Abstract
Many cell lines derived from solid cancers can form spheroids, which recapitulate tumor cell clusters and are more representative of the in vivo situation than 2D cultures. During spheroid formation, a small proportion of a variety of different colon cancer cell lines did not integrate into the sphere and lost cell-cell adhesion properties. An enrichment protocol was developed to augment the proportion of these cells to 100% purity. The basis for the separation of spheroids from non-spheroid forming (NSF) cells is simple gravity-sedimentation. This protocol gives rise to sub-populations of colon cancer cells with stable loss of cell-cell adhesion. SW620 cells lacked E-cadherin, DLD-1 cells lost α-catenin and HCT116 cells lacked P-cadherin in the NSF state. Knockdown of these molecules in the corresponding spheroid-forming cells demonstrated that loss of the respective proteins were indeed responsible for the NSF phenotypes. Loss of the spheroid forming phenotype was associated with increased migration and invasion properties in all cell lines tested. Hence, we identified critical molecules involved in spheroid formation in different cancer cell lines. We present here a simple, powerful and broadly applicable method to generate new sublines of tumor cell lines to study loss of cell-cell adhesion in cancer progression.
Similar content being viewed by others
Introduction
The use of cancer cell lines grown on 2D plastic surfaces as a basic model to study cancer biology and a preclinical drug testing system is limited due to lack of structural architecture. 3D aggregates, known as multicellular tumor spheroids, have been developed to overcome these limitations1. Spheroids much better recapitulate the in vivo situation of tumors than cell monolayers, as they are composed of proliferating, non-proliferating, well-oxygenated, hypoxic and necrotic cells2,3 (reviewed in ref.4). Furthermore, 3D growth of cells in spheroids influences cell behavior, cell shape, polarity5, gene expression6,7, proliferation5,7, cell motility8, differentiation9 and drug sensitivity as well as radiation resistance10 (reviewed in refs1,4).
Multicellular spheroid formation depends on homotypic cell adhesion, which in epithelial cells is primarily mediated via the adherens junction (AJ) protein E-cadherin (CDH1)11. AJs are associated with the filamentous (F-) actin cytoskeleton and are crucial for epithelial sheet formation12. The cytoplasmic domain of classical cadherins can bind ß-catenin, which can interact via α-catenins and vinculin as well as other molecules with the actin cytoskeleton13. In this way, force or tension can be sensed and transduced in epithelial structures ultimately leading to altered linkage of AJs to the F-actin network14. E-cadherin is essential for the establishment of AJs. However, the depletion of E-cadherin in confluent epithelial sheets had little effect on the localization or function of established AJs. Differential E-cadherin expression levels have been associated with altered spheroid formation in head and neck carcinoma cell lines15. Differential E-cadherin expression was also associated with compact spheroid formation in hepatocellular carcinoma cell lines16,17 and in renal cell carcinoma18. In addition spheroid models were used to identify cooperative roles for E-cadherin and the desmosome proteins DSG2 and DSC2 in colon and breast carcinoma cell lines19. Cells lacking the linker protein α-catenin are unable to associate tightly, despite sufficient cadherin expression20,21,22. Even in established epithelial monolayers depletion of α-catenin is essential for the maintenance of AJs23. In vivo, forced E-cadherin expression represses cancer development and loss of E-cadherin24 or cadherin switching25 during epithelial to mesenchymal transition (EMT) is associated with increased mobility and tumor progression as well as metastasis26. Loss of α-catenin induces the formation of premalignant squamous cell cancer in skin. In addition, α-catenin acts as an invasion repressor27 and in colorectal cancer (CRC), reduced α-catenin levels are associated with tumor progression28,29.
However, this is a simplified concept focusing on two major components of the AJ complex in carcinogenesis. The reality in human cancer is by far more complex. For example in human CRC expression changes in a plethora of other cell adhesion genes have been reported, including induction of P-cadherin (CDH3)30, OB-cadherin (CDH11), claudins (CLDN1/2) and desmosomal proteins (DSC3, DSG2) and downregulation of CLDN5/8/23, DSC2, and protocadherins (PCDHB7/14)31. Interestingly, P-cadherin displayed a promotion of invasion whenever E-cadherin was also present in mammary32,33, pancreatic34 and ovarian35 cancer models. In contrast, in HT29 colon cancer cells loss of P-cadherin in the presence of E-cadherin induced a more invasive phenotype30. However, when expressed alone, P-cadherin was invariantly invasion suppressive36,37,38. In breast cancer P-cadherin overexpression is correlated with reduced tumor growth in mice and bad prognosis in human patients when E-cadherin is co-expressed in the tumor, whereas the expression of P-cadherin or E-cadherin alone displayed reduced cancer growth. Loss of both cadherins was associated with an intermediate phenotype39.
Thus, determining the predominant molecule mediating homotypic cell adhesion in cancer cells and loss of adhesion variants in these cells is of vital importance to study and better understand cell adhesion and cell dissemination in human cancer. Here we applied the simple method of spheroid formation to identify subclones of several spheroid-forming colon cancer cell lines, which have lost the ability to integrate into the spheroid and thus lacked the capacity of homotypic cell adhesion. These cells displayed increased migration and invasion in vitro. Expression profiling revealed which adhesion molecule was lost in the non-spheroid-forming (NSF) subclones. Knockdown of candidate genes in the spheroid-forming parental cells by siRNA was a fast, robust and reliable way to functionally test the involvement of the deregulated adhesion molecules in homotypic cell-cell adhesion and spheroid formation ability.
Results
Formation of colon cancer spheroids consistently display single cells separated from compact cell aggregates, which can be enriched by differential sedimentation
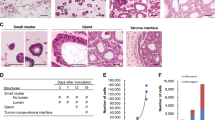
The colon cancer cell lines HCT116, DLD-1 and SW620 were analyzed for spheroid formation capacity in ultra low attachment (ULA) round bottom 96-well plates. We recognized that four days after tumor spheroid formation a small amount of cancer cells did not integrate into the compact spheres (Fig. 1A,B,C). These cells were visible as round, single cells at the border of the spheroids. We hypothesized that these non-spheroid forming (NSF) cells lost the ability to integrate into the spheres and aimed to identify the reason for the exclusion. For molecular analysis the enrichment and separation from the spheroid forming (SF) cells was mandatory. To augment for these NSF cells, spheroids in their culture medium were transferred into 15 ml centrifuge tubes. The spheroids were separated by three minutes of gravitational sedimentation (spheres sank to the bottom of the tube) from NSF cells, which remained - during this short time period - in the supernatant. The supernatants and pellets were collected; the cells were plated on conventional 2D culture plates and were expanded. Subsequent they were subjected for two further rounds of spheroid formation, gravity sedimentation and NSF cell harvesting procedure (Fig. 1D) to yield high purity NSF as well as SF populations, which were also used in further experiments. After three rounds of this enrichment, pure NSF and SF populations were further analyzed. Importantly, these cells displayed normal attachment to plastic surfaces in conventional 2D cultures.
Spheroid formation as a tool to select non spheroid forming (NSF) cells. (A) HCT116 spheroid formation was induced by seeding tumor cells in ULA plates. After 4 days spheroids were formed from the vast majority of cells but a small amount of cancer cells were not incorporated into the compact cell aggregates (arrowheads). (B) DLD-1 cells were treated as in A and photographed. Non-incorporated single cells (NSF) at the periphery of the spheroid surface are indicated by arrowheads. (C) SW620 cells were treated as in A and photographed. Non-incorporated single cells (NSF) at the periphery of the spheroid surface are indicated by arrowheads. Scale bars in overviews: 100 µm. (D) Scheme of workflow to enrich NSF cells from spheroid forming (SF) ones. Spheroids were generated by seeding tumor cells into non-adhesive, U-shaped, 96-well plates. After 4 days tumor spheroids were harvested and transferred in their culture medium from 96-well plates into 15 ml centrifuge tubes. Within 3 min, gravity forced the cancer spheroids to sink to the bottom of the centrifuge tubes and the supernatant, which contained the NSF cells, was removed and further used. This procedure was repeated two times to highly enrich the NSF cells and generate stable sublines.
Enrichment of non-spheroid integrating cells leads to complete loss of spheroid formation capacity
After the three-round separation protocol, the same number (1 × 103) of HCT116-SF and HCT116-NSF cells was seeded into each well of ULA 96-well plates and cell morphology was investigated for 3 days. After 72 h, compact multicellular spheroids were formed by HCT116-SF. In contrast, HCT116-NSF cells were neither capable of producing spheroids nor displaying spheroid-like partial cell clusters after 72 h (Fig. 2A). At higher magnification it was evident that the layer of HCT116-NSF cells was composed of cells with round morphology without any obvious close cell-cell adhesion (Fig. 2A, inset). They displayed a one-cell layer appearance in the round bottom ULA 96-well plate, forced by the U-shaped form of the wells to sit next to each other. Therefore, the NSF cell-mass (expanding only in two dimensions) appeared bigger in diameter albeit the same cell numbers seeded (Fig. 2A) than the SF cells, which formed spheres (compacting and expanding in three dimensions). The cell-covered area was determined and served as a measure for spheroid formation (small area) or loose cell layers (large area). This measurement of projected cell covered areas (see schemes in Fig. 2A) was used to quantitatively describe and distinguish spheroid formation versus cell assemblage at the bottom of the U-well. The projected area was significantly increased in the NSF cells (Fig. 2B).
Comparison of the phenotype of HCT116-SF and SW620-SF cells versus their NSF sublines. (A) Time course of spheroid formation of HCT-116-SF cells compared to HCT-116-NSF. Lack of spheroid formation in the case of HCT116-NSF is evident from the formation of loosely associated cells at the base of the round well bottom. For clarity boarders of cell covered areas (either spheroid projected areas or loose cell assemblages, see schematic side view) are indicated by white dotted lines. (B) Quantification of projected spheroid versus cell assemblage areas of after 72 hours, n = 7 for SF and NSF each. (C) Time course of spheroid formation of SW620-SF cells compared to HCT116-NSF. Lack of spheroid formation in the case of SW620-NSF is evident from the formation of loosely associated cells at he base of the round well bottom. For clarity boarders of cell covered areas (either spheroids or loose cell assemblages) are indicated by white dotted lines. (D) Quantification of projected spheroid versus cell assemblage areas of after 72 h, n = 5 each. Boxes represent the interquartile range; the horizontal line in the box indicates the median; whiskers extend to min and max; p-values are indicated. Scale bars: 100 µm.
SW620, another colon cancer cell line, which forms more loosely aggregated spheroids (see Fig. 1A) when compared to HCT116 cells after 72 h in ULA plates were also analyzed. We wanted to investigate if the SF/NSF enrichment protocol could yield, on the one hand, more compact spheroids (SF) and, on the other hand, if it was possible to obtain NSF cells also of this cancer cell line. The aggregates generated from SW620-SF did not display any change in spheroid morphology (Fig. 2C) as compared to parental SW620. However, SW620-NSF cells showed an even looser aggregation phenotype and were present as copious sheets. These structures were composed of rounded cells without signs of tight cell-cell interaction with their neighbors (Fig. 2C). The projected cell-covered area was increased twofold in NSF vs SF (Fig. 2D). Hence, NSF subtypes can even be isolated from a cell line displaying aggregate behavior rather than spheroid appearance.
The NSF phenotype is reproducibly generated in DLD-1 and SW620 colon cancer cells
Next, we examined the reproducibility of NSF formation using the above-described SW620 and DLD-1 cells. We aimed to collect individual DLD-1-NSF and SW620-NSF sublines from single wells of ULA round bottom 96-well plates. For this, 2 × 102 DLD-1 or SW620 cells were seeded into each well and were subjected to long-term culture (2 weeks) with partial medium changes every third day to supply the spheres with fresh medium. After 14 days of expansion spheroids were grown and interestingly non-integrated NSF cells were also significantly expanded in number (Supplemental Figure S1A). Spheroids were separated from single cells by sedimentation in microcentrifuge tubes and cells were expanded in 2D without further selection. Four SW620 (Supplemental Figure S1B) and six DLD-1 single cell phenotype sub-lines were further analyzed. All of them stably displayed the loss of spheroid formation phenotype (three of the DLD-1 sublines are shown in Fig. 3A) and kept it also after several freeze and thaw cycles. This was also quantitatively verified by assessing the cell-covered area (Fig. 3B).
DLD-1-NSF cells can be reliably obtained and display no chromosomal aberrations compared to their parental origin. (A) DLD-1 cells were seeded into ULA round bottom 96-well plates. After 2 weeks spheroids were separated from non-integrated NSF-cells by sedimentation (see Supplemental Figure S1), cultured in 2D and 6 individual DLD-1-NSF sublines were generated. DLD-1-NSF sublines were analyzed for spheroid formation capacity. Representative images of DLD-1 SF and DLD-1 NSF sublines #3, #5 and #6 after 24 hours, 48 hours and 72 hours at spheroid inducing conditions. Scale bars: 100 µm. (B) Quantification of projected spheroid versus cell assemblage areas of after 72 hours (cell covered areas), DLD-1 SF n = 6, DLD-NSF #3 n = 5, #5 n = 4, #6 n = 4. Boxes represent the interquartile range; the horizontal line in the box indicates the median; whiskers extend to min and max. P-values are indicated. (C) Karyogramms of DLD-1-SF and NSF sublines #3, #5, #6.
The near diploid chromosome set of parental DLD-1 was retained in all DLD-1-NSF sub-lines, as shown by lack of aberrations in metaphase chromosome spreads (Fig. 3C). This indicates that despite the microsatellite instability (MSI) phenotype of DLD-1 there is, at the chromosome level, no selection for grossly aberrant clones by the selection procedure.
The NSF phenotype is associated with increased migratory potential and augmented invasion
In a further set of experiments, we asked whether the loss of the spheroid formation phenotype resulted in further phenotypic differences. In all three lines cell migration was significantly increased in the NSF setting. DLD-1-NSF, HCT116-NSF and SW620-NSF displayed a fourfold, 2.5 fold and 1.3 fold higher migratory ability, respectively, than their corresponding SF counterparts (Fig. 4A). Similarly, invasion through a collagen I matrix was significantly elevated in all three NSF cultures as compared to their corresponding SF sublines (Fig. 4B). Moreover, when seeded as single cells in a collagen I gel, HCT116-SF and SW620-SF formed small compact round colonies after 6 days of culture. In sharp contrast, HCT116-NSF displayed loosely, irregularly formed colonies with cells protruding into the gel matrix. SW620-NSF produced larger colonies than the SF cells and showed spindle like cells invading into the collagen gel (Fig. 4C, Supplemental Figure S2A, B). Interestingly, DLD-1-SF cells died when seeded as single cells into the matrix, whereas DLD-1-NSF displayed irregular colonies with rounded cells and occasional spindle like protrusions (Fig. 4C, Supplemental Figure S2C).
Increased migration and invasion of NSF sublines. (A) For assessment of migration a transwell migration assay was performed and the number of migrated cells was determined after 20 h. In case of HCT116 and DLD-1 cells eight transwell inserts were evaluated; for SW620 cells n = 4. (B) A transwell invasion assay was used and cells invaded through collagen I covered membranes were counted after 24 hours; n = 4 for each individual experiment. Boxes represent the interquartile range; the horizontal line in the box indicates the median; whiskers extend to min and max. P values are indicated. (C) Representative phase contrast pictures of colonies grown from single cells seeded into collagen I gels (CG). Pictures were taken after 6 days (HCT116 and SW620 lines) and 13 days (DLD-1 lines). White asterisks indicate single cells or invasive structures protruding into the matrix. Scale bars: 50 µm.
Quantitative PCR array expression profiling identifies loss of molecules involved in cell adhesion in SW620-NSF, HCT116-NSF and DLD-1-NSF cells
Using two qPCR arrays (RT2 profiler, Qiagen) we aimed to identify the molecular reasons for loss of cell-cell adhesion in the NSF sublines of DLD-1, HCT116 and SW620 cells. These arrays revealed the mRNA expression levels of 162 cell-to-cell and cell-to-matrix adhesion genes and adhesion-related molecules in SW620-SF, SW620-NSF, DLD-1-SF, DLD-1-NSF and HCT116-SF as well as HCT116-NSF (Heatmaps, Fig. 5A,B; full dataset in Supplementary Table 1). The mRNAs most effectively lost in SW620-NSF, HCT116-NSF and DLD-1-NSF compared to their respective SF counterparts were E-cadherin (CDH1), P-cadherin (CDH3) and α-catenin (CTNNA1), respectively; see Fig. 5A–E. A complete list of all genes analyzed including Ct values, negative and positive controls as well as fold changes are given in Supplementary Table 1. Loss of these molecules in the NSF cells was confirmed at the protein level in Western blot experiments. E-cadherin was lost only in SW620-NSF but was maintained in the other NSF lines, whereas HCT116-NSF showed selective lack of P-cadherin and DLD-1-NSF displayed unique loss of expression of α-catenin (Fig. 5F). This lack of α-catenin was consistently seen in any individually generated DLD-1-NSF subline, whereas E- and P-cadherin displayed constant high expression (Fig. 5G).
Detection of molecular changes in NSF cells versus SF cells using RT2 profiler qPCR arrays. (A) SW620, DLD-1 and HCT116 SF-and NSF-cells were subjected to sphere forming conditions and mRNA was isolated and analyzed with RT2 PCR profiling to obtain expression patterns for 160 adherens junction, extracellular matrix and adhesion molecules. Heat maps indicating the level of expression representing are shown (red: high expression, yellow: intermediate expression, white low or absent). Full dataset available in Supplementary Table 1. (C–E) Quantification of CDH1, CDH3 and CTNNA1 mRNA expression of the data from (A). Maximal fold changes (FC) between SF and NSF are indicated in red. (F) E-(CDH1), P-cadherin (CDH3) and α-catenin (CTNNA1) protein expression was assessed by Western blotting. GAPDH is shown as loading control. (G) Western blot analysis of α-catenin as well as E- and P-cadherin expression in the DLD1-NSF subclones #1–6. GAPDH and α-tubulin are shown as controls.
Lack of cell-cell adhesion by loss of one adhesion protein in the NSF phenotype is not associated with changes in expression and distribution of other adhesion markers
The single loss of either E-cadherin, P-cadherin or α-catenin could potentially effect the expression or (sub)cellular distribution of other important adhesion molecules. To test this, spheroids were immunofluorescently stained with the cell adhesion markers E-cadherin, P-cadherin, epithelial cell adhesion molecule (EpCAM) and F-actin (Fig. 5A) and optical section were recorded with a confocal microscope. Loss of E-cadherin was evident in SW620-NSF cells (Fig. 6A) as already seen in Western blots (compare Fig. 5F), but did not affect EpCAM, P-cadherin of F-actin expression or distribution (Fig. 6A, Supplemental Figure S3). The same was evident when DLD-1-SF and DLD-1-NSF cells (Supplemental Figure S4) as well as HCT116-SF and HCT116-NSF cells were analyzed (Supplemental Figure S5). Taken together, these data imply that no gross alterations of other cell adhesion molecules were induced upon the loss of one specific molecule in the distinct cell types.
Distribution of adhesion markers and functional assessment of loss of E-cadherin, P-cadherin and α-catenin for spheroid formation by RNA interference. (A) IF analysis of E-cadherin, EpCAM, P-cadherin (all green) and F-actin (red) expression in SW620-SF and SW620-NSF. Cell nuclei are stained with DAPI (blue). Small inset: IgG control. Scale bars: 25 µm. B left panel, representative phase contrast micrographs of SW620 NTC vs SW620-CDH1-KD. Scale bars: 100 µm. (C) IF analysis of E-cadherin, EpCAM, P-cadherin (all green) and F-actin (red) expression in SW620-NTC and SW620-CDH1 KD cells. Cell nuclei are stained with DAPI (blue) Scale bars: 25 µm. Representative phase contrast pictures of (D) DLD-1-NTC vs DLD-1-CTNNA-KD, (E) HCT116-NTC vs HCT-CDH3KD (left panels). Cell covered areas are encircled with a white dotted line for clarity. Scale bars: 100 µm. (B,D,E) Middle panels Quantification of spheroid versus cell assemblage areas after 72 h (projected areas), SW620 n = 4 each; DLD-1-NTC n = 9, DLD-1-CTNNA1-KD n = 8; HCT116, n = 8 each. Boxes represent the interquartile range; the horizontal line in the box indicates the median; whiskers extend to min and max. P-values are indicated. (B,D,E) right panels Knockdown efficiency was determined by Western blotting.
Knockdown of E-cadherin, P-cadherin or α-catenin lead to a NSF phenotype in SW620, HCT116 or DLD-1, respectively
Next we tested, whether loss of the respective molecules identified above in the NSF colon cancer cells were causally responsible for the lack of cell adhesion during spheroid formation. For this the molecules were knocked down by siRNA in the corresponding parental cell lines. Indeed loss of E-cadherin in SW620 cells (SW620-CDH1-KD) phenocopied the SW620-NSF cells in respect to the incapability of spheroid formation, whereas non-targeting controls (SW620-NTC) displayed proper cell aggregation like parental SW620 (Fig. 6B, compare to Fig. 2A). In accordance to the loss of E-cadherin in the NSF cells, CDH1 knockdown in NTC siRNA transfected parental SW620 cells did not change levels or localization of other junction proteins like P-cadherin, EpCAM or the junction associated protein F-actin as demonstrated by immunofluorescence analysis (Fig. 6C, Supplemental Figure S6).
Similarly, DLD-1-CTNNA1-KD (Fig. 6D) and HCT116-CDH3-KD (Fig. 6E) lost spheroid formation ability. Strikingly, these knockdown cells displayed exactly the same phenotype as the respective NSF sublines (compare to Figs 2A and 3A). NTC controls formed spheres indistinguishable from non-modified DLD-1 or HCT116 cells. High knockdown efficiency was verified by Western blotting (Fig. 6B–D, right panel).
Functional assessment of loss of E-cadherin, P-cadherin and α-catenin for spheroid formation
E-cadherin is the major homotypic cell-cell adhesion molecule for epithelial cells and it was expressed at comparable levels in all three colon cancer cell lines analyzed. However, downregulation was only seen in SW620-NSF cells. Therefore, we were interested, whether forced loss of E-cadherin expression in HCT116 and DLD-1 cells would interfere with spheroid formation. Interestingly, in HCT116 cells knockdown of E-cadherin (CDH1) did not disrupt spheroid establishment at all (Fig. 7A), whereas in DLD-1 loss of E-cadherin impaired spheroid formation (Fig. 7B). In both cell types E-cadherin was efficiently reduced at the protein level (Fig. 7C). In two other colon cancer cell lines (LS174T and HT29), which were not included in the initial study, ablation of E-cadherin expression also induced a NSF phenotype (Supplemental Figure S7).
Phenotypes induced by loss of E-cadherin, P-cadherin and α-catenin by RNA interference. (A,D,G) Phase contrast micrographs of cells cultivated under spheroid forming conditions for 48 hours. Cell covered areas are encircled with a white dotted line for clarity. Scale bars: 100 µm. (A) CDH1 knockdown in HCT116 and DLD-1 cells. (D) CDH3 knockdown in DLD-1 and SW620 cells. (G) CTNNA1 knockdown in HCT116 and SW620 cells. (B,E,H) Quantification of projected spheroid versus cell assemblage areas of after 48 hours (cell covered areas). Boxes represent the interquartile range; the horizontal line in the box indicates the median; whiskers extend to min and max. P-values are indicated and non-significant comparisons are labeled with ns. (B) HCT116-NTC, HCT116-CDH1-KD n = 4 each; DLD-1-NTC, DLD-1-CDH1-KD n = 6 each; E DLD-1-NTC, DLD-1-CDH3-KD n = 6 each; SW620-NTC, SW620-CDH3-KD n = 6 each. H HCT116-NTC, HCT116-CTNNA1-KD n = 6 each; SW620-NTC n = 6, SW620-CTNNA1-KD n = 5. (C,F,I) Knockdown efficiency was determined by Western blotting.
DLD-1 cells expressed also high levels of P-cadherin (CDH3) in addition to E-cadherin (see Fig. 5F). Therefore, we were interested whether depletion of P-cadherin might have an effect on proper spheroid formation in these cells. Again, siRNA-mediated knockdown of CDH3 was performed in parental DLD-1 cells and spheroid formation was monitored. No difference in compact spheroid formation was detectable between DLD-1-NTC and DLD-1-CDH3 (Fig. 7D,E), in spite of highly effective knockdown as verified by almost complete loss of P-cadherin protein expression in the siRNA treated cells (Fig. 7F). As SW620 cells did not express P-cadherin (see Figs 4 and 7F) no change of phenotype was expected when these cells were treated with CDH3 siRNA. Indeed, there was no difference in spheroid formation detectable and NTC and CDH3 siRNA transfection of SW620-SF cells led to the same spheroid formation phenotype as seen in parental SW620-SF cells (Fig. 7D,E). This experiment also demonstrated that the transfection approach with a targeting siRNA per se, did not alter the spheroid formation ability.
Due to its proposed central mechanical role as a tension transducer in the development of adherens junctions14 and its essential role for cell adhesion20,21,22,40,41 we speculated that α-catenin (CTNNA1) might have a major role in spheroid formation in all our models. Indeed, knockdown of α-catenin in all three cell lines tested (DLD-1, SW620 and HCT116) completely impaired spheroid formation (Figs 6D and 7G,H and Supplemental Figure S8). Depletion of CTNNA1 was highly efficient at the protein level (Figs 6D and 7I).
Double knockdown of E- and P-cadherin results in a severe loss of cell adhesion phenotype in DLD-1 but not in HCT116
Next, we examined whether in HCT116 and DLD-1, which express E-cadherin and P-cadherin simultaneously, double knockdown of both cell adhesion molecules would result in a more severe phenotype. This would suggest that both adhesion molecules are participating in cell-cell adhesion in these cells leading to residual cell to cell binding activity, if only one is knocked out. Results from above indicated that E-cadherin is the major factor mediating strong cell-cell adhesion and spheroid formation in DLD-1, whereas in HCT116 P-cadherin has this function.
In HCT116 double knockdown of CDH1 and CDH3 had no surplus effect on the phenotype compared to HCT116-CDH3 KD. In relation to HCT116-NTC, the increase in cell-covered area remained the same in HCT116-CDH1-CDH3-KD as compared to HCT116-CDH3-KD (Fig. 8A). Of note, E-cadherin knockdown alone again had no effect on spheroid formation at all. All molecules were efficiently depleted in the respective knockdowns or knockdown combinations as demonstrated by Western blotting (Fig. 8B).
Effect of double versus single knockdowns in HCT116 and DLD-1 cells. (A,C) Quantification of projected spheroid versus cell assemblage areas of after 48 h (cell covered areas). Boxes represent the interquartile range; the horizontal line in the box indicates the median; whiskers extend to min and max. P-values are indicated and non significant comparisons are labeled with ns. HCT116-NTC (n = 12), HCT116-CDH1-KD (n = 12); HCT116-CDH3-KD (n = 10), HCT116-CDH1/3-KD (n = 13); DLD-1-NTC (n = 17), DLD-1-CDH1-KD (n = 12), DLD-1-CDH3-KD (n = 15), DLD-1-CDH1/3-KD (n = 17), DLD-1-CTNNA1-KD (n = 8). (B,D) Knockdown efficiency was determined by Western blotting.
Interestingly, in DLD-1, double knockdown of CDH1 and CDH3 resulted in significantly increased cell-covered areas as compared to DLD-1-CDH1-KD (Fig. 8C). This indicated that, when E-cadherin is lost, there is still residual low cell-cell binding capacity present, which is mediated by P-cadherin in these cells. Surprisingly, depletion of α-catenin resulted in a phenotype similar to E-cadherin loss alone (Fig. 8C,right panel), suggesting that unexpectedly residual P-cadherin mediated residual cell adhesion might be independent of α-catenin. Again, proper knockdown was verified by Western blotting (Fig. 8D).
Discussion
Cancer progression is often associated with a change of cell adhesion properties such as loss of tight cell-cell interaction24, cadherin switching during EMT25, loss of E-cadherin or induction of cell adhesion molecules being not expressed in the normal epithelial cells of the corresponding tissue31,42. We report here a simple, rapid and cost efficient way to select for cancer cells with diminished homotypic cell-cell interaction. The exclusion of subpopulations of cells into multicellular spheroids could be used as a highly reproducible and robust way to select for loss of cell adhesion variants in the allegedly homogenous cell population of various established colon cancer cell lines (HCT116, DLD-1, SW620). Of note, this selection procedure was applicable in cell lines displaying a wide range of different spheroid phenotypes. Spheroid morphology ranged from tightly associated, compact and dense spheroids with established tight junctions (DLD-143) via an intermediate phenotype (HCT116) to rather loose cell aggregates (SW620). Despite this variation, the gravity based selection process (see Fig. 1C) was effective to produce non spheroid forming (NSF) sublines from these three cancer cell lines. There are several ways of spheroid formation (ultralow attachment plates, presence of methylcellulose to avoid attachment of the cells to plastic, hanging drops or specialized plates for hanging drop like spheroid formation for example from InSphero) or stirring. These are different technical approaches to obtain the same principle: loss of attachment to the culture plate and therefore induction of cell-cell adhesion and spheroid formation. For isolating NSF cells by gravity all these methods would be feasible, however, a 96-well round bottom setting is essential for visual inspection of the appearance of NSF cells nearby the spheroid.
Moreover, as demonstrated for DLD-1 and SW620, parallel selection rounds always gave rise to multiple NSF sublines (Supplemental Figure S1) which displayed no changes in chromosome content (see Fig. 3) compared to the parental cells. These independently generated NSF sublines all displayed the same NSF phenotype and at the molecular level consistently lost α-catenin expression, whereas E- and P-cadherin levels did not change. Thus, these results implicate that the enrichment protocol for NSF cells might be a widely applicable and consistent novel method to isolate loss of cell adhesion mutants from other cancer cell lines of different origin or presumably also from primary patient material when grown as cancer fragment explants44 or as organoids45.
Molecular profiling of the NSF population and comparison to the parental or SF counterpart using qPCR profiling (as done in our approach with RT2 profiler PCR arrays for cell junction and adhesion molecules) readily identified candidate genes, which were significantly different in the SF vs NSF variants. More global analysis by full genome profiling (arrays or RNAseq) would be another possibility. Of note, the majority of these genes were not regulated, indicating that the protocol did not select for or even induced grossly abnormal cancer cell subtypes but rather identified highly specific changes in the NSF sublines. This was further supported by the unchanged DLD-1 karyotype in the NSF versus the parental cells as discussed above.
The identification of target genes allowed rapid functional testing by siRNA-mediated knockdown with an easy and fast readout. For this, selective knockdown of the identified target in the parental (i.e. spheroid forming) population and subsequent culture of these cells under spheroid forming conditions is performed. If the identified target is indeed responsible for homotypic cell-cell adhesion mediated spheroid formation, the knockdown cells are no longer able to form spheroids as compared to cells transfected with non-targeting control siRNA. Thus, simple functional experiments with a highly sensitive biological readout (spheroid formation versus loss of spheroid formation) rapidly validated identified target genes. In our experiments we identified three different molecules being responsible for the loss of spheroid formation ability in three different colon cancer cell sublines, respectively. This mechanistic testing demonstrated that in SW620-NSF loss of E-cadherin was responsible for the inability to form spheroids, whereas in HCT116-NSF lack of P-cadherin and the absence of α-catenin in DLD-1-NSF were the reasons for impaired spheroid formation.
HCT116 cells express both E-cadherin and P-cadherin proteins in parallel (see Figs 5F and 7C,I). In HCT116 a frameshift mutation of CDH1 (c.357delG, p.H121fs*94) at one allele has been reported by Efstathiou et al.46 and Cosmic database mining with the CanSAR website47, which leads to heterozygous deletion of the majority of the E-Cadherin protein encoded by this allele. The other allele is unaffected and full length E-cadherin is detected in Western blots and is located at cell-cell contacts (our results and Tay et al.48). Whether the heterozygous mutation leads to a dominant negative form has not been investigated so far. However, we show here for the first time that HCT116 spheroid formation is independent from E-cadherin expression but critically dependent on the presence of P-cadherin. Of note, P-cadherin expression in HCT116 is known and a specific antibody against P-cadherin (PF-03732010) disrupts cell aggregate formation in HCT116 and has anti-metastatic effects in vivo in mice HCT116 xenograft experiments49. Thus, we conclude that in HCT116 a subpopulation is steadily emanating that loses P-cadherin expression leading to the loss of cell-cell adhesion phenotype. In line with this, even the selected SF sublines of HCT116 produce NSF cells. The molecular reason for the P-cadherin loss is not known so far and additional experiments are necessary to further evaluate the phenotype in vitro and in vivo.
In contrast, in DLD-1 forced depletion of E-cadherin but not P-cadherin resulted in the loss of spheroid formation. However, loss of E-cadherin was not detected in the naturally occurring NSF variants isolated by the NSF selection protocol despite analysis of seven independent attempts. In DLD-1 α-catenin was consistently lost in the NSF subclones. Natural round-shaped variants of DLD-1 cells deficient for α-catenin were reported earlier and these cells also displayed impaired cell aggregation ability21. Cell-cell adhesion could be restored by re-expressing wildtype α-catenin in these cells50 leading to decreased proliferation in 3D. Strikingly, loss of α-catenin was shown for a subpopulation of HCT-8 cells, which displayed a round morphology phenotype28. The colon cancer cell line HCT-8 derived from the same patient and is identical to DLD-127 as well as HCT-15 and HRT-1851. This was further validated by STR profiling, RNAseq, mutational analysis and drug response pattern52. The CTNNA1 gene is heterozygously mutated in DLD-1/HCT-8/HCT-15/HRT-18. Due to genetic instability as a consequence of a mutation in the HMSH6 mismatch repair gene, round-shaped cell variants occur spontaneously, all carrying either a mutation or exon skipping in the second CTNNA1 allele27. These mutants lacking α-catenin expression were shown to be more invasive in a chick heart invasion assay27. Thus, these data clearly demonstrate that two totally different assays based on phenotypic appearance (round appearance versus exclusion from spheroid formation) could identify the same mutant subpopulations of cells. The spheroid assay might be of advantage for high throughput screening to identify loss of cell-cell adhesion in any parental spheroid-forming cell line and less experienced researchers in cell biology might find it easier to identify and isolate such variants by the spheroid assay than by assessing rather subtle morphological differences in 2D culture.
Interestingly, despite the reported dysfunctional mismatch repair in DLD-1, we could not identify lack of E-cadherin expression in any of the DLD-1-NSF sublines; it was always α-catenin affected. However, artificial depletion of E-cadherin by siRNA-mediated knockdown displayed the same NSF phenotype as loss or mutation of CTNNA1. Either depletion of E-cadherin or α-catenin led to the very same phenotype and the cell-covered area was indistinguishable under non-attachment conditions. Loss of P-cadherin in DLD-1 did not disrupt the spheroid forming ability at all. Unexpectedly however, depletion of E-cadherin and P-cadherin by double knockdown resulted in an even stronger loss of cell-cell adhesion, and a significantly increased cell-covered area than NSF phenotypes induced by E-cadherin or α-catenin downmodulation alone (see Fig. 7). This was not the case for HCT116 cells, indicating a specific effect. We speculate that loss of both E- and P-cadherin result in total loss of cell adhesion, whereas there is obviously a residual weak cell adhesion activity left in cells devoid of either E-cadherin or α-catenin, and which is facilitated by P-cadherin. Thus, P-cadherin is able to mediate a weak but measurable cell-cell adhesion effect, which is clearly independent of α-catenin.
Taken together, on the one hand we provide here a novel protocol to conveniently and reliably isolate subpopulations of cancer cell lines with altered cell-cell adhesion properties leading to a non-spheroid forming phenotype. On the other hand, we identified the molecular reason for this loss of adhesion in three colon cancer cell lines using expression profiling and functional testing with a siRNA approach. This is valid to all cell lines of different cancers with spheroid forming capacity. Thus, as changes in cell-cell adhesion properties of cancer cells are critically involved in cancer progression and dissemination, our method provides a broadly applicable protocol and our results on the molecular reasons for the loss of homotypic cancer cell adhesion build the basis for further studies on cell adhesion phenotypes and will shed new light on the impact of these changes on cancer cell growth, invasion and metastasis.
Methods
Cell culture
Human colon cancer cell lines DLD-1 (ATCC®# CCL-221TM), HCT116 (ATCC®# CCL-247 TM) and SW620 (ATCC®#CCL-227 TM) were obtained from the American Type Culture Collection (ATCC® Rockville, MD, USA) and were cultured in Dulbecco’s modified Eagle’s medium (DMEM), high glucose (4.5 g/l), or in RPMI respectively, supplemented with 10% fetal calf serum, 2 mM L-Glutamine and antibiotics (60 mg/l penicillin, 100 mg/l streptomycin sulfate) at 37 °C and 5% CO2.
Spheroid formation
Spheroid formation was induced as described53. In brief, 3000 colon cancer cells were seeded in 100 µl cell culture medium per well in an ultra-low attachment (ULA) 96-well plates (Nunclon Sphera, round bottom, Thermo Scientific). The plates were centrifuged for 15 min at 300 g in order to ensure immediate close contact of cells and thereafter spheroids were formed for 3 days and grown in DMEM high glucose (4.5 g/l), supplemented with 5% fetal calf serum, 2 mM L-Glutamine and antibiotics (60 mg/l penicillin, 100 mg/l streptomycin sulfate). Centrifugation is not essential for spheroid formation but mediates more rapid spheroid formation.
Metaphase Spreads (Karyogram analysis)
Metaphase preparation was carried out by standard methods. By adding 0.1 μg/ml colcemid (Gibco, ThermoFisher) dividing cells were locked in metaphase for 60 minutes. Then cells were treated for 15 minutes with hypotonic solution and fixed using methanol and acetic acid. Afterwards cells were dropped onto slides, dried at 42 °C for 30 minutes and then incubated at 60 °C over night. Slides were used for Giemsa-trypsin banding of chromosomes. Karyotyping was done using the “MetaSystems Ikaros” software version 5.3.18.
Expression profiling using RT2 profiler arrays
For gene expression analysis, ‘Human Adherens Junctions’ and ‘Human Extracellular Matrix and Adhesion Molecules’ RT2 Profiler PCR Arrays (Qiagen) were used. These RT2 Arrays profile simultaneously the expression of 84 genes involved in cell-cell contact and cell adhesion, within one sample. In brief, cells were harvested and mRNA was isolated using the ReliaPrepTM RNA Cell Miniprep System (Promega). The mRNA concentration was measured with a NanoDrop Fluorospectrometer (Thermo Fisher Scientific, Inc.; Waltham, MA, USA). For cDNA synthesis the RT2 First Strand Kit was used. A total of 0.5 µg RNA was reverse transcribed and the cDNA was amplified by the RT2 SYBR Green Master mix. Data were evaluated using the Qiagen PCR Array Data Analysis Web portal. Gene expression was normalized to the mean expression of the reference genes (GAPDH, ACTB, B2M HPRT and RPLP0) and was calculated using the ∆∆CT method.
siRNA-mediated knockdown
For siRNA-mediated knockdown, cells were seeded in 6-well plates and were grown overnight to 20% confluence. For transfection, a total of 100 pM siRNA (ON-Target plus Smart pools, Dharmacon; for double knockdown 50 pM of siRNA for target 1 and for target 2), and 5 μl of Lipofectamine RNAiMAX were mixed with Opti-MEM I and incubated for 20 min at room temperature. Then the formed siRNA/lipid complexes were carefully added per well (to a final concentration of 50 nM siRNA). Cells were incubated for 72 hours and grown under normal conditions to 90% confluence. Subsequently, the transfected cells were used for spheroid formation and Western blotting.
Western blotting
For Western blotting whole cell lysates were prepared from spheroids. Briefly, spheroids were washed in PBS and extracted in RIPA lysis buffer (50 mM Tris pH 7.6, 150 mM NaCl, 1% TritonX-100, 0.1% SDS, 0.5% sodium deoxycholate, 1 mM PMSF, 4 µg/ml aprotinin, 4 µg/ml leupeptin, 0.6 µg/ml benzamidinchloride, 20 µg/ml trypsin inhibitor). Samples (10–15 µg of total protein content) were mixed with 4 × loading dye (200 mM Tris pH 6.8, 400 mM DTT, 8% SDS, 0.4% bromophenol blue, 40% glycerol), denatured at 95 °C for 5 min, subjected to SDS-PAGE and transferred to a nitrocellulose membrane. Blocked membranes were probed with primary antibodies at 4 °C overnight and with horseradish peroxidase-conjugated secondary antibodies (anti-mouse IgG-heavy and light chain antibody and anti-rabbit IgG-heavy and light chain antibody, Bethyl Laboratories Inc., Montgomery, TX) at RT for 1 h. Signals were detected using enhanced chemiluminescence. Primary Antibodies used were: anti-E-Cadherin (clone HECD1, mouse monoclonal, 1:1000, Abcam ab1416), anti-P-Cadherin (clone OTI2D5, mouse monoclonal, 1:4000, OriGene TA506403), anti α-Catenin (clone 1G5, mouse monoclonal, 1:500, Thermo Scientific MA1-2000), anti GAPDH (rabbit, 1:10000, Trevigen 2275-PC-100), anti α-Tubulin (clone DM1A, mouse monoclonal, 1:1000, Merck-Millipore, Calbiochem CP06).
Immunofluorescence
Immunofluorescence staining was performed on whole mount spheroids as described54. In brief, spheroids were fixed in PBS containing 4% PFA and 1% Triton X-100 for 3 h at 4 °C and washed in PBS. Spheroids were then dehydrated in an ascending series of methanol and subsequently rehydrated. After blocking in PBST (0.1% Triton X-100 in PBS)/3% Bovine Serum Albumin (MP Biomedicals, Illkirch, France) o/n at 4 °C and washing in PBST spheroids were incubated with primary antibodies in PBST at 4 °C for 48 h and washed followed by incubation with Alexa Fluor conjugated secondary antibodies for 24 h. Cell nuclei were counterstained with DAPI. 5 µm optical sections were recorded on a on a Leica-SP8 with a 20× immersion objective (NA = 1.3; Leica Microsystems, Wetzlar, Germany). Antibodies used were: E-cadherin (HECD-1, ab1416, Abcam, 1:150), P-Cadherin (TA506403, OriGene, 1:100), EpCAM (ab8601, Abcam, 1:50), phalloidin-Alexa Fluor 594 (A 22283, Thermo Scientific, Waltham, MA, 1:40). Anti-mouse-IgG-heavy-and-light-chain (Bethyl Laboratories Inc., Montgomery, TX) was used as IgG control. Alexa Fluor 488-conjugated secondary antibody (Thermo Scientific) was used at 1:500.
Migration and invasion assays
For migration 2.5 × 104 cells were seeded in serum-free medium in the upper chamber of transwell migration inserts (8 µm, Sigma-Aldrich). The cells were allowed to migrate towards complete medium. After 20 hours, non-migrated cells were removed and migrated cells were fixed (Roti-Histofix 4%) and stained for 30 min with 0.006% (w/v) crystal violet-solution (Sigma-Aldrich). Five random images were taken and membrane coverage was determined (ImageJ64) or cells were counted manually. The invasion assay was performed similar to the migration assay except that the transwell inserts were prior covered with collagen I gel and the experiment was analyzed after 24 hours. For collagen gel invasion assays 200 single cells were embedded in Collagen I gel as described55. The medium was changed every second day. After 6–13 days pictures of the formed colonies were taken.
Statistical analysis
Quantitative experiments were conducted at least in biological triplicates and statistical significance was calculated using the Student’s T-test or one way ANOVA. P-values are indicated. The number of replicates is always indicated in the Figure legends.
References
Kim, J. B. Three-dimensional tissue culture models in cancer biology. Semin Cancer Biol 15, 365–377, https://doi.org/10.1016/j.semcancer.2005.05.002 (2005).
Hirschhaeuser, F. et al. Multicellular tumor spheroids: an underestimated tool is catching up again. J Biotechnol 148, 3–15, https://doi.org/10.1016/j.jbiotec.2010.01.012 (2010).
Frieboes, H. B. et al. An integrated computational/experimental model of tumor invasion. Cancer Res 66, 1597–1604, https://doi.org/10.1158/0008-5472.CAN-05-3166 (2006).
Yamada, K. M. & Cukierman, E. Modeling tissue morphogenesis and cancer in 3D. Cell 130, 601–610, https://doi.org/10.1016/j.cell.2007.08.006 (2007).
Liu, H., Radisky, D. C., Wang, F. & Bissell, M. J. Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. J Cell Biol 164, 603–612, https://doi.org/10.1083/jcb.200306090 (2004).
Streuli, C. H., Bailey, N. & Bissell, M. J. Control of mammary epithelial differentiation: basement membrane induces tissue-specific gene expression in the absence of cell-cell interaction and morphological polarity. J Cell Biol 115, 1383–1395 (1991).
Weaver, V. M. et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol 137, 231–245 (1997).
Friedl, P., Hegerfeldt, Y. & Tusch, M. Collective cell migration in morphogenesis and cancer. Int J Dev Biol 48, 441–449, https://doi.org/10.1387/ijdb.041821 (2004).
Amatangelo, M. D., Bassi, D. E., Klein-Szanto, A. J. & Cukierman, E. Stroma-derived three-dimensional matrices are necessary and sufficient to promote desmoplastic differentiation of normal fibroblasts. Am J Pathol 167, 475–488, https://doi.org/10.1016/S0002-9440(10)62991-4 (2005).
Desoize, B. & Jardillier, J. Multicellular resistance: a paradigm for clinical resistance? Crit Rev Oncol Hematol 36, 193–207 (2000).
van Roy, F. & Berx, G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci 65, 3756–3788, https://doi.org/10.1007/s00018-008-8281-1 (2008).
Tsukita, S., Furuse, M. & Itoh, M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol 2, 285–293, https://doi.org/10.1038/35067088 (2001).
Vite, A., Li, J. & Radice, G. L. New functions for alpha-catenins in health and disease: from cancer to heart regeneration. Cell Tissue Res 360, 773–783, https://doi.org/10.1007/s00441-015-2123-x (2015).
Yonemura, S., Wada, Y., Watanabe, T., Nagafuchi, A. & Shibata, M. alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol 12, 533–542, https://doi.org/10.1038/ncb2055 (2010).
Schmidt, M., Scholz, C. J., Polednik, C. & Roller, J. Spheroid-based 3-dimensional culture models: Gene expression and functionality in head and neck cancer. Oncol Rep 35, 2431–2440, https://doi.org/10.3892/or.2016.4581 (2016).
Pomo, J. M., Taylor, R. M. & Gullapalli, R. R. Influence of TP53 and CDH1 genes in hepatocellular cancer spheroid formation and culture: a model system to understand cancer cell growth mechanics. Cancer Cell Int 16, 44, https://doi.org/10.1186/s12935-016-0318-1 (2016).
Lin, R. Z., Chou, L. F., Chien, C. C. & Chang, H. Y. Dynamic analysis of hepatoma spheroid formation: roles of E-cadherin and beta1-integrin. Cell Tissue Res 324, 411–422, https://doi.org/10.1007/s00441-005-0148-2 (2006).
Shimazui, T. et al. Role of complex cadherins in cell-cell adhesion evaluated by spheroid formation in renal cell carcinoma cell lines. Oncol Rep 11, 357–360 (2004).
Saias, L., Gomes, A., Cazales, M., Ducommun, B. & Lobjois, V. Cell-Cell Adhesion and Cytoskeleton Tension Oppose Each Other in Regulating Tumor Cell Aggregation. Cancer Res 75, 2426–2433, https://doi.org/10.1158/0008-5472.CAN-14-3534 (2015).
Watabe, M., Nagafuchi, A., Tsukita, S. & Takeichi, M. Induction of polarized cell-cell association and retardation of growth by activation of the E-cadherin-catenin adhesion system in a dispersed carcinoma line. J Cell Biol 127, 247–256 (1994).
Ozawa, M. Identification of the region of alpha-catenin that plays an essential role in cadherin-mediated cell adhesion. J Biol Chem 273, 29524–29529 (1998).
Maeno, Y. et al. alpha-catenin-deficient F9 cells differentiate into signet ring cells. Am J Pathol 154, 1323–1328 (1999).
Capaldo, C. T. & Macara, I. G. Depletion of E-cadherin disrupts establishment but not maintenance of cell junctions in Madin-Darby canine kidney epithelial cells. Mol Biol Cell 18, 189–200, https://doi.org/10.1091/mbc.E06-05-0471 (2007).
Cavallaro, U. & Christofori, G. Cell adhesion in tumor invasion and metastasis: loss of the glue is not enough. Biochim Biophys Acta 1552, 39–45 (2001).
Cavallaro, U., Schaffhauser, B. & Christofori, G. Cadherins and the tumour progression: is it all in a switch? Cancer Lett 176, 123–128 (2002).
Onder, T. T. et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res 68, 3645–3654, https://doi.org/10.1158/0008-5472.CAN-07-2938 (2008).
Vermeulen, S. J. et al. The alphaE-catenin gene (CTNNA1) acts as an invasion-suppressor gene in human colon cancer cells. Oncogene 18, 905–915, https://doi.org/10.1038/sj.onc.1202348 (1999).
Vermeulen, S. J. et al. Transition from the noninvasive to the invasive phenotype and loss of alpha-catenin in human colon cancer cells. Cancer Res 55, 4722–4728 (1995).
Raftopoulos, I., Davaris, P., Karatzas, G., Karayannacos, P. & Kouraklis, G. Level of alpha-catenin expression in colorectal cancer correlates with invasiveness, metastatic potential, and survival. J Surg Oncol 68, 92–99 (1998).
Van Marck, V., Stove, C., Jacobs, K., Van den Eynden, G. & Bracke, M. P-cadherin in adhesion and invasion: opposite roles in colon and bladder carcinoma. Int J Cancer 128, 1031–1044, https://doi.org/10.1002/ijc.25427 (2011).
Bujko, M. et al. Expression changes of cell-cell adhesion-related genes in colorectal tumors. Oncol Lett 9, 2463–2470, https://doi.org/10.3892/ol.2015.3107 (2015).
Paredes, J. et al. P-cadherin is up-regulated by the antiestrogen ICI 182,780 and promotes invasion of human breast cancer cells. Cancer Res 64, 8309–8317, https://doi.org/10.1158/0008-5472.CAN-04-0795 (2004).
Ben Hamida, A. et al. Markers of subtypes in inflammatory breast cancer studied by immunohistochemistry: prominent expression of P-cadherin. BMC Cancer 8, 28, https://doi.org/10.1186/1471-2407-8-28 (2008).
Taniuchi, K. et al. Overexpressed P-cadherin/CDH3 promotes motility of pancreatic cancer cells by interacting with p120ctn and activating rho-family GTPases. Cancer Res 65, 3092–3099, https://doi.org/10.1158/0008.5472.CAN-04-3646 (2005).
Cheung, L. W. et al. P-cadherin cooperates with insulin-like growth factor-1 receptor to promote metastatic signaling of gonadotropin-releasing hormone in ovarian cancer via p120 catenin. Oncogene 30, 2964–2974, https://doi.org/10.1038/onc.2011.7 (2011).
Van Marck, V. et al. P-cadherin promotes cell-cell adhesion and counteracts invasion in human melanoma. Cancer Res 65, 8774–8783, https://doi.org/10.1158/0008-5472.CAN-04-4414 (2005).
Sarrio, D. et al. Functional characterization of E- and P-cadherin in invasive breast cancer cells. BMC Cancer 9, 74, https://doi.org/10.1186/1471-2407-9-74 (2009).
Bauer, K., Dowejko, A., Bosserhoff, A. K., Reichert, T. E. & Bauer, R. J. P-cadherin induces an epithelial-like phenotype in oral squamous cell carcinoma by GSK-3beta-mediated Snail phosphorylation. Carcinogenesis 30, 1781–1788, https://doi.org/10.1093/carcin/bgp175 (2009).
Ribeiro, A. S. et al. P-cadherin functional role is dependent on E-cadherin cellular context: a proof of concept using the breast cancer model. J Pathol 229, 705–718, https://doi.org/10.1002/path.4143 (2013).
Ozawa, M. & Kemler, R. Altered cell adhesion activity by pervanadate due to the dissociation of alpha-catenin from the E-cadherin.catenin complex. J Biol Chem 273, 6166–6170 (1998).
Imamura, Y., Itoh, M., Maeno, Y., Tsukita, S. & Nagafuchi, A. Functional domains of alpha-catenin required for the strong state of cadherin-based cell adhesion. J Cell Biol 144, 1311–1322 (1999).
van Roy, F. Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat Rev Cancer 14, 121–134, https://doi.org/10.1038/nrc3647 (2014).
Walzl, A. et al. The Resazurin Reduction Assay Can Distinguish Cytotoxic from Cytostatic Compounds in Spheroid Screening Assays. J Biomol Screen 19, 1047–1059, https://doi.org/10.1177/1087057114532352 (2014).
Cheung, K. J., Gabrielson, E., Werb, Z. & Ewald, A. J. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 155, 1639–1651, https://doi.org/10.1016/j.cell.2013.11.029 (2013).
Prasetyanti, P. R. et al. Regulation of stem cell self-renewal and differentiation by Wnt and Notch are conserved throughout the adenoma-carcinoma sequence in the colon. Molecular cancer 12, 126, https://doi.org/10.1186/1476-4598-12-126 (2013).
Efstathiou, J. A. et al. Mutated epithelial cadherin is associated with increased tumorigenicity and loss of adhesion and of responsiveness to the motogenic trefoil factor 2 in colon carcinoma cells. Proc Natl Acad Sci USA 96, 2316–2321 (1999).
Tym, J. E. et al. canSAR: an updated cancer research and drug discovery knowledgebase. Nucleic Acids Res 44, D938–943, https://doi.org/10.1093/nar/gkv1030 (2016).
Tay, P. N. et al. Palladin, an actin-associated protein, is required for adherens junction formation and intercellular adhesion in HCT116 colorectal cancer cells. Int J Oncol 37, 909–926 (2010).
Zhang, C. C. et al. PF-03732010: a fully human monoclonal antibody against P-cadherin with antitumor and antimetastatic activity. Clin Cancer Res 16, 5177–5188, https://doi.org/10.1158/1078-0432.CCR-10-1343 (2010).
Matsubara, S. & Ozawa, M. Expression of alpha-catenin in alpha-catenin-deficient cells results in a reduced proliferation in three-dimensional multicellular spheroids but not in two-dimensional monolayer cultures. Oncogene 23, 2694–2702, https://doi.org/10.1038/sj.onc.1207423 (2004).
Vermeulen, S. J. et al. Did the four human cancer cell lines DLD-1, HCT-15, HCT-8, and HRT-18 originate from one and the same patient? Cancer Genet Cytogenet 107, 76–79 (1998).
Medico, E. et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun 6, 7002, https://doi.org/10.1038/ncomms8002 (2015).
Korff, T. & Augustin, H. G. Integration of endothelial cells in multicellular spheroids prevents apoptosis and induces differentiation. J Cell Biol 143, 1341–1352 (1998).
Weiswald, L. B. et al. In situ protein expression in tumour spheres: development of an immunostaining protocol for confocal microscopy. BMC Cancer 10, 106, https://doi.org/10.1186/1471-2407-10-106 (2010).
Dolznig, H. et al. Modeling colon adenocarcinomas in vitro a 3D co-culture system induces cancer-relevant pathways upon tumor cell and stromal fibroblast interaction. Am J Pathol 179, 487–501, https://doi.org/10.1016/j.ajpath.2011.03.015 (2011).
Author information
Authors and Affiliations
Contributions
M.S., M.Sch., S.W. and H.D. designed the experiments and analyzed the data. M.S., M.Sch., S.W., S.H., K.P., A.R., N.K., C.U. performed the experiments. B.W. and J.N. provided the chromosome analysis. M.S., M.H. and H.D. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stadler, M., Scherzer, M., Walter, S. et al. Exclusion from spheroid formation identifies loss of essential cell-cell adhesion molecules in colon cancer cells. Sci Rep 8, 1151 (2018). https://doi.org/10.1038/s41598-018-19384-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-19384-0
This article is cited by
-
The ‘bIUreactor’: An Open-Source 3D Tissue Research Platform
Annals of Biomedical Engineering (2024)
-
Bioinformatic analysis of key pathways and genes shared between endometriosis and ovarian cancer
Archives of Gynecology and Obstetrics (2022)
-
Challenges of applying multicellular tumor spheroids in preclinical phase
Cancer Cell International (2021)
-
Rosuvastatin inhibit spheroid formation and epithelial–mesenchymal transition (EMT) in prostate cancer PC-3 cell line
Molecular Biology Reports (2020)
-
Proteome profiling of low grade serous ovarian cancer
Journal of Ovarian Research (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.