates that lack erosion of maternal epithelia throughout pregnancy e.g.32,35,55.
In S. crassicaudata, we also observe a large proportion of immune genes upregulated pre-implantation (14% of the top 50, Table 1). In contrast to other marsupial studies, we did not see a change in interleukin-6 gene expression15,18, even though interleukin-6 is expressed in other tissues in S. crassicaudata56. The differences may be because our study focussed on preimplantation pregnancy. In M. domestica, immune genes are upregulated at implantation, including a range of inflammatory and wound-healing markers18. There is increasing recognition of the importance of the presence of maternal immune factors in the eutherian uterus for embryo implantation and uterine remodelling; the maternal immune response must be precisely regulated for successful mammalian pregnancy57,58. Our results allow comparison of both major lineages of marsupials, Australididelphia (S. crassicaudata, here) and Didelphimorphia (M. domestica15,18), and suggest that a delicate balance of up- and down-regulated immune factors was a feature of the pregnant uterus of the most recent common ancestor of therian mammals, exapted for the evolution of viviparity in this lineage. Immune genes of stable expression in M. domestica18 across pregnancy display the same pattern in S. crassicaudata (CD3D, CD3D, CD3G, CD4, CD68, CD8B, IL4R). Further examination of gene expression at late stage pregnancy in S. crassicaudata is necessary to draw conclusions about the precise immunogenic changes that facilitate implantation and placentation in the dunnart, and whether these mirror the changes seen in the Didelphimorphia. Finally, immune factors prevent pathogenic infection in vertebrate gestational tissues32,57, and our dataset identifies several candidate genes responsible for immune defence in the pregnant dunnart uterus (BPI, BPIFB1, GZMA and PRF1) (Table 5).
Remodelling of the pregnant uterus
Differentially regulated S. crassicaudata genes are significantly enriched for a number of GO categories related to tissue proliferation, tissue remodelling, and cell membrane components (Supplementary Table 1). The cell adhesion molecule pathway is significantly downregulated as identified by KEGG pathway analysis (Table 4), and more than one third of the top 50 downregulated genes have putative functions associated with cytoskeleton and remodelling (Table 2). Alterations to both cell adhesion and remodelling are expected during the period of receptivity in preparation for implantation, and embryonic implantation in S. crassicaudata involves significant morphological and molecular remodelling12,24,26. Our findings demonstrate that, as for eutherian mammals42,59 and viviparous skinks35,41,60, remodelling involves expression changes of cathepsins (CTSL), cadherins (e.g. CDH11, CDH20), and numerous protocadherins (Tables 5 and 6).
Similar expression patterns of remodelling genes across diverse viviparous groups suggest a common suite of molecules is required in preparing the uterus for implantation in live-bearing taxa60. Down-regulation of cell adhesion molecules occurs in S. crassicaudata, including JAM2, which is associated with tight junctions61,62. Embryonic attachment in S. crassicaudata is invasive, yet unlike many eutherian mammal species with invasive placentation, the invasion involves embryonic erosion of an originally intact uterine epithelium, rather than a loss of cellular adhesion to facilitate invasion12,24. In viviparous skinks, reduced lateral cell adhesion makes the uterus more plastic and likely facilitates remodelling63. Down-regulation of the cell adhesion pathway may play a similar role in preparing the S. crassicaudata uterus for implantation of the embryo.
Several genes that function in angiogenesis and vascular morphogenesis are downregulated in the S. crassicaudata uterus during pregnancy (e.g. ADGRA2, ADGRB2, ANGPTL1, EPHB4, ISM1, PDZRN3, RHOJ, TNMD, VEGFD; Table 6). This result was unexpected, given the upregulation of angiogenic genes such as EPAS1, HIF1A and VEGFA during pregnancy in skinks and rats e.g.35,64,65,66; however several of these genes are inhibitors, rather than promoters, of angiogenesis e.g. ISM167. Their downregulation in S. crassicaudata uterus during pregnancy may simply reflect temporality of our sampling: the transcriptome comes from uteri prior to the development of extensive vascularisation during placental formation, and it is possible that embryos do not require much oxygen at this early developmental stage.
Extracellular matrix molecules are down-regulated during early pregnancy in S. crassicaudata, including laminin (LAMA3), collagens (COL7A1, COL15A1), fibulin (FBLN7), fibronectins (FLRT2, FLRT3) and receptors (ITGA4), keratins (KRT22), and elastins (EMILIN1) (Table 6). We suggest that uterine receptivity in S. crassicaudata involves significant remodelling of the extracellular matrix. Increased expression of laminins68,69,70, fibronectin71 and fibronectin receptor ITGA472 is associated with uterine receptivity in eutherian mammals. The opposite trend for these molecules in S. crassicaudata is unexpected, yet could be explained by differences in alterations to the uterine stroma in marsupial and eutherian pregnancy. In eutherian mammals, increased expression of extracellular matrix molecules is related to cellular differentiation of uterine stromal fibroblasts to decidual cells (decidualisation)73,74. This cellular transformation does not occur in S. crassicaudata, as marsupials lack decidual cells73. In addition, the uterine stroma of S. crassicaudata and other marsupials is relatively cell-poor, and uterine receptivity involves a significant reduction in stromal cell abundance12,27. Thus, the specific markers of uterine receptivity may differ between viviparous amniotes, as they relate to species-specific uterine cellular processes. Additionally, reduction in extracellular matrix leading up to implantation may help to reduce the diffusion distance between maternal blood vessels and the uterine epithelium. In marsupials, reduction of this diffusion distance is a critical step in preparation for haemotrophic nutrient transfer37.
Uterine receptivity and quiescence
A number of genes differentially expressed in the dunnart uterus are similar to mediators of uterine receptivity in humans. Estrogen and progesterone are the key hormones controlling receptivity of the uterus to an implanting embryo22, and our data reveal differential expression of genes binding to and effecting action of these hormones (PAQR7; PRDM2) in the dunnart uterus just prior to implantation (Table 5). These hormones coordinate morphological and physiological changes in the uterus to promote receptivity, and a number of potential markers of uterine receptivity in eutherians22 are differentially expressed in the S. crassicaudata uterus. Mucins, which are apically located glycoproteins in the epithelium of the uterus, have anti-adhesive properties, and must be removed from the site of attachment before implantation can take place; dysregulation of mucin expression affects eutherian fertility22,75,76. A similar situation is present in marsupials, given that the mucin MUC5AC is the most highly downregulated gene in pre-implantation dunnart pregnancy (Table 2), and that MUC1 increases in the grey opossum uterus after breach of the shell coat18. Mucins are also downregulated in the uterus during pregnancy in a viviparous skink34. A number of other genes involved in uterine receptivity in humans and mice are also differentially expressed in the dunnart pre-implantation uterus, including the homeobox genes HOXA10 and HOXA11, and phospholipases (PLA2G10, PLA2G3)22,77.
Maintaining quiescence of the uterus (i.e. preventing uterine contraction) is another key requirement for progress of a successful pregnancy. Two of the most significantly downregulated genes in the pregnant dunnart uterus are the prostaglandin receptors PTGER3 and PTGFR (Table 2). The products of these genes likely bind prostaglandins to stimulate myometrial contractions78.
Similarities in early pregnancy between Australididelphia and Didelphimorphia
We identified 97% of the genes that were differentially expressed between non-pregnant and pre-implantation M. domestica uterus18 in the S. crassicaudata uterine transcriptome. This result indicates a substantial overlap in the range of expressed genes between the two species, as expected given that these species derive from a single origin of viviparity. There are many shared genes that are differentially expressed in M. domestica and S. crassicaudata (at the same stages of pregnancy: non-pregnant uterus compared to pre-implantation uterus) (Supplementary Tables 3 and 4). The overlap indicates that many of the uterine functions identified in S. crassicaudata are shared across both major marsupial lineages. For example, remodelling of the uterus is a shared characteristic, with genes involved in extracellular matrix (e.g. cadherin-related genes FAT4, CDH11, CDH19 and PCDH11X down in pregnancy; laminin-related genes EGFLAM, COL15A1 down in pregnancy), cellular motility (e.g. FGF1, NRG1, SEMA5B down in pregnancy; RAB25, FGFR1, HBEGF up in pregnancy) and cell adhesion (e.g. ITGA4, PTK7, TRIP6 up in pregnancy) differentially regulated in both S. crassicaudata and M. domestica. Histotrophic function is also shared across early pregnancy in marsupials: genes involved in lysosomal transport are upregulated in pregnancy in both M. domestica and S. crassicaudata (e.g. ATP6V1B2, AP3D1, TMEM165, TMEM79), and pathway analysis indicates an overrepresentation of pregnancy-upregulated genes of protein processing and export, secretion, and lysosome function in the shared gene lists between the two species (Supplementary Table 7).
Of the top 50 genes of M. domestica that are upregulated during pregnancy, 20% are also upregulated in S. crassicaudata early pregnancy. These genes include ELF5 (ESE2), an epithelium-specific transcription factor thought to regulate gene expression in glandular epithelium79 and which we postulate may be important in supporting gene expression for glandular secretions; CTAGE5, involved in exporting collagen from the endoplasmic reticulum80, and therefore possibly important for remodelling of the extracellular matrix; FGFBP1, which mediates cellular proliferation and migration81; and LVRN, which in humans is a trophoblast-specific factor82 that may regulate molecules at the interface of maternal and embryonic tissue to facilitate the development of a placenta83. The expression of LVRN in uterine tissues during early pregnancy in both major marsupial lineages suggests that this molecule may also be involved in initiating placentation at the maternal tissue interface, although further research is required to explore this hypothesis. Of the top 50 M. domestica genes downregulated during early pregnancy, 14% are also downregulated in S. crassicaudata early pregnancy. These genes include transcription factors (CBX2, SOX4); the motor-protein encoding gene KIF26B; VTCN1 (B7-H4), which negatively regulates T-cell immune responses84; and IGFBP5, which regulates the action of the insulin-like growth factors that mediate cell growth and also has apoptotic action85. Interestingly, transgenic mice that overexpress IGFBP5 display reduced female fertility85, suggesting that the downregulation of this gene may be essential to early pregnancy across mammals.
Conclusions
Genomic and transcriptomic methods are valuable tools for examining the physiology and evolution of marsupial pregnancy15,17,18,86,87. While the M. domestica transcriptome identified the importance of immune modulation for successful implantation and placentation in the marsupial uterus18, a range of other physiological changes is also required to support the internal incubation of the embryo prior to placentation. Our transcriptome study highlights the importance of such processes, including remodelling of the pre-implantation uterus, uterine quiescence, and nutrient provision via histotrophy prior to the development of the placenta; many of the genes underpinning these functions are shared across the dunnart and the opossum. The S. crassicaudata dataset is an ideal complement to the transcriptome of the opossum15,18, because these animals represent both major clades of marsupials (Australididelphia and Didelphimorphia, which diverged ~75 Mya88), and the cladistic derivation of both groups is similar (within-clade divergence of Dasyuridomorphia and Didelphimorphia both ~30 Mya88).
This transcriptome analysis reveals the importance of histotrophic nutrient transport prior to embryo implantation, before nutrient transport function is supplanted by the complex, nutritive placenta. Early pregnancy is a critical time for successful reproduction, and disruption to histotrophy could disrupt embryonic development. 40–50% of human pregnancies fail in the first trimester21, most of which is prior to the development of the definitive chorioallantoic placenta89. The putative gene functions identified here are similar to those in the pregnant uterus in other amniotes34,35,90. The conservation of genes underpinning pre-placental nutrient transport, gestational tissue remodelling, and uterine quiscence in amniote pregnancy is remarkable given that mammals and reptiles represent multiple independent origins of viviparity. Conserved elements underpinning aspects of early eutherian and marsupial pregnancy may provide new information for understanding human pregnancy disorders91,92, which is important given the difficulties in studying the human uterus in vivo22. This work furthers our understanding of the mechanisms underlying the survival of early embryos in our earliest live bearing mammalian ancestors, and highlights the importance of histotrophic nutrition to the embryo prior to the development of the nutritive placenta.
Methods
Tissue collection
Animals were held at a temperature-controlled breeding colony at the University of Sydney (in accordance with approved University of Sydney Animal Ethics Committee Protocol 704). Animals were housed either singly or in pairs, in plastic cages, and were provided with nesting boxes, nesting material, and enrichment material. Animals were held under the natural photocycle for Sydney (33°52’ S, 151°12’ E) and fed commercial cat food daily; water was provided ad libitum. Vaginal epithelial cells in smears of the urogenital sinus were examined microscopically to monitor estrous cycling of females93,94. A large number of cornified epithelial cells in the urine and a sharp increase in body mass defined the peak of oestrous93,95,96. Females were then paired with males, and the first day that sperm were detected in urine of the female was designated day 1 after mating25,95. Paired females were monitored for signs of pregnancy, including an increase in pouch area and vascularisation, loss of the furred pouch lining, and increase in body mass93,96.
Early pregnant (n = 3) and non-pregnant (n = 3) females were euthanised by CO2 inhalation, followed by immediate decapitation. The presence of embryos in excised uteri confirmed gestation, and the stage of pregnancy was determined by comparing size and morphology of embryos to the timetable of embryonic development12. We specifically targeted early-pregnant animals between days 6–8 of pregnancy, prior to implantation and placentation12, the stage of pregnancy where the shelled egg is present in the uterus.
Transcriptome sequencing and annotation
Uterine samples were homogenised using the 3 mm steel bead TissueLyser II system (Qiagen, Hilden Germany) and QiaShredder (Qiagen). Total RNA was extracted using an RNeasy Plus Mini Kit (Qiagen), which includes an in-built DNAse treatment. RNA concentration and integrity were assessed using a Bioanalyzer (Agilent, Santa Clara CA) and only high quality RNA (RIN > 8) was used for downstream analysis. Samples for transcriptomics were sequenced after Truseq RNA sample prep with on an Illumina HiSeq 2500 with 100 bp paired-end sequencing, at the Ramaciotti Centre for Genomics, Sydney, Australia. Reads from all samples were combined in a de novo assembly with Trinity v2.0.428, using the default parameters and the–trimmomatic and–min_kmer_cov 2 options. To assess the assembly completeness we used BUSCO v2.0.129 with the default parameters in the transcriptome mode (-m tran), and searched against the tetrapod set of orthologs (tetrapoda_odb9). We used Kallisto30 to estimate abundance and DESeq231 to call differential expression as implemented in the Trinity pipeline. We assessed correlation of gene expression between samples using the PtR script in Trinity. We annotated transcripts and assigned GO terms using the default parameters of the Trinotate pipeline v3.0.228; which allowed us to identify particular gene functions on which to focus our analyses. Graphical representation of enriched GO terms was carried out using the cateGOrizer tool97. KEGG pathway analysis of annotated genes was carried out using DAVID version 6.8 (available: http://david.abcc.ncifcrf.gov/home.jsp, last accessed June 2017)98, using EASE score of 0.1 and M. domestica as background. P-values were Benjamini-Hochberg corrected to account for multiple hypothesis testing.
Differentially expressed genes between non-pregnant and pre-implantation uterus in M. domestica were compared to the S. crassicaudata uterine gene expression data using discontiguous megablasts optimised for cross-species comparison, using the –task dc-megablast option and the default parameters. Monodelphis domestica transcripts18 identified as differentially expressed between non-pregnant and mid-gravid (pre-implantation) uterus (adjusted P < 0.001) were searched against the S. crassicaudata uterine transcriptome assembly, and the results compared to the S. crassicaudata differential gene expression results from DESeq2. Differentially expressed genes shared between the two species were analysed using the DAVID functional annotation tool version 6.8 (available: http://david.abcc.ncifcrf.gov/home.jsp, last accessed November 2017)33, with GO_ALL biological process, cellular component and molecular function terms, using M. domestica as background. The Functional Annotation Clustering option was used to group significantly enriched GO terms using a modified Fisher’s Exact Test by function and the DAVID Fuzzy clustering algorithm33. Grouping was performed using DAVID settings for highest stringency and P-values were Benjamini-Hochberg corrected to account for multiple hypothesis testing. KEGG pathway analysis using DAVID was carried out using an EASE score of 0.1 and Benjamini-Hochberg corrected P-values.
Data availability statement
All sequence data have been uploaded to GenBank (BioProject ID PRJNA399240).
Change history
22 July 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Renfree, M. B. Marsupials: Placental Mammals with a Difference. Placenta 31, S21–S26, https://doi.org/10.1016/j.placenta.2009.12.023 (2010).
Enders, A. C. & Schlafke, S. Cytological aspects of trophoblast-uterine interaction in early implantation. American Journal of Anatomy 125, 1–29, https://doi.org/10.1002/aja.1001250102 (1969).
Roberts, C. T. & Breed, W. G. Placentation in the dasyurid marsupial, Sminthopsis crassicaudata, the fat-tailed dunnart, and notes on placentation of the didelphid, Monodelphis domestica. Journal of Reproduction and Fertility 100, 105–113, https://doi.org/10.1530/jrf.0.1000105 (1994).
Zeller, U. & Freyer, C. Early ontogeny and placentation of the grey short-tailed opossum, Monodelphis domestica (Didelphidae: Marsupialia): contribution to the reconstruction of the marsupial morphotype. J. Zool. Syst. Evol. Res. 39, 137–158, https://doi.org/10.1046/j.1439-0469.2001.00167.x (2001).
Hughes, R. L. Morphological studies on implantation in marsupials. J Reprod Fertil 39, 173–186 (1974).
Renfree, M. B. Proteins in the uterine secretions of the marsupial Macropus eugenii. Dev. Biol. 32, 41–49, https://doi.org/10.1016/0012-1606(73)90218-2 (1973).
Roberts, C. T., Breed, W. G. & Mayrhofer, G. Origin of the oocyte shell membrane of a dasyurid marsupial: an immunohistochemical study. J. Exp. Zool. 270, 321–331, https://doi.org/10.1002/jez.1402700311 (1994).
Roberts, C. T. & Breed, W. G. Variation in ultrastructure of mucoid coat and shell membrane secretion of a dasyurid marsupial. Reprod. Fertil. Dev. 8, 645–648 (1996).
Hughes, R. L. & Shorey, C. D. Observations on the permeability properties of the egg membranes of the marsupial. Trichosurus vulpecula. Journal of Reproduction and Fertility 32, 25–32, https://doi.org/10.1530/jrf.0.0320025 (1973).
Selwood, L. Marsupial egg and embryo coats. Cells Tissues Organs 166, 208–219 (2000). doi:16733.
Cruz, Y. P. & Selwood, L. Histological differences between gravid and non-gravid uteri in the dasyurid marsupial, Sminthopsis macroura (Spencer). Journal of Reproduction and Fertility 111, 319–325, https://doi.org/10.1530/jrf.0.1110319 (1997).
Laird, M. K., Thompson, M. B., Murphy, C. R. and McAllan, B. M. Uterine epithelial cell changes during pregnancy in a marsupial (Sminthopsis crassicaudata; Dasyuridae). J. Morphol. 275, 1081–1092, https://doi.org/10.1002/jmor.20282 (2014).
Renfree, M. B. Influence of the Embryo on the Marsupial Uterus. Nature 240, 475–477 (1972).
Tyndale-Biscoe, H. and Renfree, M. Reproductive physiology of marsupials. (Cambridge University Press, 1987).
Hansen, V. L., Schilkey, F. D. & Miller, R. D. Transcriptomic Changes Associated with Pregnancy in a Marsupial, the Gray Short-Tailed Opossum Monodelphis domestica. PLoS ONE 11, e0161608, https://doi.org/10.1371/journal.pone.0161608 (2016).
Ager, E. et al. Insulin is imprinted in the placenta of the marsupial. Macropus eugenii. Dev. Biol. 309, 317–328, https://doi.org/10.1016/j.ydbio.2007.07.025 (2007).
Guernsey, M. W., Chuong, E. B., Cornelis, G., Renfree, M. B. and Baker, J. C. Molecular conservation of marsupial and eutherian placentation and lactation. Elife 6, https://doi.org/10.7554/eLife.27450 (2017).
Griffith, O. et al. The evolutionary origins of embryo implantation: eutherian implantation is homologous to the attachment reaction in the opossum. in press (2017).
Selwood, L. Marsupial oocytes, fertilization and embryonic development can provide useful tools to study developmental mechanisms. IUBMB Life 59, 617–621, https://doi.org/10.1080/15216540701606934 (2007).
Cindrova-Davies, T. et al. RNA-seq reveals conservation of function among the yolk sacs of human, mouse, and chicken. PNAS, https://doi.org/10.1073/pnas.1702560114 (2017).
Norwitz, E. R., Schust, D. J. & Fisher, S. J. Implantation and the survival of early pregnancy. N. Engl. J. Med. 345, 1400–1408, https://doi.org/10.1056/NEJMra000763 (2001).
Zhang, S. et al. Deciphering the molecular basis of uterine receptivity. Mol. Reprod. Dev. 80, 8–21 (2013).
Bennett, J., Breed, W., Hayman, D. & Hope, R. Reproductive and Genetic Studies With a Laboratory Colony of the Dasyurid Marsupial Sminthopsis crassicaudata. Aust. J. Zool. 37, 207–222, https://doi.org/10.1071/ZO9890207 (1989).
Roberts, C. T. & Breed, W. G. Embryonic-maternal cell interactions at implantation in the fat-tailed dunnart, a dasyurid marsupial. The Anatomical Record 240, 59–76, https://doi.org/10.1002/ar.1092400107 (1994).
Roberts, C. & Breed, W. Changes in structure of the trophectoderm of a marsupial in mid-pregnancy up to the time of implantation. Reproduction, fertility and development 8, 797–802 (1996).
Dudley, J. S., Murphy, C. R., Thompson, M. B. & McAllan, B. M. Desmoglein-2 during pregnancy and its role in the evolution of viviparity in a marsupial (Sminthopsis crassicaudata; Dasyuridae). J. Morphol. 276, 261–272, https://doi.org/10.1002/jmor.20333 (2015).
Laird, M. K., Turancova, M., McAllan, B. M., Murphy, C. R. and Thompson, M. B. Uterine focal adhesion dynamics during pregnancy in a marsupial (Sminthopsis crassicaudata; Dasyuridae). The Anatomical Record300, 1150–1159, https://doi.org/10.1002/ar.23535 (2017).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protocols 8, 1494–1512, https://doi.org/10.1038/nprot.2013.084 (2013).
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212, https://doi.org/10.1093/bioinformatics/btv351 (2015).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527, https://doi.org/10.1038/nbt.3519 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 15, 550, https://doi.org/10.1186/s13059-014-0550-8 (2014).
Whittington, C. M., Griffith, O. W., Qi, W., Thompson, M. B. & Wilson, A. B. Seahorse Brood Pouch Transcriptome Reveals Common Genes Associated with Vertebrate Pregnancy. Mol. Biol. Evol. 32, 3114–3131, https://doi.org/10.1093/molbev/msv177 (2015).
Huang, D. W. et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8, R183–R183, https://doi.org/10.1186/gb-2007-8-9-r183 (2007).
Griffith, O. W., Brandley, M. C., Belov, K. & Thompson, M. B. Reptile Pregnancy Is Underpinned by Complex Changes in Uterine Gene Expression: A Comparative Analysis of the Uterine Transcriptome in Viviparous and Oviparous Lizards. Genome Biol. Evol. 8, 3226–3239, https://doi.org/10.1093/gbe/evw229 (2016).
Brandley, M. C., Young, R. L., Warren, D. L., Thompson, M. B. & Wagner, G. P. Uterine gene expression in the live-bearing lizard, Chalcides ocellatus, reveals convergence of squamate reptile and mammalian pregnancy mechanisms. Genome Biol. Evol. 4, 394–411, https://doi.org/10.1093/gbe/evs013 (2012).
Wooding, P. and Burton, G. J. Comparative Placentation: Structures, functions and evolution. (Springer, 2008).
Freyer, C., Zeller, U. & Renfree, M. B. The marsupial placenta: A phylogenetic analysis. Journal of Experimental Zoology Part A: Comparative Experimental Biology 299A, 59–77, https://doi.org/10.1002/jez.a.10291 (2003).
Renfree, M. B. & Shaw, G. Diapause. Annu. Rev. Physiol. 62, 353–375, https://doi.org/10.1146/annurev.physiol.62.1.353 (2000).
Baardman, M. E. et al. The Role of Maternal-Fetal Cholesterol Transport in Early Fetal Life: Current Insights. Biol. Reprod. 88, 24, https://doi.org/10.1095/biolreprod.112.102442 (2013).
Miele, L., Cordella-Miele, E. & Mukherjee, A. B. Uteroglobin: structure, molecular biology, and new perspectives on its function as a phospholipase A2 inhibitor. Endocr. Rev. 8, 474–490, https://doi.org/10.1210/edrv-8-4-474 (1987).
Van Dyke, J. U., Brandley, M. C. & Thompson, M. B. The evolution of viviparity: molecular and genomic data from squamate reptiles advance understanding of live birth in amniotes. Reproduction 147, R15–R26, https://doi.org/10.1530/rep-13-0309 (2014).
Satterfield, M. C. et al. Tight and Adherens Junctions in the Ovine Uterus: Differential Regulation by Pregnancy and Progesterone. Endocrinology 148, 3922–3931, https://doi.org/10.1210/en.2007-0321 (2007).
Song, G. et al. Cathepsin B, cathepsin L, and cystatin C in the porcine uterus and placenta: potential roles in endometrial/placental remodeling and in fluid-phase transport of proteins secreted by uterine epithelia across placental areolae. Biol. Reprod. 82, 854–864, https://doi.org/10.1095/biolreprod.109.080929 (2010).
Bazer, F. W. Uterine protein secretions: Relationship to development of the conceptus. J. Anim. Sci. 41, 1376–1382 (1975).
Biazik, J. M., Thompson, M. B. & Murphy, C. R. Lysosomal and alkaline phosphatase activity indicate macromolecule transport across the uterine epithelium in two viviparous skinks with complex placenta. J. Exp. Zool. B 312B, 817–826, https://doi.org/10.1002/jez.b.21297 (2009).
Dantzer, V. Scanning Electron Microscopy of Exposed Surfaces of the Porcine Placenta. Cells Tissues Organs 118, 96–106 (1984).
Renfree, M. B. Uterine proteins in the marsupial, Didelphis marsupialis virginiana, during gestation. Journal of Reproduction and Fertility 42, 163–166, https://doi.org/10.1530/jrf.0.0420163 (1975).
Stroband, H. W. J., Taverne, N., Langenfeld, K. & Barends, P. M. G. The ultrastructure of the uterine epithelium of the pig during the estrous cycle and early pregnancy. Cell and Tissue Research 246, 81–89, https://doi.org/10.1007/bf00219003 (1986).
Zhang, S. et al. Physiological and molecular determinants of embryo implantation. Mol. Asp. Med. 34, 939–980, https://doi.org/10.1016/j.mam.2012.12.011 (2013).
Marti, N. et al. Genes and proteins of the alternative steroid backdoor pathway for dihydrotestosterone synthesis are expressed in the human ovary and seem enhanced in the polycystic ovary syndrome. Mol. Cell. Endocrinol. 441, 116–123, https://doi.org/10.1016/j.mce.2016.07.029 (2017).
Miller, W. L. & Auchus, R. J. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr. Rev. 32, 81–151, https://doi.org/10.1210/er.2010-0013 (2011).
DuSell, C. D., Umetani, M., Shaul, P. W., Mangelsdorf, D. J. & McDonnell, D. P. 27-hydroxycholesterol is an endogenous selective estrogen receptor modulator. Mol. Endocrinol. 22, 65–77, https://doi.org/10.1210/me.2007-0383 (2008).
Hylemon, P. B. et al. Bile acids as regulatory molecules. J. Lipid Res. 50, 1509–1520, https://doi.org/10.1194/jlr.R900007-JLR200 (2009).
Lindau, R. et al. IL-34 at the human feta and maternal interface. J. Reprod. Immunol. 111, 11–12, https://doi.org/10.1016/j.jri.2015.06.075 (2015).
Hendrawan, K., Whittington, C. M., Brandley, M. C., Belov, K. and Thompson, M. B. The Regulation of Uterine Proinflammatory Gene Expression during Pregnancy in the Live-Bearing Lizard, Pseudemoia entrecasteauxii. J. Exp. Zool. B, 328, 334-346. https://doi.org/10.1002/jez.b.22733 (2017).
Borthwick, C. R., Young, L. J., McAllan, B. M. & Old, J. M. Identification of the mRNA encoding interleukin-6 and its receptor, interleukin-6 receptor alpha, in five marsupial species. Dev. Comp. Immunol. 65, 211–217, https://doi.org/10.1016/j.dci.2016.07.008 (2016).
Racicot, K., Kwon, J.-Y., Aldo, P., Silasi, M. & Mor, G. Understanding the Complexity of the Immune System during Pregnancy. Am. J. Reprod. Immunol. 72, 107–116, https://doi.org/10.1111/aji.12289 (2014).
Sisti, G., Kanninen, T. T. & Witkin, S. S. Maternal immunity and pregnancy outcome: focus on preconception and autophagy. Genes Immun. 17, 1–7, https://doi.org/10.1038/gene.2015.57 (2016).
Rawn, S. M. & Cross, J. C. The Evolution, Regulation, and Function of Placenta-Specific Genes. Annu. Rev. Cell Dev. Biol. 24, 159–181, https://doi.org/10.1146/annurev.cellbio.24.110707.175418 (2008).
Wu, Q., Thompson, M. B. & Murphy, C. R. Changing distribution of cadherins during gestation in the uterine epithelium of lizards. J. Exp. Zool. B 316B, 440–450, https://doi.org/10.1002/jez.b.21419 (2011).
Tsukita, S., Furuse, M. & Itoh, M. Structural and signalling molecules come together at tight junctions. Curr. Opin. Cell Biol. 11, 628–633, https://doi.org/10.1016/S0955-0674(99)00016-2 (1999).
Balda, M. S. & Matter, K. Transmembrane proteins of tight junctions. Semin. Cell Dev. Biol. 11, 281–289, https://doi.org/10.1006/scdb.2000.0177 (2000).
Biazik, J. M., Thompson, M. B. & Murphy, C. R. Desmosomes in the Uterine Epithelium of Noninvasive Skink Placentae. The Anatomical Record: Advances in Integrative Anatomy and Evolutionary Biology 293, 502–512, https://doi.org/10.1002/ar.21093 (2010).
Whittington, C. M., Grau, G. E., Murphy, C. R. & Thompson, M. B. Unusual angiogenic factor plays a role in lizard pregnancy but is not unique to viviparity. J. Exp. Zool. B 324, 152–158, https://doi.org/10.1002/jez.b.22615 (2015).
Whittington, C. M., Danastas, K., Grau, G. E., Murphy, C. R. & Thompson, M. B. Expression of VEGF 111 and other VEGF-A variants in the rat uterus is correlated with stage of pregnancy. Journal of Comparative Physiology B 187, 353–360, https://doi.org/10.1007/s00360-016-1040-y (2017).
Murphy, B. F., Belov, K. & Thompson, M. B. Evolution of viviparity and uterine angiogenesis: vascular endothelial growth factor (VEGF) in oviparous and viviparous skinks. J. Exp. Zool. B 314B, 148–156, https://doi.org/10.1002/jez.b.21317 (2010).
Xiang, W. et al. Isthmin is a novel secreted angiogenesis inhibitor that inhibits tumour growth in mice. J. Cell. Mol. Med. 15, 359–374, https://doi.org/10.1111/j.1582-4934.2009.00961.x (2011).
Korgun, E. T., Cayli, S., Asar, M. & Demir, R. Distribution of laminin, vimentin and desmin in the rat uterus during initial stages of implantation. J. Mol. Hist. 38, 253–260, https://doi.org/10.1007/s10735-007-9095-4 (2007).
Blankenship, T. N. & Given, R. L. Loss of laminin and type IV collagen in uterine luminal epithelial basement membranes during blastocyst lmplantation in the mouse. The Anatomical Record 243, 27–36, https://doi.org/10.1002/ar.1092430105 (1995).
Glasser, S. R., Lampelo, S., Munir, M. I. & Julian, J. Expression of desmin, laminin and fibronectin during in situ differentiation (decidualization) of rat uterine stromal cells. Differentiation 35, 132–142 (1987).
Rider, V., Carlone, D. L., Witrock, D., Cai, C. & Oliver, N. Uterine fibronectin mRNA content and localization are modulated during implantation. Dev. Dyn. 195, 1–14, https://doi.org/10.1002/aja.1001950102 (1992).
Basak, S., Dhar, R. & Das, C. Steroids Modulate the Expression of α4 Integrin in Mouse Blastocysts and Uterus During Implantation1. Biol. Reprod. 66, 1784–1789, https://doi.org/10.1095/biolreprod66.6.1784 (2002).
Wagner, G. P., Kin, K., Muglia, L. & Pavlicev, M. Evolution of mammalian pregnancy and the origin of the decidual stromal cell. Int. J. Dev. Biol. 58, 117–126, https://doi.org/10.1387/ijdb.130335gw (2014).
Kin, K., Maziarz, J. & Wagner, G. P. Immunohistological Study of the Endometrial Stromal Fibroblasts in the Opossum, Monodelphis domestica: Evidence for Homology with Eutherian Stromal Fibroblasts. Biol. Reprod. 90, 111–112, https://doi.org/10.1095/biolreprod.113.115139 (2014).
Meseguer, M., Pellicer, A. & Simon, C. MUC1 and endometrial receptivity. Mol. Hum. Reprod. 4, 1089–1098 (1998).
Wagener, K. et al. Endometrial mRNA expression of selected pro-inflammatory factors and mucins in repeat breeder cows with and without subclinical endometritis. Theriogenology 90, 237–244, https://doi.org/10.1016/j.theriogenology.2016.12.013 (2017).
Cha, J., Sun, X. & Dey, S. K. Mechanisms of implantation: strategies for successful pregnancy. Nat. Med. 18, 1754–1767 (2012).
Sykes, L., MacIntyre, D. A., Teoh, T. G. & Bennett, P. R. Anti-inflammatory prostaglandins for the prevention of preterm labour. Reproduction 148, R29–R40, https://doi.org/10.1530/rep-13-0587 (2014).
Oettgen, P. et al. Characterization of ESE-2, a novel ESE-1-related Ets transcription factor that is restricted to glandular epithelium and differentiated keratinocytes. J. Biol. Chem. 274, 29439–29452 (1999).
Tanabe, T., Maeda, M., Saito, K. & Katada, T. Dual function of cTAGE5 in collagen export from the endoplasmic reticulum. Mol. Biol. Cell 27, 2008–2013, https://doi.org/10.1091/mbc.E16-03-0180 (2016).
Tassi, E. et al. Enhancement of fibroblast growth factor (FGF) activity by an FGF-binding protein. J. Biol. Chem. 276, 40247–40253, https://doi.org/10.1074/jbc.M104933200 (2001).
Fujiwara, H. et al. Human extravillous trophoblasts express laeverin, a novel protein that belongs to membrane-bound gluzincin metallopeptidases. Biochem. Biophys. Res. Commun. 313, 962–968, https://doi.org/10.1016/j.bbrc.2003.12.024 (2004).
Fujiwara, H. et al. New regulatory mechanisms for human extravillous trophoblast invasion. Reproductive Medicine and Biology 4, 189–195, https://doi.org/10.1111/j.1447-0578.2005.00104.x (2005).
Sica, G. L. et al. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity 18, 849–861 (2003).
Salih, D. A. et al. Insulin-like growth factor-binding protein 5 (Igfbp5) compromises survival, growth, muscle development, and fertility in mice. Proc. Natl. Acad. Sci. USA 101, 4314–4319, https://doi.org/10.1073/pnas.0400230101 (2004).
Johnson, R. N. et al. Koala genome reveals adaptations to a eucalyptus diet, genomic integration of the koala retrovirus and an ancient population decline. (In revision).
Renfree, M. B. et al. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 12, https://doi.org/10.1186/gb-2011-12-8-r81 (2011).
Meredith, R. W. et al. Impacts of the Cretaceous Terrestrial Revolution and KPg Extinction on Mammal Diversification. Science 334, 521–524, https://doi.org/10.1126/science.1211028 (2011).
Jones, C. J. & Jauniaux, E. Ultrastructure of the materno-embryonic interface in the first trimester of pregnancy. Micron 26, 145–173 (1995).
Hernández-Díaz, N., Torres, R. and Ramírez-Pinilla, M. P. Proteomic Profile of Mabuya sp. (Squamata: Scincidae) Ovary and Placenta During Gestation. J. Exp. Zool. B 328, 371–389, https://doi.org/10.1002/jez.b.22739 (2017).
McGary, K. L. et al. Systematic discovery of nonobvious human disease models through orthologous phenotypes. PNAS 107, 6544–6549, https://doi.org/10.1073/pnas.0910200107 (2010).
Kosova, G., Stephenson, M. D., Lynch, V. J. and Ober, C. Evolutionary forward genomics reveals novel insights into the genes and pathways dysregulated in recurrent early pregnancy loss. Human Reproduction, https://doi.org/10.1093/humrep/deu355 (2015).
McAllan, B. M., Feay, N., Bradley, A. J. & Geiser, F. The influence of reproductive hormones on the torpor patterns of the marsupial Sminthopsis macroura: bet-hedging in an unpredictable environment. Gen. Comp. Endocrinol. 179, 265–276, https://doi.org/10.1016/j.ygcen.2012.08.024 (2012).
Pollock, K. et al. Oestrus in the Julia Creek dunnart (Sminthopsis douglasi) is associated with wheel running behaviour but not necessarily changes in body weight, food consumption or pouch morphology. Anim. Reprod. Sci. 117, 135–146, https://doi.org/10.1016/j.anireprosci.2009.03.005 (2010).
Selwood, L. & Woolley, P. A. A timetable of embryonic development, and ovarian and uterine changes during pregnancy, in the stripe-faced dunnart, Sminthopsis macroura (Marsupialia: Dasyuridae). Journal of Reproduction and Fertility 91, 213–227, https://doi.org/10.1530/jrf.0.0910213 (1991).
Woolley, P. Reproduction in Sminthopsis macroura (Marsupialia, Dasyuridae) .1. The Female. Aust. J. Zool. 38, 187–205, https://doi.org/10.1071/ZO9900187 (1990).
Hu, Z.-L., Bao, J. & Reecy, J. M. CateGOrizer: A Web-Based Program to Batch Analyze Gene Ontology Classification Categories. Online Journal of Bioinformatics 9, 108–112 (2008).
Huang, D., Sherman, B. & Lempicki, R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 http://www.nature.com/nprot/journal/v4/n1/suppinfo/nprot.2008.211_S1.html (2009).
Acknowledgements
The authors are grateful for the laboratory assistance of Jacqueline Herbert, and we thank Matthew Brandley for useful discussions on data analysis and interpretation. RNA integrity analysis was carried out in the Bosch Institute Molecular Biology Facility at the University of Sydney. This work was supported by a University of Sydney Animal and Veterinary Biosciences Research Fellowship to CMW from the estate of Mabs Melville; Australian Research Council Grants DP130101589 to BMM and MBT and DP180103370 to CMW and MBT; and CMW is also supported by a L’Oreal-UNESCO for Women in Science Fellowship.
Author information
Authors and Affiliations
Contributions
C.M.W., M.B.T., K.B. and B.M.M. conceived the experiment; C.M.W. and D.O. analysed the data; C.M.W., M.K.L. and B.M.M. wrote the manuscript; all authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Whittington, C.M., O’Meally, D., Laird, M.K. et al. Transcriptomic changes in the pre-implantation uterus highlight histotrophic nutrition of the developing marsupial embryo. Sci Rep 8, 2412 (2018). https://doi.org/10.1038/s41598-018-20744-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-20744-z
This article is cited by
-
MicroRNA expression profile analysis in sperm reveals hsa-mir-191 as an auspicious omen of in vitro fertilization
BMC Genomics (2020)
-
Genome-wide association study to identify genomic regions and positional candidate genes associated with male fertility in beef cattle
Scientific Reports (2020)
-
A comparison of uterine contractile responsiveness to arginine vasopressin in oviparous and viviparous lizards
Journal of Comparative Physiology B (2020)
-
Didelphis albiventris: an overview of unprecedented transcriptome sequencing of the white-eared opossum
BMC Genomics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.