Abstract
Silver nanomaterials have been mainly developed as antibacterial healthcare products worldwide, because of their antibacterial activity. However, there is little data regarding the potential risks and effects of large amounts of silver nanomaterials on plants. In contrast, N-glycans play important roles in various biological phenomena, and their structures and expressions are sensitive to ambient environmental changes. Therefore, to assesse the effects of silver nanomaterials, we focused on the correlation between N-glycans and the effects of silver nanomaterials in plants and analyzed N-glycan structures in Oryza sativa seedlings exposed to silver nanocolloids (SNCs). The phenotype analysis showed that the shoot was not affected by any SNC concentrations, whereas the high SNC exposed root was seriously damaged. Therefore, we performed comparative N-glycan analysis of roots. As a result, five of total N-glycans were significantly increased in SNC exposed roots, of which one was a free-N-glycan with one beta-N-acetylglucosamine residue at the reducing end. Our results suggest that the transition of plant complex type N-glycans, including free-N-glycans, was caused by abnormalities in O. sativa development, and free-N-glycan itself has an important role in plant development. This study originally adapted glycome transition analysis to environmental toxicology and proposed a new category called “Environmental glycobiology”.
Similar content being viewed by others
Introduction
Nanomaterial is a general term for small substances that are 1–100 nm in diameter. Nanomaterials have many unique electrical, chemical, and physical properties and are used in electronics, medicine, and healthcare fields. Silver nanomaterials have been developed and mainly used for their antibacterial activities in clothes, appliances, cosmetics, and plastics. However, there have been concerns that silver nanomaterials are likely released into the aquatic environment through factory and household wastewater on a large scale, and there are also concerns regarding the effects of silver nanomaterials on aquatic organisms and ecological systems. The toxicity of silver nanomaterials in the embryos of aquatic organisms such as medaka and zebrafish has been reported1,2. The toxicity affected the expression of morphogenesis- and cell proliferation-related genes and induced the increase of severe development abnormalities and mortality. The toxicity of silver nanomaterials was dependent on the particle size, shape, and capping materials3,4. Toxicity of silver nanomaterials has also been reported in plants, with the toxicity affecting germination, development, and photosynthetic efficiency because of the induction of oxidative stress, cytotoxicity, and genotoxicity5,6. For example, silver nanoparticle exposure significantly reduced root elongation, shoot and root fresh weights, and total chlorophyll and carotenoid contents7. Colman et al. showed that low silver nanoparticle concentrations caused a decrease in biomass8. Furthermore, the toxicity of silver nanoparticles affects the expression of several proteins that are mainly involved in primary metabolism and cell defense in wheat seedlings9. At a gene level, silver nanoparticles activate gene expression involved in plant cellular events, including cell proliferation, metabolism, and hormone signaling pathways10. The above-mentioned studies showed that silver nanomaterials have high toxicity in plants. Therefore, the risk assessment of silver nanomaterials in plants is important.
Asparagine (N)-linked glycans (N-glycans) are comprised several types of monosaccharides, forming complex compositions and linkage types. N-Glycan has a trimannosyl core structure [Man alpha1–6(Man alpha1-3)Man beta1-4GlcNAc beta1-4GlcNAc-Asn], which is a common feature in eukaryotes. Many N-glycan structures are linked to proteins or peptides and are closely involved in all life phenomena, such as development, signaling, and cell-to-cell recognition. In plants, N-glycan structures are categorized into three main types: high-mannose, complex, and paucimannose types; except for hybrid types. A characteristic of plant-specific N-glycans is the addition of beta1,2-xylose and alpha1,3-fucose to the trimannosyl core structure. High-mannose type N-glycans are synthesized in endoplasmic reticulum (ER), and other type N-glycans are synthesized in the Golgi apparatus. Paucimannose type N-glycans are linked to vacuole proteins and complex type N-glycans are linked to secretory proteins. In addition, cell alterations are reflected in gene expressions through cell signaling, whereas N-glycan is synthesized as a result of the integral expression of glycosyltransferase genes, and N-glycan structure is sensitive to slight environmental changes11. Therefore, N-glycan structural analysis is valuable for the risk assessment of silver nanomaterial toxicity in plants. However, there is little data regarding the toxicity of silver nanomaterials in glycobiology. In this study, to assesse the effects of silver nanomaterials, we focused on the correlation of N-glycan structures and the effect of silver nanomaterials in Oryza sativa and analyzed the N-glycan structures in SNC exposed O. sativa seedlings.
Results and Discussion
Phenotype analysis of O. sativa seedling exposed to silver nanocolloids (SNCs)
To observe the effect of SNCs on O. sativa seedlings, O. sativa seeds were grown with and without SNC exposure. Germination rate was 95% (control), 100% (SNCs 0.5 mg/L), 100% (SNCs 1.0 mg/L), 95% (SNCs 1.5 mg/L), 100% (SNCs 3.0 mg/L), 95% (SNCs 5.0 mg/L), 90% (SNCs 10 mg/L), 100% (SNCs 25 mg/L) after 48 h incubation. The result shows that there is no effect on germination rate at any concentration of SNC for 48 h exposure in O. sativa. Figure 1A shows the results of root and shoot elongation in O. sativa exposed to SNCs at 0 (control), 0.5, 1.0, 1.5, 3.0, 5.0, 10, and 25 mg/L for 96 h. Shoot and root length of control was 1.46 ± 0.08 cm and 0.98 ± 0.08 cm, respectively. The shoot elongation was not affected at any SNC concentration, whereas the root length increased from 0.5 to 10 mg/L based on Fig. 1A; however, in roots exposed to 25 mg/L SNCs, the lengths were two times lower than those of the control. Representative images of control and 25 mg/L SNC exposed O. sativa seedlings are shown in Fig. 1B. From the phenotype analysis, root length was seriously affected by 25 mg/L of SNC exposure.
The effect of SNCs was also observed in other plants. SNCs also have a significant effect on Arabidopsis thaliana and poplar development12. SNCs are mainly present in two forms: silver nanoparticles and free ions (Ag+), which are derived from silver nanoparticles. Free Ag+ is more poisonous than SNCs because of its oxidative potency. Wang et al.12 demonstrated that free Ag+ tends to accumulate in A. thaliana roots. Previous studies also showed that silver nanoparticles or free Ag+ inhibited the growth of O. sativa roots13, and these materials affect cell metabolism-related proteins14. In addition, the effects of SNCs or free Ag+ occur in ER- and vacuole-localized proteins of Eruca sativa15. Nair et al. reported that total sugar levels are decreased in SNC exposed O. sativa seedlings7. From these reports and our observation, SNCs and SNC derived molecules may affect N-glycans that are synthesized in the ER and Golgi apparatus of O. sativa roots. Therefore, we focused on the correlation of N-glycan structures and silver nanomaterials in O. sativa and analyzed the N-glycan structures in SNC exposed O. sativa roots.
N-Glycan analysis of SNC exposed O. sativa roots
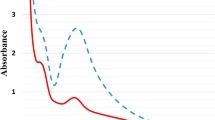
N-Glycans were prepared by hydrazinolysis, N-acetylation, and pyridylamination (PA). The resulting PA-N-glycans were separated according to their degree of saccharide polymerization by size-fractionation HPLC. Then, N-glycans of control and 25 mg/L of SNC exposed roots were compared (Fig. 2I, control and II, SNC treatments). Thirteen peaks were detected (indicated by bars). The areas of peaks B, E, and M increased in SNC exposed roots, and in particular, peak B was extremely increased in SNC exposed roots. Therefore, to identify each N-glycan structure in detail, reversed phase HPLC was performed, and branched N-glycan isomers were separated. Reversed phase HPLC analysis revealed three major peaks (peaks E1, E2, and E3) of peak E (Fig. 3). Comparing the HPLC elution times with known-position N-glycans, peaks E1, E2, and E3 coincided with those of GNM3FX, GNM3FX, and GN2M3X, respectively. Similarly, one major peak, M1, was detected in reversed phase HPLC analysis, and peak M1 coincided with the elution position of Gal2F2GN2M3FX.
Results of size-fractionation HPLC analysis of PA-N-glycans derived from O. sativa seedlings. (I) Control, (II) SNCs exposure, PA-N-glycans were applied to a Cosmosil 5NH2-MS column (4.6 ID × 150 mm). Arrowheads 5–12 indicate the degree of polymerization of PA-isomaltooligomer. The opened circle, closed square, opened triangle, closed star, closed circle represent mannose, N-acetylglucosamine, fucose, xylose, and galactose residues, respectively.
Table 1 shows the identified N-glycan structures and ratios derived from peaks E1, E2, E3, and M1, and each N-glycan is shown in terms of percentage proportion relative to GN2M3FX (peak G). The largest N-glycan Gal2F2GN2M3FX was increased three fold after SNC exposure. For other plant complex type N-glycans, GNM3FX, GNM3FX, and GN2M3X, SNC exposure caused up to five- or six-fold higher accumulations than the control. In general, complex type N-glycans are formed in from the cis-Golgi to medial Golgi apparatus, and higher complex modifications occur downstream in the synthetic pathway. Recent reports showed that protein-linked complex type N-glycans are related to proper targeting and functioning of linking proteins16,17 and also showed that complex type N-glycans play an important role in resistance to external stresses such as salt stress11,18,19. These reports showed that the Golgi-localized N-glycan synthetic enzymes are related to plant growth and development, and their defect inhibited growth and caused abnormalities. Therefore, the transition of complex type N-glycans may be related to the disorder of Golgi-localized N-glycan synthetic enzymes and genes in SNC exposed roots. The relatively small N-glycans GNM3FX, GNM3FX, and GN2M3X were affected by SNCs more significantly. These results imply that the upstream part of the N-glycan complex pathway was affected by SNCs; therefore, the downstream part of the synthetic pathway was less affected for more complicated N-glycans such as Gal2F2GN2M3FX. These results may provide evidence that the intermediate complex type N-glycans play an important role in plants under excessive stress conditions.
Free-N-glycan analysis of SNC exposed O. sativa roots
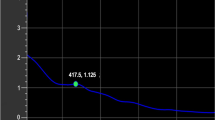
Reversed phase HPLC analysis revealed that peak B1 were eluted at around 3 min; therefore, to purify peak B1, twice reversed phase HPLC was performed (Fig. 4A). Peak B1 was further analyzed using MALDI-TOF mass spectrometry and sequential enzyme digestion. Mass spectrometry analysis of peak B1 showed that the m/z ratio was 1143.90 (Na+), which corresponded to (Hex)3(HexNAc)2(Pent)1-PA. The m/z ratio and elution position on reversed phase HPLC revealed that the N-glycan structure of peak B1 was predicted to be a free-GNM3X structure [GlcNAc1Man3Xyl1GlcNAc1-PA] with one GlcNAc residue at the reducing end. To ascertain the N-glycan structure of peak B1, peak B1 was enzymatically digested with two exoglycosidases. Peak B1 [GlcNAc1Man3Xyl1GlcNAc1-PA] was converted to Man3Xyl1GlcNAc1-PA, releasing one GlcNAc residue by beta-N-acetylhexosaminidase (Fig. 4B-II), and Man3Xyl1GlcNAc1-PA was further converted to Man1Xyl1GlcNAc1-PA, releasing two mannose residues by alpha-mannosidase (Fig. 4B-III). Thus, peak B1 was assigned as free-GNM3X structure with one GlcNAc residue at the reducing end (Fig. 4B-I).
Structural analysis of peak B1. (A) Result of second reversed phase HPLC analysis of peak B1. (B) Sequential enzyme digestions of peak B1, I: peak B1, II: beta-N-acetylhexosaminidase digestion of I, III: alpha-mannosidase digestion of II. The peaks marked by the asterisks indicated non-specific peaks.
Peak G is a major component of O. sativa roots (Fig. 2). As a result of reversed phase HPLC, two major peaks were detected; as a result of N-glycan two-dimensional mapping, the retention time of peak G2 corresponded to that of known position N-glycan, GN2M3FX20, and the other one was eluted at around 3 min (Fig. 5A, peak G1). In a similar way to peak B1, peak G1 was analyzed using MALDI-TOF mass spectrometry and exoglycosidase digestion. Mass spectrometry analysis showed that the m/z ratio was 1456.67 (Na+), which corresponded to (Hex)7 (HexNAc)1-PA. The mass value and elution position of reversed phase HPLC revealed that peak G1 was predicted to be a free-M7 structure [Man7GlcNAc1-PA]. To confirm the N-glycan structure, peak G1 was enzymatically digested with alpha-mannosidase. As a result, peak G1 [Man7GlcNAc1-PA] was converted to Man4GlcNAc1-PA, Man3GlcNAc1-PA, Man2GlcNAc1-PA, and Man1GlcNAc1-PA, releasing from three to six mannose residues by alpha-mannosidase (Fig. 5B-II); therefore, peak G1 was assigned as free-M7 structure.
As shown in Table 2, the proportion of free-GNM3X (peak B1) increased six fold after SNC exposure. In contrast to free-GNM3X, free-M7 increased three fold after SNC exposure (Table 2). To date, high-mannose type free-N-glycans have been detected in plant during development21,22, and other complex type free-N-glycans have been detected in the culture broth of rice cultured cells23 and Egeria densa24, and most of them have Lewis a structure [Gal beta1-3(Fuc alpha1-4)GlcNAc beta1-] that is characteristic of the N-glycan of extracellular glycoproteins. Maeda et al. discussed that a mechanism responsible for the production of complex type free-N-glycans is present under special or artificial conditions and native plant tissues24. In animal cells, the accumulation of sialyl free-N-glycans is caused by a decline in free-N-glycan metabolism by basal autophagy25. Mkhikian et al. demonstrated that alternative N-glycan structures were generated under unusual growth conditions26. These observations suggest that the occurrence of specific N-glycan structures is involved in cell conditions under excessive stress conditions.
The elongation of O. sativa shoots was unaffected by SNC exposure (Fig. 1A), and N-glycan structures were also unaffected. We suggested that a root defense mechanism serves to protect shoot development from SNC toxicity, and O. sativa roots may have a defense mechanism against soil environmental changes. Although the generation of high-mannose type free-N-glycans is generally caused by the deglycosylation from misfolded glycoproteins in ER-associated degradation system27, there is almost no information about the biological significance of complex type free-N-glycan. Our results showed that the increase of complex type free-N-glycans was caused by the effect of SNCs, suggesting that complex type free-N-glycan itself in O. sativa roots has an important role for resistance mechanism against excessive environmental changes. N-Glycan is one of the post-translational modifications, and the structure and quantity are sensitive to ambient environment. In addition, N-glycans are generated by the result of the integrated multiple gene expression, and the biosynthesis is strictly controlled by many glycosyltransferases and glycosidases in each specific organelle. Though, it is difficult to identify which genes are specifically affected under stress condition, N-glycan analysis can derive the related genes and synthetic regions. Therefore, N-glycan analysis can predict to the initial response to environmental changes at comprehensively, and the transition is valuable for studying the relationship between environmental changes and biological response. As the case for monitoring the signaling pathway of plant under environmental changes, novel plant nanobionics approach has been reported28. Our results revealed a correlation between free-N-glycans and plant development under excessive stress conditions and also demonstrated that free-N-glycan transitions are valuable as stress markers for assessing trace environmental changes. This is the first report of the relationship between “environmental changes and glycome transition”, and the present study originally adapted glycome transition to environmental toxicology and proposed a new category called “Environmental glycobiology”.
Materials and Methods
Chemicals
Purified SNCs (28.4 ± 8.5 nm, release 81.1% Ag+ suspended in distilled water29) were purchased from Utopia Silver Supplements (Utopia, TX, USA). Cosmosil packed columns, 5NH2-MS (4.6 ID × 150 mm) and 5C18-P (4.6 ID × 150 mm), were purchased from Nacalai Tesque (Kyoto, Japan). Known-position PA-sugar chains were purchased from Takara Bio Inc. (Shiga, Japan). alpha-Mannosidase (from jack bean) was purchased from Sigma (MO, USA). beta-N-acetylhexosaminidase (from jack bean) was purchased from Prozyme (CA, USA).
Plant materials and sample preparation
Seeds of O. sativa (Koshihikari) were supplied by Itakura Agricultural Cooperative Society, Japan. Before each experiment, seeds were washed five times with water and then washed twice with deionized water. The washed twelve seeds were then placed in a plate and immersed in 7 mm depth of water (control) or each concentration of SNCs suspension (0.5, 1.0, 1.5, 3.0, 5.0, 10.0, and 25.0 mg/L of SNCs). The plates were placed in a controlled environmental chamber, and kept at 37 °C for 96 h in the dark. The resulting seedlings were washed and then dried.
Preparation of pyridylaminated N-glycans from O. sativa seedlings
N-Glycan preparation was performed according to the method of Natsuka et al.30. Dried shoots and roots were ground in a mortar at room temperature, and a ten milligram sample was used. N-Glycans were prepared by hydrazinolysis, N-acetylation. The reducing ends of the liberated N-glycans were then tagged with a fluorophore, 2-aminopyridyne (pyridylaminated N-glycans; PA-N-glycans), as described in previous paper20. These preparations were performed following details in Hase et al.31 with minor modifications.
Separation of PA-N-glycans
Size-fractionation HPLC was performed in a Cosmosil 5NH2-MS column (4.6 ID × 150 mm) at a flow rate of 0.8 mL/min at 40 °C. PA-N-glycans were detected with a fluorescence spectrophotometer at 310 nm excitation and 380 nm emission. Reversed phase HPLC was performed on a Cosmosil 5C18-P column (4.6 ID × 150 mm) at a flow rate of 1.5 mL/min at 40 °C. The detection of PA-N-glycans performed by use of fluorescence spectrophotometer at 315 nm excitation and 400 nm emission. Each HPLC conditions were described in previous paper20.
Mass spectrometry analysis of PA-glycans
MALDI-TOF mass spectrometry analysis was then performed using an AXIMA resonance instrument (Shimadzu) in reflector mode. Sample preparation was described in previous paper20.
Glycosidase digestion of PA-N-glycans
A two picomoles of PA-N-glycans was prepared in 1 microL of accessory reaction buffer (5 mM CaCl2, 10 mM ammonium acetate buffer, pH 4.5) and 2 microL of D. D. W, 1 microL of beta-N-acetylhexosaminidase (0.05 units/microL) was added, and the mixture was incubated at 37 °C for 4 h, and then 1 microL of jack bean alpha-mannosidase (19 units/mg) and 5 microL of 10 mM ammonium acetate buffer (pH 4.5) were added, and the mixture was incubated at 37 °C for 1 h. alpha-Mannosidase digestion was described in previous paper20. To stop all reactions, the mixtures were boiled for 5 min at 95 °C, and the mixture was analyzed by size-fractionation HPLC.
References
Kashiwada, S. et al. Silver nanocolloids disrupt medaka embryogenesis through vital gene expressions. Environ. Sci. Technol. 46, 6278–6287 (2012).
Bar-Ilan, O., Albrecht, R. M., Fako, V. E. & Furgeson, D. Y. Toxicity assessments of multisized gold and silver nanoparticles in zebrafish embryos. Small 5, 1897–1910 (2009).
Yeo, M. K. & Kang, M. Effects of nanometer sized silver materials on biological toxicity during zebrafish embryogenesis. Bull. Korean Chem. Soc. 29, 1179–1184 (2008).
Asharani, P., Wu, Y., Gong, Z. & Valiyaveettil, S. Toxicity of silver nanoparticles in zebrafish models. Nanotechnology 19, 255102 (2008).
Cox, A., Venkatachala, P., Sahi, S. & Sharma, N. Silver and titanium dioxide nanoparticle toxicity in plants: A review of current research. Plant Physiol. Biochem. 107, 147–163 (2016).
Rastogi, A. et al. Impact of Metal and Metal Oxide Nanoparticles on Plant: A Critical Review. Front Chem. 5, https://doi.org/10.3389/fchem.2017.00078 (2017).
Nair, P. M. & Chung, I. M. Physiological and molecular level effects of silver nanoparticles exposure in rice (Oryza sativa L.) seedlings. Chemosphere 112, 105–113 (2014).
Colman, B. P. et al. Low concentrations of silver nanoparticles in biosolids cause adverse ecosystem responses under realistic field scenario. PLoS One 8, e57189 (2013).
Vannini, C. et al. Phytotoxic and genotoxic effects of silver nanoparticles exposure on germinating wheat seedlings. J. Plant Physiol. 171, 1142–1148 (2014).
Syu, Y. Y., Hung, J. H., Chen, J. C. & Chuang, H. W. Impacts of size and shape of silver nanoparticles on Arabidopsis plant growth and gene expression. Plant Physiol. Biochem. 83, 57–64 (2014).
Kang, J. S. et al. Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. USA 105, 5933–5938 (2008).
Wang, J. et al. Phytostimulation of poplars and Arabidopsis exposed to silver nanoparticles and Ag+ at sublethal concentrations. Environ. Sci. Technol. 47, 5442–5449 (2013).
Mirzajan, F. et al. Proteomics study of silver nanoparticles toxicity on Oryza sativa L. Ecotoxicol Environ. Saf. 108, 335–339 (2014).
Hossain, Z. Mustafa, G. Sakata, K. & Komatsu, S. Insights into the proteomic response of soybean towards Al2O3, ZnO, and Ag nanoparticles stress. J. Hazard. Mater. 304, 291–305 (2016).
Vannini, C. et al. Morphological and proteomic responses of Eruca sativa exposed to silver nanoparticles or silver nitrate. PLoS One 8, e68752 (2013).
Rips, S. et al. Multiple N-glycans cooperate in the subcellular targeting and functioning of Arabidopsis KORRIGAN1. Plant Cell 26, 3792–3808 (2014).
Von Schaewen, A. Rips, S. Jeong, I. S. & Koiwa, H. Arabidopsis thaliana KORRIGAN1 protein: N-glycan modification, localization, and function in cellulose biosynthesis and osmotic stress responses. Plant Signal Behav. 10, e1024397 (2015).
Fanata, W. I. et al. N-glycan maturation is crucial for cytokinin-mediated development and cellulose synthesis in Oryza sativa. Plant J. 73, 966–979 (2013).
Von Schaewen, A., Frank, J. & Koiwa, H. Role of complex N-glycans in plant stress tolerance. Plant Signal Behav. 3, 871–873 (2008).
Horiuchi, R., Hirotsu, N. & Miyanishi, N. N-Glycan transition of the early developmental stage in Oryza sativa. Biochem. Biophys. Res. Commun. 477, 426–432 (2016).
Kimura, K., Inoue, M., Yoshie, T. & Kimura, Y. Changes in structural features of free N-glycan and endoglycosidase activity during tomato fruit ripening. Biosci. Biotechnol. Biochem. 72, 2936–2945 (2008).
Kimura, Y. & Matsuo, S. Free N-glycans already occur at an early stage of seed development. J. Biochem. 127, 1013–1019 (2000).
Maeda, M., Kimura, M. & Kimura, Y. Intracellular and extracellular free N-glycans produced by plant cells: occurrence of unusual plant complex-type free N-glycans in extracellular spaces. J. Biochem. 148, 681–692 (2010).
Maeda, M., Ebar, N., Tani, M., Vavricka, C. J. & Kimura, Y. Occurrence of complex type free N-glycans with a single GlcNAc residue at the reducing termini in the fresh-water plant, Egeria densa. Glycoconj. J. 34, 229–240 (2017).
Seino, J. et al. Basal autophagy is required for the efficient catabolism of sialyloligosaccharides. J. Biol. Chem. 288, 26898–26907 (2013).
Mkhikian, H. et al. Golgi self-correction generates bioequivalent glycans to preserve cellular homeostasis. Elife 5, e14814 (2016).
Suzuki, T. & Funakoshi, Y. Free N-linked oligosaccharide chains: formation and degradation. Glycoconj. J. 23, 291–302 (2006).
Ghorbanpour, M. & Fahimirad, S. Plant Nanobionics a Novel Approach to Overcome the Environment Challenges. Medicinal Plants and Environmental Challenges, https://doi.org/10.1007/978-3-319-68717-9_14, 247–257 (2017).
Kataoka, C., Ariyoshi, T., Kawaguchi, H., Nagasaka, S. & Kashiwada, S. Salinity increases the toxicity of silver nanocolloids to Japanese medaka embryos. Environ. Sci.: Nano 2, 94–103 (2015).
Natsuka, S., Hirohata, Y., Nakakita, S., Sumiyoshi, W. & Hase, S. Structural analysis of N-glycans of the planarian Dugesia japonica. FEBS J. 278, 452–460 (2011).
Hase, S., Ikenaka, T. & Matsushima, Y. Structure analyses of oligosaccharides by tagging of the reducing end sugars with a fluorescent compound. Biochem. Biophys. Res. Commun. 85, 257–263 (1987).
Acknowledgements
This study was partly supported by a Grant-in-Aid for Strategic Research Base Project for Private Universities, which is funded by the Ministry of Education, Culture, Sports, Science and Technology of Japan, 2014–2018 (S1411016).
Author information
Authors and Affiliations
Contributions
R.H., Y.N., S.K. and N.M. conceived and designed the experiments, R.H. and Y.N. performed sample preparation and N-glycan analysis. R.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Horiuchi, R., Nakajima, Y., Kashiwada, S. et al. Effects of silver nanocolloids on plant complex type N-glycans in Oryza sativa roots. Sci Rep 8, 1000 (2018). https://doi.org/10.1038/s41598-018-19474-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-19474-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.