Abstract
Metallo-β-lactamases (MBLs) are a group of enzymes that can inactivate most commonly used β-lactam-based antibiotics. Among MBLs, New Delhi metallo-β-lactamase-1 (NDM-1) constitutes an urgent threat to public health as evidenced by its success in rapidly disseminating worldwide since its first discovery. Here we report the biochemical and genetic characteristics of a novel MBL, ElBla2, from the marine bacterium Erythrobacter litoralis HTCC 2594. This enzyme has a higher amino acid sequence similarity to NDM-1 (56%) than any previously reported MBL. Enzymatic assays and secondary structure alignment also confirmed the high similarity between these two enzymes. Whole genome comparison of four Erythrobacter species showed that genes located upstream and downstream of elbla2 were highly conserved, which may indicate that elbla2 was lost during evolution. Furthermore, we predicted two prophages, 13 genomic islands and 25 open reading frames related to insertion sequences in the genome of E. litoralis HTCC 2594. However, unlike NDM-1, the chromosome encoded ElBla2 did not locate in or near these mobile genetic elements, indicating that it cannot transfer between strains. Finally, following our phylogenetic analysis, we suggest a reclassification of E. litoralis HTCC 2594 as a novel species: Erythrobacter sp. HTCC 2594.
Similar content being viewed by others
Introduction
Ever since the first antibiotic, penicillin, was used in the fight against infections caused by bacteria, there have been rising concerns regarding bacterial resistance to antibiotics acquired via the exposure of bacteria to sub-lethal quantities of antibiotics. As the number of different antibiotics used in clinical practice has increased, antibiotic resistance has become a serious problem worldwide. The emergence of genes encoding antibiotic-hydrolyzing enzymes, in particular, pose a major threat to human health. For example, β-lactamases enzymatically hydrolyze the β-lactam ring, the core structure of β-lactam-based antibiotics such as cephalosporins, cephamycins and carbapenems1. β-lactamases are divided into four different classes based on their nucleotide and amino acid sequences2. Due to their prevalence among clinical isolates, researches have mainly been focused on Class B β-lactamases (metallo-β-lactamases, MBLs)3,4. In addition, several MBL encoding genes have been found to be associated with mobile genetic elements (MGEs), enabling these genes to spread easily between species5,6. Another feature of MBLs is their dependence on metal ions (usually Zn2+) for catalytic activity. The requirement of metal ions as cofactors is a very common feature among enzymes7. To date, four subgroups (B1, B2, B3 and B4) of MBLs have been identified, based on their sequence homology8. Although MBLs of all the four subgroups need metal ions for their catalytic activity, they differ in the number of metal ions required. Generally, subgroups B1, B3 and B4 MBLs bind two Zn2+ in their active sites9,10,11,12,13,14, while subgroup B2 MBLs require only one Zn2+, the binding of an additional metal ion leads to inhibition of their activity8,15. However, BcII in subgroup B1 shows catalytic activity in the presence of both one and two Zn2+ 16,17,18.
The most common types of MBLs observed in clinical isolates are Imipenemase (IMP), Verona imipenemase (VIM) and New Delhi metallo-β-lactamase (NDM)19. IMP was initially discovered in 1994, in a clinical isolate of Serratia marcescens in which the enzyme conferred resistance to imipenem20. VIM was first identified in a carbapenem resistant isolate of Pseudomonas aeruginosa from an Italian patient in 199921. NDM was first reported in 2009 in a carbapenem resistant Klebsiella pneumoniae strain isolated from a Swedish patient of Indian origin22. Since their first identification, all of the above mentioned enzymes have been found in various isolates worldwide. In addition, new types of MBLs are continuously emerging in both clinical isolates and environmental isolates4,6,23,24.
New Delhi metallo-β-lactamase-1 (NDM-1) shows very low similarities to other known β-lactamases, with the highest similarity (32.4%) being to VIM-1/VIM-222. However, in our previous study, we performed a complete genome sequence analysis of a marine bacterium Erythrobacter litoralis HTCC 2594 (GenBank accession no. NC_007722) and reported an unexpectedly high similarity (56%) between NDM-1 and a β-lactamase II from E. litoralis HTCC 2594 (ElBla2)25. This report prompted Girlich et al. to characterize chromosomally encoded MBLs from five other Erythrobacter species (E. citreus, E. flavus, E. longus, E. aquimaris and E. vulgaris) in the hope to determine potential reservoirs of acquired MBLs26. However, they were unable to draw a connection between these species and plasmid-mediated carbapenemases spreading worldwide26. Interestingly, although ElBla2 was identified from E. litoralis HTCC 2594, it differed significantly from MBLs of other Erythrobacter species, sharing only 15% amino acid identity26. In addition, phylogenetic tree analysis showed that ElBla2 clustered in the same branch as clinical MBLs (NDM-1, VIM-1 and IMP-1) instead of with MBLs from its own genus members25,26. All these findings supported continued efforts into a comprehensive study on ElBla2.
In the present study, ElBla2 was first biochemically characterized to verify its ability to engender antibiotic resistance. We then aligned the amino acid sequences of ElBla2 with amino acid sequences of MBLs from different subgroups to identify the sites related to catalytic activity and metal ion binding. Whole genome comparison of four Erythrobacter species was performed and the gene arrangements around elbla2 were investigated in detail. By analyzing MGEs, we tried to estimate the gene transfer potential of elbla2. Finally, supported by the phylogenetic analysis, we propose a reclassification of the producer of ElBla2 as a novel species of genus Erythrobacter.
Results
Cloning and heterologous expression of ElBla2
Expression of ElBla2 in E. coli BL21 (DE3) conferred increased minimum inhibitory concentrations (MICs) to various β-lactams compared to the control strain (Table 1). The purification of the recombinant ElBla2 was visualized by SDS-PAGE (see Supplementary Fig. S1). The molecular weight of ElBla2 without its signal peptide was estimated to be 24.86 kDa (Expasy27), which was in accordance with the SDS-PAGE result.
Biochemical characterization
The enzymatic activity of the purified ElBla2 towards various antibiotic agents including amoxicillin, ampicillin, cefepime, meropenem, nitrocefin, cefotaxime and ceftazidime was determined by monitoring the hydrolysis of the antibiotic agents. ElBla2 reacted with amoxicillin, ampicillin, meropenem, cefotaxime and ceftazidime. Cefepime and nitrocefin, however, were not hydrolyzed by ElBla2. The comparison of kinetic parameters of ElBla2 and other MBLs (NDM-122, CphA28 AIM-113 and SPR-112) for the hydrolysis of different types of β-lactams (amoxicillin, cefotaxime, meropenem and cefepime) are summarized in Table 2.
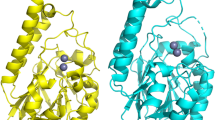
Sequence alignment and phylogenetic analysis
ElBla2 is encoded by a 786 bp ORF in the chromosome of E. litoralis HTCC 2594 and has a 56% amino acid sequence similarity to NDM-1. We used NDM-1 (Protein Data Bank code 3SPU) as a template to model the protein structure of ElBla2. The alignment and secondary structure of ElBla2 and NDM-1 are shown in Fig. 1. The alignment and comparison of ElBla2 and other MBLs are shown in Supplementary Table S1, Figs S2 and S3. In order to investigate the evolutionary relationship between ElBla2 and other MBLs, a phylogenetic tree was constructed using the amino acids sequences of various MBLs belonging to subgroups B1, B2 and B3 (Fig. 2).
Evolutionary relationships of ElBla2 and other MBLs. The evolutionary history was inferred using the Neighbor-Joining method54. The optimal tree with a total branch length of 11.09452999 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method66 and represent the number of amino acid substitutions per site. The analysis involved 25 amino acid sequences of MBLs belonging to subgroup B1 (red), B2 (green) and B3 (purple). All ambiguous positions were removed for each sequence pair. There were a total of 336 positions in the final dataset. Evolutionary analyses were conducted in MEGA755.
Comparative genomic study
Thus far, there are four complete genomes of Erythrobacter, E. atlanticus s21-N3 (CP011310 and CP015441), E. gangjinensis CGMCC1.15024 (CP018097 and CP018098), E. litoralis DSM 8509 (CP017057) and E. litoralis HTCC 2594 (NC_007722) deposited in GenBank. All the four genomes were compared using ProgressiveMauve and significant homology was observed for the region around elbla2 (Fig. 3). Interestingly, the genes located upstream and downstream of elbla2 were highly similar in the four species (Fig. 4 and Supplementary Table S2).
Progressive Mauve alignment of E. litoralis HTCC 2594, E. litoralis DSM 8509, E. atlanticus s21-N3 and E. gangjinensis CGMCC1.15024. Homologous alignments are represented as colored blocks. The gene encoding ElBla2 is located at position 1,569,105–1,569,800 bp in the genome of E. litoralis HTCC 2594.
Structure of elbla2 locus in E. litoralis HTCC 2594 compared to E. litoralis DSM 8509, E. atlanticus s21-N3 and E. gangjinensis CGMCC1.15024 as inferred from RAST. Genes annotated with the same function were labeled in the same color, elbla2 is labeled in red and genes annotated as a hypothetical protein are labeled in white.
Mobile genetic elements
In an attempt to estimate the transfer potential of elbla2, the whole genome of E. litoralis HTCC 2594 was analyzed for the presence of MGEs including prophages, genomic islands (GIs), insertion sequences (ISs) and integrons. Two prophages and 13 GIs were identified in the genome (see Supplementary Figs S4, S5, Tables S3 and S4). A total of 25 ORFs related to ISs of different families were also found (Supplementary Table S5), but no integrons were detected.
Discussion
NDM-1 has been the cause of great concern since it was first reported in 200922. Given that the enzyme only shares a few similarities with other MBLs, researchers have endeavored to find its close relatives. In our previous study, we reported an unexpectedly high similarity in amino acid sequences between NDM-1 and ElBla225. In the present work, a comprehensive analysis of both biochemical and genetic aspects of ElBla2 was performed. To our knowledge, no other MBL has been reported with such a high similarity to NDM-1.
ElBla2 was successfully expressed in E. coli BL21 (DE3) and antimicrobial susceptibility testing showed increased MICs to various β-lactams compared with the control strain (Table 1). High-level resistance to penicillins was detected in the ElBla2-producing strain, the highest MICs (≥128 mg L−1) were for piperacillin. The MICs of cephalosporins were also increased with the production of ElBla2, with the exception of cefepime. Furthermore, production of ElBla2 conferred resistance to carbapenems. MICs of ertapenem and imipemem were ≥8 mg L−1 and ≥16 mg L−1, respectively. However, the production of ElBla2 had no effect on the MIC of aztreonam. In addition, the purified ElBla2 showed enzymatic activity towards amoxicillin, ampicillin, meropenem, cefotaxime and ceftazidime, but not cefepime or nitrocefin. The inability of ElBla2 to hydrolyze nitrocefin is noteworthy since most of MBLs catalyze nitrocefin, as reported in other studies. However, there are exceptions. For example, Ferreira et al. revealed that two isolates of Staphylococcus aureus, one isolate of S. epidermidis and one isolate of S. lugdunensis which carried blaZ genes tested negative in nitrocefin tests29. Pitkala et al. found that 15 out of 19 Staphylococcus spp. isolates harboring blaZ genes showed a very weak reaction to nitrocefin30. Although the nitrocefin tests are commonly used tests for MBL detection, there are a large number of false-negative results according to different studies, indicating that not all MBLs can hydrolyze nitrocefin. ElBla2 showed lower K m values for cefotaxime than for the other two substrates, indicating that ElBla2 binds tightly to cefotaxime (Table 2). In comparison, NDM-1, closely related to ElBla2 in terms of amino acid sequence, also exhibits low K m values to most cephalosporins, especially to cefotaxime (K m = 10 μM)22. However, the turnover rate (kcat) of ElBla2 for the different substrates was lower compared to that of NDM-1. The highest turnover rate of ElBla2 was observed for meropenem (2.1 ± 0.2 s−1) whereas the turnover rate by NDM-1 for meropenem is 5.6-fold higher10. The highest catalytic efficiency (kcat/K m ) of ElBla2 (0.11 ± 0.03 s−1 μM−1) was also observed for meropenem (3.1-fold of amoxicillin and 1.6-fold of cefotaxime). This value was, however, also lower than the corresponding value of NDM-1.
The secondary protein structure of ElBla2 and NDM-1 are very similar (Fig. 1); both contain 13 β-sheets and six α-helices. Many active sites in NDM-1 are also found to be conserved in ElBla2, such as N220 which is involved in the interaction with the lactam carbonyl group and K211 which orients the negatively charged carboxylate common to β-lactam substrates31. Residues L65, M67 and W93 in NDM-1, which form a hydrophobic face that tightly interacts with the R1 phenyl group of the substrate, are not all conserved in ElBla2 since M67 is substituted by a leucine in ElBla231. The alignment of ElBla2 to MBLs representing four subgroups of Class B β-lactamases indicated that ElBla2 harbors all the six conserved residues which forms the amino acid ligands in the two metal ion (Zn1 and Zn2) binding sites (see Supplementary Fig. S2). Recently, a number of MBLs from non-pathogenic bacteria and environments that have not been impacted by human activities were reported, such as LAR-1-LAR13 from the metagenome of remote Alaskan soil32, MIM-1 from Novosphingobium pentaromativorans and MIM-2 from Simiduia agarivorans33. Characterizations of these enzymes showed that they were efficient MBLs. For example, LAR-8 was found to be active towards penicillins and carbapenems34, while LAR-12 displayed strong carbapenemase activity35. Some of these enzymes are also bifunctional. For example, the enzymes MIM-1 and MIM-2 exhibit lactonase activity besides lactamase activity36. Comparison of amino acid sequences between ElBla2 and these environmentally-derived MBLs showed that ElBla2 shares relatively lower identity with these MBLs than with clinically-derived NDM-1 (see Supplementary Table S1). Furthermore, in order to predict whether ElBla2 has lactonase activity like MIM-1 and MIM-2, amino acid sequences of these three proteins were aligned. The result showed that two of the three conserved residues were absent in ElBla2 (see Supplementary Fig. S3). These residues are known to be critical for lactonase activity and are conserved in MIM-1, MIM-2 and other N-acyl homoserine lactonases (AHLases)36. The absence of these residues indicates that ElBla2 likely has no lactonase activity.
It has previously been reported that NDM-1 only shares low identity with other MBLs and forms a unique branch within subgroup B122. However, with the discovery of ElBla2 in the marine bacterium E. litoralis HTCC 2594, the branch in subgroup B1 is populated by both NDM-1 and ElBla2 (Fig. 2). The similarity in amino acid sequences between ElBla2 and NDM-1 was first discovered by Zheng et al.25 and this finding instigated the study of chromosomally encoded MBLs from the genus Erythrobacter26. However, these endeavors failed to identify any MBLs from Erythrobacter spp. sharing high similarity with ElBla2, NDM-1 or any other MBL of clinical importance26; making ElBla2 unique. In recent years, the boom in whole genome sequencing projects has made the data pool of bacterial genomes more extensive. Even as an increasing number of species are being sequenced, no MBL with higher sequence similarity to NDM-1 than ElBla2 has yet been found. Following whole genome comparison analysis of four species of Erythrobacter, we observed that the gene arrangements around elbla2 were highly conserved in all of the Erythrobacter species included in the study (Fig. 4), suggesting that elbla2 may have been lost during evolution.
Many genes, especially those contributing to virulence and antibiotic resistance in bacteria, can be transmitted via various types of MGEs37. As a result, bacteria are able to acquire virulence and antibiotic resistance via horizontal gene transfer (HGT). Although numerous MGEs were identified in the genome of E. litoralis HTCC 2594 (see Supplementary Figs S4, S5 and Tables S3–S5), elbla2 was not located in or near any of these MGEs, indicating that it may unable to transfer among strains. Contrastingly, blaNDM-1 has always been reported to be located on a plasmid or embedded in the chromosome along with MGEs and can thus spread widely among different strains, making it an important public health issue22,38.
Generally, bacteria of the same species share high similarity in genome sequences. However, elbla2 was only located in the chromosome of E. litoralis HTCC 2594. In fact, another strain of E. litoralis, DSM 8509, does not encode any MBL that shows sequence similarity to elbla2. Therefore, a blast-analysis was performed between these two strains and surprisingly, many differences between their genomes were observed (see Supplementary Fig. S6). For this reason, the 16S rRNA gene sequences of HTCC 2594 and DSM 8509 were subsequently compared using BLASTN and the result showed a similarity of 98%. Though the original standard for 16 S rRNA gene sequence at which prokaryotic species delineate is a similarity of 97%39, a more relaxed cut-off value of 98.7–99% has recently been proposed40. The low similarity (98%) of 16 S rRNA gene sequence between HTCC 2594 and DSM 8509 may suggest that they belong to different species. To further test this assumption, we calculated the average nucleotide identity (ANI) values between them. ANI has been widely used to compare two prokaryotic genome sequences when classifying and identifying bacteria and ANI values of about 95–96% are considered to be the species boundary41,42,43. In accordance with the 16 S rRNA gene sequence similarity, the ANI value between HTCC 2594 and DSM 8509 (74.23%) is much lower than the species boundary. Supported by these results, we can conclude that HTCC 2594 and DSM 8509 are not of the same species. DSM 8509 is the type strain of E. litoralis species and the original publication reporting its isolation and identification emphasized its distinct phylogenetic position and phenotypic characteristics44. The publications related to HTCC 2594 on the other hand, only report the sequence of its whole genome and a characterization of its functional genes45,46. For these reasons, we propose to reclassify E. litoralis HTCC 2594 as a novel species: Erythrobacter sp. HTCC 2594.
In conclusion, we have successfully characterized a novel MBL, named ElBla2, from a marine bacterium. This novel enzyme is the MBL with the highest degree of amino acid sequence similarity to NDM-1 discovered thus far. Subsequent to the biochemical characterization of ElBla2, its secondary structure was aligned with that of NDM-1 to compare the active sites. The alignment of ElBla2 to MBLs representing the four subgroups of the Class B family indicated that ElBla2 harbors all the conserved residues related to the binding of two metal ions. Furthermore, gene arrangements of four species of Erythrobacter were compared and the results showed that the genes located upstream and downstream of elbla2 were highly conserved, indicating that elbla2 may have been lost during evolution. Also, elbla2 did not locate in or near MGEs, suggesting that it is unable to be transferred among strains. Finally, supported by the phylogenetic analysis we propose a reclassification of the producer of ElBla2, E. litoralis HTCC 2594, as a novel species: Erythrobacter sp. HTCC 2594.
Methods
Cloning of elbla2
DNA was synthesized according to the nucleic acid sequence of the gene encoding β-lactamase II in E. litoralis HTCC 2594 (elbla2) (GenBank accession NC_007722). The full-length of elbla2 was amplified from the synthesized DNA by PCR using the primers EL-F1 and EL-R (Table 3). However, expression of the full-length ElBla2 did not yield any detectable protein. Therefore, we predicted the presence of signal peptide cleavage sites in ElBla2 using SignalP 4.1 Server47 and found a potential cleavage site between position 28 and 29. According to the position of the cleavage site in ElBla2, the mature-length elbla2 was amplified using primers EL-F2 and EL-R (Table 3). Cloning of the mature-length elbla2 into a pET28a vector (Invitrogen) was performed as previously described48. The recombinant plasmid pET28a-elbla2 was verified by DNA sequencing for the presence of the elbla2 gene. Recombinant plasmid pET28a-elbla2 was transformed into E. coli BL21 (DE3) cells for production of ElBla2.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing of E. coli BL21 harboring pET28a-elbla2 was determined using the VITEK 2 system employing panel AST-GN-13 (bioMerieux, France). The results were interpreted using the standards of the Clinical and Laboratory Standards Institute. E. coli BL21 harboring pET28a were used as controls.
Enzymatic assay
ElBla2 was purified using the Ni-NTA affinity chromatography column (Qiagen, Germany) according to the manufacturer’s protocol. The purified protein was verified by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and the concentration of the protein was estimated using Bradford reagent49. The enzymatic activity of ElBla2 towards various β-lactam antibiotic agents was determined at 35 °C in Tris-HCl buffer (pH 7.4) using a Coulter DU 800 spectrophotometer (BecKman) as described elsewhere48. The details of reaction conditions are shown in the Supplementary Methods. The enzyme kinetics analysis for ElBla2 was performed by measuring enzyme velocity towards different concentrations of the substrates. The substrates and the concentrations used were amoxicillin (ranging from 20 μM to 500 μM), cefotaxime (ranging from 10 μM to 100 μM) and meropenem (ranging from 10 μM to 200 μM). Nonlinear regression curves for each substrate were drawn using the substrate concentrations and the enzyme velocity. Then the Lineweaver-Burk plot, which plots the reciprocal of substrate concentration vs. the reciprocal of enzyme velocity, was created for each substrate. The concentration of ElBla2 in the reaction was measured by the method of Bradford49 using bovine serum albumin as standard. The K m and k cat values were calculated according the Michaelis–Menten equation using GraphPad Software (GraphPad Inc., USA)50. All values were recorded in triplicate and the blank control was performed using the deactivated enzyme.
Bioinformatics analysis
Protein structure modelling was conducted using the SWISS-MODEL Workspace51. Multiple sequence alignment was performed using Clustal Omega52 and the secondary structure information from the aligned sequence was rendered using ESPript 3.053. The phylogenetic tree was constructed according to the Neighbor-Joining Method54 using Molecular Evolutionary Genetics Analysis (MEGA) 7.0 software55. BLASTN was used for the alignment of the whole genomes and the genome alignment was visualized by BLAST Ring Image Generator56. The genomic ANI was calculated using EzBioCloud57 with OrthoANIu algorithm58. Genomic comparison was conducted using ProgressiveMauve59. Genomes were annotated by Rapid Annotations using Subsystems Technology (RAST) server60. Prophage regions were predicted with the PHAge Search Tool Enhanced Release server61,62 and GIs were predicted by IslandViewer 4 using four different methods (IslandPick, IslandPath-DIMOB, SIGI-HMM, and Islander)63. Insertion sequences and integrons were predicted using ISsaga64 and IntegronFinder65, respectively.
References
Miraula, M., Whitaker, J. J., Schenk, G. & Mitic, N. Beta-Lactam antibiotic-degrading enzymes from non-pathogenic marine organisms: a potential threat to human health. J. Biol. Inorg. Chem. 20, 639–651 (2015).
Walsh, T. R., Toleman, M. A., Poirel, L. & Nordmann, P. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18, 306–325 (2005).
Maltezou, H. C. Metallo-beta-lactamases in Gram-negative bacteria: introducing the era of pan-resistance? Int. J. Antimicrob. Agents. 33(405), e401–407 (2009).
Pfennigwerth, N. et al. Genetic and biochemical characterization of HMB-1, a novel subclass B1 metallo-beta-lactamase found in a Pseudomonas aeruginosa clinical isolate. J. Antimicrob. Chemother. 72, 1068–1073 (2017).
Zheng, F., Sun, J., Cheng, C. & Rui, Y. Molecular characteristics of carbapenem-resistant gram-negative bacteria in southern China. Microb. Drug. Resist. 21, 178–185 (2015).
Gudeta, D. D. et al. The soil microbiota harbors a diversity of carbapenem-hydrolyzing beta-lactamases of potential clinical relevance. Antimicrob. Agents. Chemother. 60, 151–160 (2015).
Mitić, N. et al. Chapter Three-Catalytic mechanisms of metallohydrolases containing two metal ions. Adv. Protein Chem. Struct. Biol. 97, 49–81 (2014).
Phelan, E. K. et al. Metallo-β-lactamases: a major threat to human health. Am. J. Mol. Biol. 4, 89 (2014).
Crowder, M. W., Spencer, J. & Vila, A. J. Metallo-β-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc. Chem. Res. 39, 721–728 (2006).
Bebrone, C. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 74, 1686–1701 (2007).
Heinz, U. & Adolph, H. W. Metallo-ß-lactamases: two binding sites for one catalytic metal ion? Cell. Mol. Life Sci. 61, 2827–2839 (2004).
Vella, P. et al. Identification and characterization of an unusual metallo-beta-lactamase from Serratia proteamaculans. J. Biol. Inorg. Chem. 18, 855–863 (2013).
Leiros, H. K. et al. Crystal structure of the mobile metallo-beta-lactamase AIM-1 from Pseudomonas aeruginosa: insights into antibiotic binding and the role of Gln157. Antimicrob. Agents Chemother. 56, 4341–4353 (2012).
Costello, A., Periyannan, G., Yang, K.-W., Crowder, M. W. & Tierney, D. L. Site-selective binding of Zn (II) to metallo-β-lactamase L1 from Stenotrophomonas maltophilia. J. Biol. Inorg. Chem. 11, 351–358 (2006).
Hernandez Valladares, M. et al. Kinetic and spectroscopic characterization of native and metal-substituted β-lactamase from Aeromonas hydrophila AE036. FEBS Lett. 467, 221–225 (2000).
Badarau, A. & Page, M. I. Loss of enzyme activity during turnover of the Bacillus cereus β-lactamase catalysed hydrolysis of β-lactams due to loss of zinc ion. J. Biol. Inorg. Chem. 13, 919 (2008).
Llarrull, L. I., Tioni, M. F. & Vila, A. J. Metal content and localization during turnover in B. cereus metallo-β-lactamase. J. Am. Chem. Soc. 130, 15842–15851 (2008).
Rasia, R. M. & Vila, A. J. Exploring the role and the binding affinity of a second zinc equivalent in B. cereus metallo-β-lactamase. Biochemistry 41, 1853–1860 (2002).
Nordmann, P., Naas, T. & Poirel, L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17, 1791–1798 (2011).
Osano, E. et al. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38, 71–78 (1994).
Lauretti, L. et al. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43, 1584–1590 (1999).
Yong, D. et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054 (2009).
Wachino, J. et al. SMB-1, a novel subclass B3 metallo-beta-lactamase, associated with ISCR1 and a class 1 integron, from a carbapenem-resistant Serratia marcescens clinical isolate. Antimicrob. Agents Chemother. 55, 5143–5149 (2011).
Pollini, S. et al. FIM-1, a new acquired metallo-beta-lactamase from a Pseudomonas aeruginosa clinical isolate from Italy. Antimicrob. Agents Chemother. 57, 410–416 (2013).
Zheng, B. et al. An unexpected similarity between antibiotic-resistant NDM-1 and beta-lactamase II from Erythrobacter litoralis. Protein Cell 2, 250–258 (2011).
Girlich, D., Poirel, L. & Nordmann, P. Diversity of naturally occurring Ambler class B metallo-beta-lactamases in Erythrobacter spp. J. Antimicrob. Chemother. 67, 2661–2664 (2012).
Gasteiger, E. et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic. Acids Res. 31, 3784–3788 (2003).
Segatore, B., Massidda, O., Satta, G., Setacci, D. & Amicosante, G. High specificity of cphA-encoded metallo-beta-lactamase from Aeromonas hydrophila AE036 for carbapenems and its contribution to beta-lactam resistance. Antimicrob. Agents Chemother. 37, 1324–1328 (1993).
Ferreira, A. M., Martins, K. B., Silva, V. R., Mondelli, A. L. & Cunha, M. L. Correlation of phenotypic tests with the presence of the blaZ gene for detection of beta-lactamase. Braz. J. Microbiol. 48, 159–166 (2017).
Pitkala, A., Salmikivi, L., Bredbacka, P., Myllyniemi, A. L. & Koskinen, M. T. Comparison of tests for detection of beta-lactamase-producing staphylococci. J. Clin. Microbiol. 45, 2031–2033 (2007).
King, D. & Strynadka, N. Crystal structure of New Delhi metallo-β-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 20, 1484–1491 (2011).
Allen, H. K., Moe, L. A., Rodbumrer, J., Gaarder, A. & Handelsman, J. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J. 3, 243–251 (2009).
Miraula, M., Brunton, C. S., Schenk, G. & Mitić, N. Identification and preliminary characterization of novel B3-type metallo-β-lactamases. Am. J. Mol. Biol. 3, 198 (2013).
Pedroso, M. M. et al. Characterization of a highly efficient antibiotic-degrading metallo-β-lactamase obtained from an uncultured member of a permafrost community. Metallomics 9, 1157–1168 (2017).
Rodriguez, M. M. et al. Crystal structure and kinetic analysis of the class B3 di-zinc metallo-beta-lactamase LRA-12 from an Alaskan soil metagenome. PLoS One 12, e0182043 (2017).
Miraula, M., Schenk, G. & Mitić, N. Promiscuous metallo-β-lactamases: MIM-1 and MIM-2 may play an essential role in quorum sensing networks. J. Inorg. Biochem. 162, 366–375 (2016).
Olaitan, A. O., Diene, S. M., Assous, M. V. & Rolain, J.-M. Genomic plasticity of multidrug-resistant NDM-1 positive clinical isolate of Providencia rettgeri. Genome Biol. Evol. 8, 723–728 (2015).
Shen, P. et al. Detection of an Escherichia coli sequence type 167 strain with two tandem copies of blaNDM-1 in the chromosome. J. Clin. Microbio. 55, 199–205 (2017).
Stackebrandt, E. & Goebel, B. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 44, 846–849 (1994).
Stackebrandt, E. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 6, 152–155 (2006).
Goris, J. et al. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91 (2007).
Richter, M. & Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. 106, 19126–19131 (2009).
Chun, J. & Rainey, F. A. Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 64, 316–324 (2014).
Yurkov, V. et al. Phylogenetic positions of novel aerobic, bacteriochlorophyll a-containing bacteria and description of Roseococcus thiosulfatophilus gen. nov., sp. nov., Erythromicrobium ramosum gen. nov., sp. nov., and Erythrobacter litoralis sp. nov. Int. J. Syst. Evol. Microbiol. 44, 427–434 (1994).
Oh, H. M., Giovannoni, S. J., Ferriera, S., Johnson, J. & Cho, J. C. Complete genome sequence of Erythrobacter litoralis HTCC2594. J. Bacteriol. 191, 2419–2420 (2009).
Woo, J. H. et al. Cloning and characterization of three epoxide hydrolases from a marine bacterium, Erythrobacter litoralis HTCC2594. Appl. Microbiol. Biot. 76, 365–375 (2007).
Nielsen, H. Predicting secretory proteins with SignalP. Methods Mol. Biol. 1611, 59–73 (2017).
Zheng, B., Jiang, X., Xu, Z., Fang, Y. & Li, L. Characterization of a novel metallo-beta-lactamases fold hydrolase from Pelagibacterium halotolerans, a marine halotolerant bacterium isolated from East China Sea. Extremophiles 20, 37–44 (2016).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Lamoureaux, T. L., Vakulenko, V., Toth, M., Frase, H. & Vakulenko, S. B. A novel extended-spectrum beta-lactamase, SGM-1, from an environmental isolate of Sphingobium sp. Antimicrob. Agents Chemother. 57, 3783–3788 (2013).
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Alikhan, N.-F., Petty, N. K., Zakour, N. L. B. & Beatson, S. A. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402 (2011).
Yoon, S. H. et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617 (2017).
Lee, I., Kim, Y. O., Park, S. C. & Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103 (2015).
Darling, A. E., Mau, B. & Perna, N. T. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS one 5, e11147 (2010).
Brettin, T. et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365 (2015).
Arndt, D. et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21 (2016).
Zhou, Y., Liang, Y., Lynch, K. H., Dennis, J. J. & Wishart, D. S. PHAST: a fast phage search tool. Nucleic Acids Res. 39, W347–W352 (2011).
Bertelli, C. et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35 (2017).
Varani, A. M., Siguier, P., Gourbeyre, E., Charneau, V. & Chandler, M. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 12, R30 (2011).
Cury, J., Jové, T., Touchon, M., Néron, B. & Rocha, E. P. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 44, 4539–4550 (2016).
Zuckerkandl, E. & Pauling, L. Evolutionary divergence and convergence in proteins. Evol. Genes Prot. 97, 97–166 (1965).
Acknowledgements
We thank Dr. Björn Berglund for his linguistic assistance during the preparation of this manuscript. This study was funded by grants from the National Key Research and Development Program of China (Nos 2016YFD0501105); the National Natural Science Foundation of China (Nos 41406140 and 81301461); the Zhejiang Provincial Natural Science Foundation of China (No. LY17H190003); and the opening foundation of the Key Laboratory of Marine Ecosystem and Biogeochemistry, Second Institute of Oceanography, State Oceanic Administration (LMEB201504).
Author information
Authors and Affiliations
Contributions
X.J. and B.Z. designed the study. X.J. and X.C. performed experiments. H.C., Y.H. and L.X. analysed data. Y.W., W.L. and F.T. contributed reagents, materials and analysis tools. X.J., H.C. and B.Z. wrote the manuscript that was revised by all co-authors.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jiang, XW., Cheng, H., Huo, YY. et al. Biochemical and genetic characterization of a novel metallo-β-lactamase from marine bacterium Erythrobacter litoralis HTCC 2594. Sci Rep 8, 803 (2018). https://doi.org/10.1038/s41598-018-19279-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-19279-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.