Abstract
Hevea brasiliensis Müll. Arg. is one of the most frequently wounded plants worldwide. Expelling latex upon mechanical injury is a wound response of rubber trees. However, JA-mediated wound responses in rubber trees are not well documented. In this work, three JAZ-interacting MYC transcription factors of H. brasiliensis (termed HbMYC2/3/4) were identified by yeast two-hybrid screening. HbMYC2/3/4 each showed specific interaction profiles with HbJAZs. HbMYC2/3/4 each localized in the nucleus and exhibited strong transcriptional activity. To identify the target genes potentially regulated by HbMYC2/3/4, cis-elements interacting with HbMYC2/3/4 were first screened by yeast one-hybrid assays; the results indicated that HbMYC2/3/4 each could bind G-box elements. Additional analysis confirmed that HbMYC2/3/4 bound the HbPIP2;1 promoter, which contains five G-box cis-elements, and regulated the expression of reporter genes in yeast cells and in planta. HbMYC2/3/4 were induced by exogenous JA treatment but suppressed by ethylene (ET) treatment; in contrast, HbPIP2;1 was positively regulated by ET but negatively regulated by JA treatment. Given that HbPIP2;1 is involved in latex drainage, it could be proposed that HbMYC2/3/4 are involved in the regulation of HbPIP2;1 expression as well as latex drainage, both of which are coordinated by the JA and ET signalling pathways.
Similar content being viewed by others
Introduction
Over 2,000 plant species produce rubber (cis-1–4-polyisoprene). Because of its high rubber productivity and rubber quality, Hevea brasiliensis Müll. Arg. is the sole commercial source of natural rubber. Rubber is produced and accumulates in latex in the laticifer network of H. brasiliensis. The laticifers consist of contiguous anastomosis cells forming a network structure arranged in rings parallel to the vascular cambium, which allow the drainage of latex from a large area of bark by a single tapping1. As the cytoplasm of laticifers, latex is harvested by farmers by regularly cutting bark at intervals of 2–3 days. As such, rubber trees become one of the most frequently wounded plants worldwide. Previous reports have shown that mechanical wounding induces laticifer differentiation and latex production. Three decades ago, 2–3-fold more laticifer rings were observed by light microscopy in the exploited trees than in the unexploited trees2,3. Additionally, secondary laticifer differentiation can be induced in the stem of epicormic shoots by treatment with exogenous jasmonic acid (JA) or its derivatives1. The induction of laticifers could serve as an excellent indicator of the wounding response and latex biosynthesis regulated by JA in rubber trees. Recent reports have shown that the differentiation of secondary laticifers was prevented when the wounding site of epicormic shoots was wrapped immediately after wounding. Wounding-induced laticifer differentiation has been proposed to be correlated with JA accumulation, reactive oxygen species, as well as dehydration at the wounding site4. Our recent report also confirmed that local tissue dehydration was a key signal for laticifer differentiation. Dehydration-related genes, such as HbDHNs and HbNAC1, are differentially expressed on wrapped and exposed wounding sites. Furthermore, HbNAC1 was shown to bind to the cis-element CACG in the promoter region of the gene encoding the small rubber particle protein (SRPP)5. Arabidopsis overexpressing HbDHNs show higher activity of antioxidant enzymes and accumulate fewer reactive oxygen species (ROS)6. Given that ROS have been proposed to represent a key signal for laticifer differentiation4, HbDHNs might act as ROS scavengers, directly or indirectly affecting laticifer differentiation. However, how JA signalling is involved in laticifer differentiation and latex biosynthesis is less known.
In Arabidopsis, JA regulates many developmental and metabolic processes, such as vegetable growth, stamen development, senescence, trichome patterning, and anthocyanin biosynthesis7,8,9. JA is also involved in the response to a number of biotic/abiotic stresses, such as necrotrophic pathogens, herbivores, mechanical wounding, UV radiation, ozone, and salinity10,11. The JA signalling pathway has been well elucidated in Arabidopsis. Dissection of JA signalling was predominantly dependent on the identification of mutants that are deficient in JA synthesis or perception via a forward genetics approach, among which coi1 (coronatine insensitive 1) is the most important gene. COI1 encodes an F-box protein that associated with other proteins, including SKP1 and CULLIN, to form the SCFCOI1 ubiquitin–ligase complex12,13. The SCFCOI1 complex binds to target proteins, which are then polyubiquitinated and subsequently degraded by the 26 S proteasome. Another major advance in study of the molecular mechanism of JA signalling was made possible by the identification of the first SCFCOI1 targets, which compose the jasmonate ZIM-domain (JAZ) protein family14,15. JAZ proteins function as repressors of JA signalling to interact with JA-responsive transcription factors (e.g., MYC2) and inhibit their transcription14,16. After the perception of wounding signals or developmental cues, JA accumulates and is conjugated with isoleucine, which serves as active form to mediate the interaction between COI1 and JAZ repressors, leading to the ubiquitination of JAZ proteins. The degradation of JAZs results in the release of downstream transcription factors, activating the JA response. The COI1-JAZ-MYC2 complex was proposed to represent the core signalling module in the JA pathway17. As the SCFCOI1 complex is highly conserved in plants13,17,18, the spatially and temporally specific expression and alternative splicing as well as the differing repression of target transcription factors of individual JAZ gene members may account for the specific responses of plants to JA signals17,19.
MYC2 was the first reported TF regulated by JAZ proteins14,20. MYC2 and its closest homologues (MYC3 and MYC4) interact the most with JAZ proteins21,22,23. All three MYC proteins belong to group IIIe of the bHLH family; in the members of this family, five different domains have been identified, including a JAZ-interacting domain (JID) at the N-terminus and a conserved ACT-like domain at the C-terminus, in addition to a DNA-binding bHLH domain24. Although all three bHLH proteins have similar protein structures, they seem to regulate specific subsets of JA responses. For example, MYC2 is a positive regulator of the JA-mediated inhibition of primary root growth, anthocyanin biosynthesis, and oxidative stress tolerance but is a negative regulator of JA-mediated resistance to necrotrophic fungi20,25; on the other hand, MYC3 and MYC4 are important for JA-mediated resistance to the herbivore Spodoptera littoralis24. The JID domain is also found in several other bHLH proteins, such as GL3, EGL3, and TRANSPARENT TESTA8 (TT8; At4g09820), all of which belong to group III of the bHLH family26. GL3, EGL3, and TT8 interconnect with both WD40 proteins and R2R3 MYB proteins to form protein complexes; these complexes then regulate multiple processes, such as the biosynthesis of anthocyanins and proanthocyanidins, the development of trichomes and root hairs, and so on8,27,28,29. Interestingly, the MYC2/3/4 complex is also involved in the JA-mediated induction of anthocyanin biosynthesis20,23,25. How and whether MYC2/3/4 interact with GL3/EGL3/TT8 to regulate anthocyanin biosynthesis remains to be resolved.
In Hevea brasiliensis, members of the JAZ gene family have been globally cloned and preliminarily characterized30,31. The next key step to reveal the JA signalling pathway in H. brasiliensis is the identification of transcription factors regulated by JAZ proteins. In this work, a bait vector of HbJAZ1 was constructed, and proteins that interact with HbJAZ1 were screened using yeast two-hybrid assays. Three bHLH proteins (termed HbMYC2, HbMYC3 and HbMYC4) were identified. Further investigation revealed that these HbMYCs interact with promoters containing G-box cis-elements, e.g., HbPIP2;1, which codes for an aquaporin involved in latex drainage32. The data provided in this study might fill the gaps of the JA-mediated mechanism of latex biosynthesis and drainage in rubber trees.
Results
Identification of the HbJAZ1-interacting transcription factors of H. brasiliensis
To screen the JAZ-interacting transcription factors in H. brasiliensis, the full-length ORFs of HbJAZ1 were inserted into pGBKT7 and transformed into a Y2HGold strain to generate a bait reporter strain. After co-cultivation of the Y2HGold bait reporter strain and the Y187 strain of the Mate & Plate library of H. brasiliensis, the cell suspension was plated on QDO/A/X media (quadruple drop-out media: SD/–Ade/–His/–Leu/–Trp + AbA + X-alpha-Gal) to screen for positive colonies. From those colonies, four prey proteins were identified as putative transcription factors, including three basic helix-loop-helix (bHLH) transcription factors and one zinc-finger protein. Two other proteins were annotated as protease subunits and WD40 proteins, which seem to be components of the 26 S proteasome complex and GL3, EGL3, and TT8 protein complexes, respectively. Using in silico cloning procedures31, the full-length cDNA sequences of these genes were identified, and four TF genes were termed HbMYC2, HbMYC3, HbMYC4, and HbZF1. The full-length CDSs of the genes were fused into pGBKT7 vectors, which were then transformed into Y2HGold yeast strains. The assays of transcriptional activity showed that the protease subunit and WD40 proteins did not exhibit transcriptional activity but that HbMYC2, HbMYC3, HbMYC4, and HbZF1 exhibited strong transcriptional activity, suggesting that they are putative transcription factors (Fig. 1).
Autoactivation and toxicity test of HbJAZ-interacting proteins. The full-length Hevea HbMYC2, HbMYC3, zinc-finger, protease subunit, and WD40 genes were cloned into a pGBKT7 vector to generate bait plasmids, which were subsequently transformed into Y2HGold strains and plated onto plates containing the following media: SD/-Trp, SD/-Trp/-His/, SD/-Trp/-His/-Ade, or SD/-Trp/-His/-Ade/X-α-gal/AbA (125 ng/ml).
Phylogenetic analysis of HbMYC2/3/4
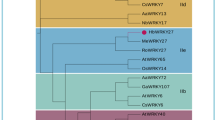
To classify the HbJAZ proteins that interact with HbJAZ1 in the yeast two-hybrid system, 23 different bHLH proteins were collected for phylogenetic analysis using Clustal Omega online software (http://www.ebi.ac.uk/Tools/msa/clustalo/). These bHLH proteins include JA-responsive MYC2/3/4, TT8, GL3 (GLABRA3) and EGL3 (ENHANCER OF GLABRA3) from Arabidopsis8,33; JAMYC2/10 from Solanum lycopersicum34; NtMYCa from tobacco35; ALC (Alcatraz, AtbHLH73), which is required for gynoecium and fruit development36; AMS (Aborted Microspores, AtbHLH21), which regulates pollen wall formation37,38,39; ABA-Inducible bHLH (AIB)40,41; SPT (Spatula, AtbHLH24), which controls the development of carpel margin tissues42,43; ILR3 (IAA-Leucine Resistant 3, AtbHLH105), which modulates iron homeostasis44; ICE1 (Inducer of CBF Expression 1, AtbHLH116), which is induced by cold45,46; ORG2 (OBP3-Responsive Gene 2, AtbHLH38), which is inducible by salicylic acid47; Phytochrome Interacting Factor 3 (PIF3, AtbHLH08) and Phytochrome Interacting Factor 3-Like 1 (PIL1, AtbHLH124)48,49,50; BEE1 (Brassinosteroid Enhanced Expression 1, AtbHLH44)51,52; RSL4 (Root Hair Defective 6-Like 4, AtbHLH54)53; RGE1 (Retarded Growth of Embryo 1, AtbHLH95); and FIT (Fe-Deficiency Induced Factor 1, AtbHLH29)54. Phylogenetic analysis showed that all JA-responsive bHLH proteins, e.g., MYC2, MYC3, MYC4, TT8, GL3, EGL3, JAMYC2, JAMYC10, and NtMYCa, clustered with HbMYC2/3/4, suggesting that these three HbMYCs might also be involved in the JA response. Other bHLH members such as AMS, SPT, ICE1, PIF3, PIL1, BEE1, RSL4, and FIT were classified together, whereas RGE1, ILR3 and ORG2 showed significantly wider genetic distance (Fig. 2a). When further comparing the protein structures, a similar distribution of the domains among MYC2/3/4 and HbMYC2/3/4 was observed. All these proteins contain a JAZ-interacting domain (JID) and an acidic domain (AD) at their N-terminus as well as a bHLH-zip domain and an ACT-like domain at their C-terminus (Fig. 2b). To identify which domain contributed to transcriptional activity, full-length and attenuated fragments of HbMYC3 were inserted into pGBKT7 vectors. These vectors were then transferred into Y2HGold yeast strains, which were subsequently plated onto SD/-Trp/-Ade/-His/X/A media. The full-length and N-terminal fragments containing the JID and AD domains exhibited strong transcriptional activity, which was indicated by strong growth in the SD/-Trp/-Ade/-His/X/A media. On the other hand, the C-terminal fragments containing the bHLH-zip domain and ACT-like domain and those containing only the JID or AD domain did not exhibit transcriptional activity, suggesting that both the JID and AD domains are essential for transcriptional activity (Fig. 2c).
Comparison of the MYC genes of Hevea brasiliensis and other bHLH-type transcription factors. (a) Phylogenetic tree of the deduced amino acid sequences of HbMYCs and other plant bHLH proteins. The phylogenetic tree was generated based on the alignment of the full-length deduced amino acid sequences of 23 bHLH proteins. Alignment was performed and the phylogenetic tree was constructed by Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/), with the default settings. (b) Domain comparisons between HbMYCs and AtMYCs, JAZ-interacting domains (JIDs), acidic domains (ADs), basic helix-loop-helix (bHLH)-zip domains and ACT-like domains were included. C. Transcriptional activity analysis of the different domains of HbMYC3 proteins. Full-length or attenuated fragments of HbMYC3 were inserted into a pGBKT7 vector, after which the vectors were transferred into Y2HGold yeast strains and then plated on SD/-Trp/-Ade/-His/X-α-gal/AbA media.
Interaction between members of the HbMYCs and HbJAZs gene families
Given that Arabidopsis MYC2/3/4 interact with different JAZ proteins, the interaction between HbMYCs and HbJAZs was investigated. Full-length HbMYCs and HbJAZs were fused to pGADT7 and pGBKT7 vectors to generate prey and bait vectors, which were further transformed into Y187 and Y2Hgold yeast strains, respectively. After co-cultivation of the Y187 and Y2Hgold strains, the mated cells were plated onto synthetic drop-out (DO) plates (SD-Trp/-Leu) and QDO/X/A plates (SD-Trp/-Leu/-Ade/-His/X/A). The yeast two-hybrid results showed that HbMYC2 interacted with HbJAZ3/6/7/8/10/11/12 and that HbMYC3 interacted with HbJAZ1/3/6/7/8/9/10/11, whereas HbMYC4 interacted with HbJAZ1/7/9/11/12. The HbMYCs each exhibited individual specific profiles of interaction with HbJAZs, although no correlation of sequence properties was observed. Interestingly, HbMYC2 was screened by the bait protein of HbJAZ1, but the full-length HbMYC2 did not interact with HbJAZ1, suggesting that some sequences of full-length HbMYC2 might inhibit the interaction between HbJAZ1 and the JID domain of HbMYC2, as HbJAZ1 interacted with the attenuated fragment of HbMYC2 in the Mate & Plate library screening (Fig. 3a).
Interaction of HbJAZs with HbMYC proteins. (a) Full-length HbMYCs and HbJAZs were fused into pGADT7 and pGBKT7 vectors to generate prey and bait vectors, respectively, which were further transformed into Y187 and Y2Hgold yeast strains, respectively. The Y187 and Y2Hgold yeast strains were then combined and cultivated. The mating cells were screened on synthetic drop-out (DO) plates (SD-Trp/-Leu), and the Y2H interactions were assessed on QDO/X/A plates (SD-Trp/-Leu/-Ade/-His/X/A). Positive control mating (+): Y2HGold [pGBKT7-53] and Y187 [pGADT7-T]; negative control mating (−): Y2HGold [pGBKT7-Lam] and Y187 [pGADT7-T]. The experiments were performed in triplicate. (b) Detection of the interaction between HbJAZ1 and HbMYCs by bimolecular fluorescence complementation. The plasmid combinations of pSPYCE-HbJAZ1 and pSPYNE-HbMYC3 or pSPYCE-HbJAZ1 and pSPYNE-HbMYC4 were co-transferred to Arabidopsis thaliana protoplasts, and the yellow fluorescence was observed by laser confocal microscopy.
To verify the confidence of the interaction between HbJAZs and HbMYCs, bimolecular fluorescence complementation (BiFC) assays were performed in Arabidopsis mesophyll cells. The combinations of pSPYCE-HbJAZ1 and pSPYNE-HbMYC3 or the combinations of pSPYCE-HbJAZ1 and pSPYNE-HbMYC4 were co-transferred to protoplasts of Arabidopsis thaliana and subsequently observed by laser confocal microscopy. No YFP fluorescence was observed when only one of the two proteins was fused to an unfolded YFP fragment; however, when pSPYNE-HbMYC3 or pSPYCE-HbJAZ1 was co-transformed with pSPYCE-HbJAZ1 into Arabidopsis protoplasts, a strong YFP signal was detected (Fig. 3b). Additionally, a YFP signal was significantly localized in the nucleus, suggesting that HbJAZ1 interacts with HbMYCs in the nucleus. The subcellular localization results also confirmed that all three HbMYCs localized in the nucleus (Fig. 4). Considering that HbMYC proteins exhibited strong transcriptional activity (Figs 1 and 2) and localized in the nucleus (Figs 3 and 4), these proteins could be regarded as transcription factors.
HbMYC2/3/4 proteins specifically bind to the G-box
To identify the target genes controlled by HbMYC2/3/4, cis-elements such as JREs, GCC boxes, ABREs, EREs, G-boxes, CACG boxes, and DREs were inserted into the MCSs of pAbAi vectors. The bait vectors were subsequently co-transformed with pGADT7-HbMYC2, pGADT7-HbMYC3, and pGADT7-HbMYC4. The yeast one-hybrid assay results showed that co-transformation of the pAbAi-G-box bait strain with pGADT7-HbMYC2, pGADT7-HbMYC3, and pGADT7-HbMYC4 significantly enhanced aureobasidin A (AbA) concentrations in the resistant cells, suggesting that HbMYC2/3/4 can bind G-box elements and activate the expression of the AUR1-C gene, an antibiotic resistance gene that provides resistance to AbA (Fig. 5). Co-transformation of HbMYCs into bait strains containing GCC boxes, ERE boxes, ABREs, and DREs did not significantly enhance AbA resistance levels, suggesting that HbMYC2/3/4 did not bind these cis-elements. Interestingly, HbMYC4 but not HbMYC2 and HbMYC3 bound JRE and evidently increased the AbA resistance level (Fig. 5).
Interaction between HbMYCs and cis-elements by yeast one-hybrid assays. pGADT7-Rec2-HbMYC2, pGADT7-Rec2-HbMYC3 and pGADT7-Rec2-HbMYC4 were transferred into Y1HGold strains containing different pAbAi-cis-element bait vectors. The strains were then plated on SD/-Leu/AbA solid media containing 0–700 ng/mL AbA to cultivate for four days at 30 °C, after which the growth of the cells was observed. Arrows indicate significant differences in the pAbAi-G-box vector.
HbMYC2/3/4 proteins bind to the promoter and regulate the expression of the HbPIP2;1 gene
To identify the targets genes of HbMYCs, we first screened which latex biosynthesis- or drainage-related gene promoters contained a G-box, which led to the identification of HbPIP2;1 as a potential target of HbMYCs. HbPIP2;1 has been proposed to be one of two aquaporins involved in ethylene stimulation during latex production by regulating the water exchange between inner liber and latex cells in Hevea brasiliensis32,55. The promoter of HbPIP2;1 contains 5 G-box core sequences, e.g., CACGTG, CAGACGTGGCA, TACGTG, CACGTC and CACATGG, which are distributed at 128 bp, 240 bp, 434 bp, 373 bp and 64 bp upstream of the ATG translation start site, respectively. The 970 bp promoter sequence upstream of the ATG translation start site of HbPIP2;1 was inserted into the MCS of the bait vector pHis2.1. Co-transformation of HbMYC2/3/4 significantly increased cell tolerance to 3-amino-1,2,4-triazole (3-AT), suggesting that HbMYC2/3/4 bound the HbPIP2;1 promoter and activated the expression of the His3 reporter gene (Fig. 6a). To further investigate whether HbMYC2/3/4 could bind the HbPIP2;1 promoter and regulate gene expression in planta, the HbPIP2;1 promoter was inserted to the MCS of the pSP-luc+NF plasmid (accession U47123). Additionally, the full-length CDSs of HbMYC2/3/4 were inserted into the MCSs of pCAMBIA1300 vectors under the control of the 35 S promoter. Two types of plasmids were co-transformed into protoplasts via PEG-mediated methods. Luciferase activity under the control of the HbPIP2;1 promoter was significantly elevated after co-transformation with 35 S::HbMYC2, 35 S::HbMYC3, or 35 S::HbMYC4, suggesting that HbMYC2/3/4 could bind the HbPIP2;1 promoter and up-regulate the expression of the reporter gene in plant cells (Fig. 6b).
HbMYC2/3/4 bind the promoter and regulate the expression of the HbPIP2;1-P gene. (a). The pGADT7-HbMYC2, pGADT7-HbMYC3, pGADT7-HbMYC4 plasmids were transformed into Y187 Gold strains containing pHis2.1-PIP-P, after which the strains were cultured in SD/-His/-Leu/-Trp + 3-AT media at 30 °C for 3–5 days. (b) Schematic diagram of the reporter and effector constructs used in the luciferase assay. The firefly luciferase (LUC) reporter was driven by the HbPIP2;1 promoter, and HbMYC2/3/4 were driven by the CaMV 35 S promoter in each of the effector constructs. c. Luciferase assay of the enhancement of the HbPIP2;1 promoter activity by the overexpression of HbMYC2, HbMYC3, and HbMYC4 in protoplasts. The pPIP2:LUC reporter and respective 35 S:MYC2/3/4 effector constructs as well as empty vector controls were co-transformed into Arabidopsis protoplasts. Luciferase activities were quantified using a dual-luciferase assay kit (Promega, USA) and detected by using a GloMax® 96 microplate luminometer (Promega, USA). The values are the means ± SDs from the results of three replicates.
Expression profiles of HbMYC2/3/4 and HbPIP2;1
To investigate how JA and ET signals affect the expression of HbMYC2/3/4 and HbPIP2;1, the leaves of the epicormic shoots of Hevea brasiliensis were sprayed with 100 µM methyl-JA or ethrel. The qRT-PCR results revealed that the expression levels of HbMYC2/3/4 were quickly induced by JA treatment and sharply suppressed by ET treatment, indicating opposite roles for JA and ET in the regulation of HbMYC2/3/4 (Fig. 7a–c). Surprisingly, the expression profile of HbPIP2;1 was not consistent with that of HbMYC2/3/4. In addition, HbPIP2;1 was negatively regulated by JA treatment but positively regulated by ET treatment (Fig. 7d), suggesting that there might be other factors coordinating with HbMYC2/3/4 to regulate the expression of HbPIP2;1.
Different expression levels of HbMYC2/3/4 and HbPIP2;1 in the MeJA- and ET-treated leaves of rubber trees. Leaves of the epicormic shoots of Hevea brasiliensis were sprayed with 100 µM methyl-JA or 100 µM ethrel. Samples were collected at 0 h, 3 h, 6 h, 14 h and 24 h after treatment. Expression levels of the HbMYC2 (a), HbMYC3 (b), HbMYC4 (c) and HbPIP2;1 (d) genes were analysed by qRT-PCR. The values are the means ± SDs from the results of three replicates. Asterisks indicate significant differences compared with those of the 0 h control (***P < 0.005, Student’s t-test).
Discussion
H. brasiliensis is a very important crop for natural rubber production. Farmers regularly harvest latex by tapping the bark at intervals of 2–3 days. The drainage and de novo biosynthesis of latex is actually a wound response of rubber trees. JA is a master phytohormone that mediates wound responses; these responses have been well elucidated in Arabidopsis and in many other plant species56. Discovery of the JAZ gene family has significantly advanced our understanding of how the JA signalling pathway operates and has reinforced the recurring theme that hormone-dependent removal of transcriptional repressors is required for the activation of various plant hormone signalling pathways57. More detailed mechanistic understanding came from the identification of JAZ targets using in vitro (e.g., yeast two-hybrid) and in planta assays7,8,21. However, JA-related wound responses have not been well documented in H. brasiliensis, and the underlying molecular mechanism is not well known. JA and mechanical wounding have been reported to induce laticifer differentiation1. In a previous study, we globally cloned the JAZ gene family of Hevea brasiliensis. In the present study, several JAZ targets, including three bHLH proteins (termed HbMYC2/3/4), were identified via yeast two-hybrid screening. All HbMYCs localized in the nucleus and exhibited transcriptional activity, suggesting that they are transcription factors (Fig. 1). Like MYC2/3/4 of Arabidopsis, HbMYC2/3/4 interacted with many HbJAZ members and exhibited specific JAZ interactions profiles (Fig. 3), indicating their specific roles in JA responses. Although two MYC transcription factors have been previously reported to be responsive to multiple treatments in Hevea brasiliensis, no evidence has shown whether they interact with JAZ proteins or are related to JA signalling58. The identification of HbMYC2/3/4 in this work provides insight into the network of JA signalling in H. brasiliensis.
Interaction between trans-factors and cis-elements is the cornerstone of gene expression regulation. To identify the targets of HbMYCs, the cis-elements that could be bound by the HbMYCs were first screened. Seven cis-elements, e.g., JREs, GCC boxes, ABREs, EREs, G-boxes, CACG boxes, and DREs, were tested for potential interaction with HbMYCs by yeast one-hybrid assays, leading to the finding that all HbMYCs could bind G-box elements (Fig. 5). In tobacco, NtMYC2 can bind the G-box elements of the promoters of the nicotine biosynthesis-related genes PMT2 (Putrescine N-Methyltransferase 2) and QPT2 (Quinolinate Phosphoribosyl Transferase 2), up-regulating their expression59,60. Many JA-responsive gene promoters, such as those of PIN2 and VSPB in potato61,62, VSP1 in Arabidopsis63, PMT1a (Putrescine N-Methyltransferase 1a) in tobacco64, LA (Leucine Aminopeptidase) of tomato34, and ORCA3 of Catharanthus65, contain G-box elements (CACGTG), which are necessary for JA responsiveness. Thus, HbMYC2/3/4 might interact with G-boxes in the promoter to regulate the expression of target genes. Additionally, HbMYC4 bound JRE boxes in the yeast one-hybrid assays (Fig. 4). JRE was first identified in Catharanthus; that JRE included a qualitative controlling element (AAACGTGCCTTT) and a quantitative controlling element (CAATAAAATATT). The bHLH transcription factor CrMYC2 could bind the JRE and activate the expression of ORCA366. Using bioinformatics tools, we can identify which gene promoters contain G-boxes and JRE boxes, enabling us to predict in silico the target genes of HbMYCs.
By primarily screening the promoters of genes involved in latex biosynthesis and drainage, HbPIP2;1 was predicted as a potential target of HbMYCs due to several G-box elements existing in the core region of its promoter. As mature laticifers are devoid of plasmodesmata, the rapid exchange of water with surrounding liber cells is dependent on aquaporins embedded in the cell membrane. HbPIP2;1 is a plasma membrane-intrinsic protein. HbPIP2;1 was up-regulated in both liber tissues and laticifers in response to bark ethrel treatment and has been proposed to play a key role in ethylene stimulation of latex yield by regulating water exchange between inner liber cells and latex cells in H. brasiliensis32,55. To determine whether HbPIP2;1 was also the target of HbMYCs, the interaction between HbMYCs and the promoter of HbPIP2;1 was analysed. The yeast one-hybrid assay results showed that HbMYC2/3/4 each could bind the promoter of HbPIP2;1 (Fig. 6a). Furthermore, the dual-luciferase assay results also confirmed that HbMYC2/3/4 bound the HbPIP2;1 promoter and up-regulated the expression of a reporter gene in planta (Fig. 6b and c). However, HbPIP2;1 was negatively regulated by JA treatment and positively regulated by ET treatment (Fig. 7), which strongly suggests that there might be other ET-responsive factors that coordinate with HbMYC2/3/4 to regulate the expression of HbPIP2;1. This proposal requires additional investigation.
Methods
Plant materials
The H. brasiliensis cultivar Reyan 7-33-97 used in this study was planted at the experimental farm of Hainan University. The plants were pruned each year, and epicormic shoots grew from the dormant buds on the pruned branches. RNA from the bark, latex and leaves was isolated as described to analyse the expression of genes in different tissues67.
Yeast two-hybrid screening
The total RNA was isolated from the latex, bark, leaves, roots, and flowers of H. brasiliensis. Different RNA samples were mixed together into an RNA pool, which was used to create a “Mate & Plate™” library in accordance with the protocol of the Matchmaker™ Gold Yeast Two-Hybrid System (Clontech, USA). Additionally, HbJAZ1 was PCR-amplified by using a primer set (Supplementary Table S1-A). The PCR products were digested by EcoRI and SalI and then ligated into pGBKT7 to generate a pGBKT7-HbJAZ1 bait vector. The yeast two-hybrid process was carried out in accordance with the Matchmaker™ Gold Yeast Two-Hybrid System (Clontech, USA).
Transcriptional activity analysis
The positive colonies of the yeast two-hybrid screening were further cultivated on QDO/X/A media three times, after which the colonies were inoculated in QDO fluid medium for plasmid isolation. The isolated plasmids were further transformed into E. coli DH5a strains for sequencing. The full-length cDNAs of proteins that interacted with HbJAZ1 in the Y2H assays were obtained by in silico cloning procedures as previously described31. The coding sequences of the full-length cDNAs were cloned into a pGBKT7 vector (Clontech Inc., USA), which was further transformed into a Y2HGold yeast strain. Transcriptional activity was examined by streaking the yeast Y2HGold transformants onto SD/-Trp/-His/-Ade/X/A media (Clontech Inc., USA).
Protoplast preparation
Protoplasts of Arabidopsis were isolated as previously described68. Four-week-old Arabidopsis rosette leaves were cut by a razor into 0.5–1 mm pieces and then incubated with an enzymatic hydrolysate [0.15% (w/v) pectolyase Y-23 (Yakult, Japan), 0.35% (w/v) cellulose RS (Yakult, Japan), 0.4 M mannitol, 20 mM 2-(N-morpholine)-ethanesulphonic acid (MES), 20 mM KCl and 10 mM CaCl2] for 2–3 hours. The protoplast was harvested by filtrating with a 45 μm syringe filter (Pall, USA) and centrifuging at 100 g at 4 °C for 8 min.
Subcellular localization analysis
The full-length coding sequences of the HbMYC genes were inserted into pCAMBA1300 vectors to generate pCAMBA1300-HbMYC-GFP vectors. The reading frames of the HbMYCs and GFP were under the control of CaMV 35S promoter. The primers used are listed in Supplementary Table S1-B. The constructs and negative controls (pCAMBA1300-GFP) were transformed into Arabidopsis protoplasts as previously described68. The GFP fluorescence signal was visualized and imaged with a laser scanning confocal microscope (FluoView FV1000, Olympus, Japan).
Bimolecular fluorescence complementation (BIFC)
To verify the interaction between HbJAZ1 and HbMYC3, the ORFs of HbJAZ1 and HbMYC3 were amplified by PCR and inserted into multiple clone sites (MCSs) of pSPYNE and pSPYCE to generate pSPYCE-HbJAZ1, pSPYNE-HbMYC3 and pSPYNE-HbMYC4 BIFC vectors in accordance with previously described methods69. The primers used are listed in Supplementary Table S1-C. Combinations of pSPYCE-HbJAZ1 and pSPYNE-HbMYC3 vectors or combinations of pSPYCE-HbJAZ1 and pSPYNE-HbMYC4 vectors were co-transformed into Arabidopsis protoplasts in accordance with previously described methods68. YFP fluorescence was visualized and imaged by a laser scanning confocal microscope (FluoView FV1000, Olympus, Japan).
Quantitative real-time PCR (qRT-PCR)
All RNA samples were treated with RQ1 RNase-free DNase I (Promega) to remove DNA contamination, and the quality and concentration of the DNaseI-treated total RNA were both checked by agarose gel electrophoresis and measured by spectrophotometry. Two micrograms of DNase I-treated total RNA was used as template for first-strand cDNA synthesis in accordance with the manufacturer’s instructions (RevertAid™ First Stand cDNA Synthesis Kit, Fermentas, LT-2028 Vilnius, Lithuania). The qRT-PCR assays were performed using an ABI-7500 Real-Time PCR apparatus with SYBR Green I dye (Takara). The cDNA encoding 18 S rRNA was chosen as a reference gene using GeNorm software. The efficiency of each primer pair was evaluated before PCR. The primers used are listed in Supplementary Table S1-E. PCR was performed as follows: 3 min at 95 °C, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 58 °C for 15 s, and extension at 72 °C for 20 s. The relative abundance of transcripts was automatically calculated using the 2−△△CT method by the ABI-7500 software using the 18S rRNA gene as an internal standard. All experiments were performed with three independent biological replicates and three technical repetitions. SE calculations and ANOVA were used for statistical and significance analyses, respectively.
Yeast two-hybrid assays
The cloned full-length CDSs of the HbJAZ genes were PCR-amplified and inserted into a pGBKT7 vector to generate a bait vector. The bait vector was first transformed into a yeast Y2H gold strain to test for toxicity and autotranscription activity as described by the manufacturer (Cat. No. 630489,Clontech, Inc., USA). Subsequently, the CDSs of the HbMYC2/3/4 genes were further fused into a pGADT7 vector to generate prey plasmid. The primers used are listed in Supplementary Table S1-A. The bait vector and prey vector were subsequently transformed into Y187 and Y2Hgold strains, respectively. The yeast two-hybrid process was performed by mating together the Y187 and Y2Hgold strains, after which they were plated onto QDO/X/A (SD-Trp/-Leu/-Ade/-His/X/A) selection media.
Yeast one-hybrid assays
The HbMYC2/3/4-interacting cis-elements were screened using a Matchmaker Gold Y1H Screening system (Clontech). Forward and reverse nucleotide oligos for each cis-element, e.g., JREs (JA-responsive elements), GCC boxes, ABREs (ABA-responsive elements), ERE (ethylene-responsive elements), G-boxes, CACG boxes, and DREs (dehydration-responsive elements), were synthesized (listed in Supplementary Table S1-D). Each pair of oligo sequence was annealed and ligated into a pAbAi vector. The resulting pAbAi-bait plasmid was transformed into a Y1HGold strain to generate a bait reporter strain. The full-length CDSs of the HbMYC2/3/4 genes were then amplified with gene-specific primers (Supplementary Table S1-A). The PCR products were subsequently cloned into the pGADT7 prey vectors (Clontech), and the resulting prey vectors were transferred into the aforementioned bait reporter strains. The transformed cells were then grown on SD/–Leu plates at 30 °C for 3 days, after which time the cells were collected. The resuspended cells were then plated onto SD/–Leu media containing different AbA concentrations.
Dual-luciferase assays
The promoter sequence of HbPIP2;1 was amplified by PCR using the genomic DNA of cultivar Reyan 7-33-97 and was inserted into the MCSs of pSP-luc+NF plasmids. The full-length CDSs of HbMYC2/3/4 were amplified and inserted into PBI121 vectors. The specific primers used are listed in Supplementary Table S1-F. Two types of plasmids were transiently co-transferred into protoplasts by the polyethylene glycol (PEG)-mediated method. Sixteen hours after incubation in the dark at 20 °C, the protoplasts were harvested. The luciferase activities were subsequently quantified using a dual-luciferase assay kit (Promega, USA) and detected by using a GloMax® 96 microplate luminometer (Promega, USA).
References
Hao, B. Z. & Wu, J. L. Laticifer differentiation in Hevea brasiliensis: induction by exogenous jasmonic acid and linolenic acid. Ann. Bot. 85, 37–43 (2000).
Hao, B. & Wu, J. L. Effects of wound (tapping) on laticifer differentiation in Hevea brasiliensis. Acta Bot. Sin. 24, 388–391 (1982).
Hao, B., Wu, J. & Yun, C. Acceleration of laticifer differentiation in Hevea brasiliensis by latex drainage. Chin. J. Trop. Crops 5, 19–23 (1984).
Tian, W.-M., Yang, S.-G., Shi, M.-J., Zhang, S.-X. & Wu, J.-L. Mechanical wounding-induced laticifer differentiation in rubber tree: an indicative role of dehydration, hydrogen peroxide, and jasmonates. J. Plant Physiol. 182, 95–103 (2015).
Cao, Y., Zhai, J., Wang, Q., Yuan, H. & Huang, X. Function of Hevea brasiliensis NAC1 in dehydration-induced laticifer differentiation and latex biosynthesis. Planta 245, 31–44 (2017).
Cao, Y., Xiang, X., Geng, M., You, Q. & Huang, X. Effect of HbDHN1 and HbDHN2 genes on abiotic stress responses in Arabidopsis. Front. Plant Sci. 8, 470 (2017).
Song, S. et al. The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23, 1000–1013 (2011).
Qi, T. et al. The jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814 (2011).
Wang, H. et al. Defence responses regulated by jasmonate and delayed senescence caused by ethylene receptor mutation contribute to the tolerance of petunia to Botrytis cinerea. Mol. Plant Pathol. 14, 453–469 (2013).
Nickstadt, A. et al. The jasmonate-insensitive mutant jin1 shows increased resistance to biotrophic as well as necrotrophic pathogens. Mol. Plant Pathol. 5, 425–434 (2004).
Overmyer, K. Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12, 1849–1862 (2000).
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M. & Turner, J. G. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094 (1998).
Xu, L. et al. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935 (2002).
Chini, A. et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 (2007).
Thines, B. et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665 (2007).
Melotto, M. et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55, 979–988 (2008).
Chini, A., Fonseca, S., Chico, J. M., Fernández-Calvo, P. & Solano, R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59, 77–87 (2009).
Devoto, A. et al. COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32, 457–466 (2002).
Chung, H. S. et al. Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63, 613–622 (2010).
Lorenzo, O., Chico, J. M., Sánchez-Serrano, J. J. & Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950 (2004).
Cheng, Z. et al. The bHLH transcription factor MYC3 interacts with the jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4, 279–288 (2011).
Fernández-Calvo, P. et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715 (2011).
Niu, Y., Figueroa, P. & Browse, J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62, 2143–2154 (2011).
Pauwels, L. & Goossens, A. The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089–3100 (2011).
Dombrecht, B. et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245 (2007).
Heim, M. A. et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747 (2003).
Walker, A. R. et al. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1350 (1999).
Nesi, N., Jond, C., Debeaujon, I., Caboche, M. & Lepiniec, L. The Arabidopsis TT2 gene encodes and R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114 (2001).
Gonzalez, A., Zhao, M., Leavitt, J. M. & Lloyd, A. M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827 (2008).
Tian, W. W., Huang, W. F. & Zhao, Y. Cloning and characterization of HbJAZ1 from the laticifer cells in rubber tree (Hevea brasiliensis Muell. Arg.). Trees 24, 771–779 (2010).
Hong, H., Xiao, H., Yuan, H., Zhai, J. & Huang, X. Cloning and characterisation of JAZ gene family in Hevea brasiliensis. Plant Biol. 17, 618–624 (2014).
Tungngoen, K. et al. Involvement of HbPIP2;1 and HbTIP1;1 aquaporins in ethylene stimulation of latex yield through regulation of water exchanges between inner liber and latex cells in Hevea brasiliensis. Plant Physiol. 151, 843–856 (2009).
Qi, T. et al. Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26, 1118–1133 (2014).
Boter, M., Ruiz-Rivero, O., Abdeen, A. & Prat, S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18, 1577–1591 (2004).
Zhang, H.-B., Bokowiec, M. T., Rushton, P. J., Han, S.-C. & Timko, M. P. Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Mol. Plant 5, 73–84 (2012).
Groszmann, M., Paicu, T., Alvarez, J. P., Swain, S. M. & Smyth, D. R. SPATULA and ALCATRAZ, are partially redundant, functionally diverging bHLH genes required for Arabidopsis gynoecium and fruit development. Plant J. 68, 816–829 (2011).
Sorensen, A. M. et al. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33, (413–423 (2003).
Xu, J. et al. ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. Plant Cell 26, 1544–1556 (2014).
Xu, J. et al. The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22, 91–107 (2010).
Nakata, M. et al. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25, 1641–1656 (2013).
Li, H. et al. The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol. Biol. 65, 655–665 (2007).
Girin, T. et al. INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell 23, 3641–3653 (2011).
Heisler, M. G., Atkinson, A., Bylstra, Y. H., Walsh, R. & Smyth, D. R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128, 1089–1098 (2001).
Zhang, J. et al. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27, 787–805 (2015).
Kurbidaeva, A., Ezhova, T. & Novokreshchenova, M. Arabidopsis thaliana ICE 2 gene: phylogeny, structural evolution and functional diversification from ICE1. Plant Sci. 229, 10–22 (2014).
Xu, W. et al. Chinese wild-growing Vitis amurensis ICE1 and ICE2 encode MYC-type bHLH transcription activators that regulate cold tolerance in Arabidopsis. PLoS One 9, e102303 (2014).
Kang, H.-G., Foley, R. C., Oñate-Sánchez, L., Lin, C. & Singh, K. B. Target genes for OBP3, a Dof transcription factor, include novel basic helix-loop-helix domain proteins inducible by salicylic acid. Plant J. 35, 362–372 (2003).
Soy, J. et al. Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J. 71, 390–401 (2012).
Ni, W. et al. Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25, 2679–2698 (2013).
Yamashino, T. et al. A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 44, 619–629 (2003).
Petridis, A., Döll, S., Nichelmann, L., Bilger, W. & Mock, H.-P. Arabidopsis thaliana G2-LIKE FLAVONOID REGULATOR and BRASSINOSTEROID ENHANCED EXPRESSION1 are low-temperature regulators of flavonoid accumulation. New Phytol. 211, 912–925 (2016).
Friedrichsen, D. M. et al. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162, 1445–1456 (2002).
Yi, K., Menand, B., Bell, E. & Dolan, L. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 42, 264–267 (2010).
Kondou, Y. et al. RETARDED GROWTH OF EMBRYO1, a new basic helix-loop-helix protein, expresses in endosperm to control embryo growth. Plant Physiol. 147, 1924–1935 (2008).
Tungngoen, K. et al. Hormonal treatment of the bark of rubber trees (Hevea brasiliensis) increases latex yield through latex dilution in relation with the differential expression of two aquaporin genes. J. Plant Physiol. 168, 253–262 (2011).
Browse, J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205 (2009).
Kazan, K. & Manners, J. M. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17, 22–31 (2012).
Zhao, Y., Zhou, L. M., Chen, Y. Y., Yang, S. G. & Tian, W. M. MYC genes with differential responses to tapping, mechanical wounding, ethrel and methyl jasmonate in laticifers of rubber tree (Hevea brasiliensis Muell. Arg.). J. Plant Physiol. 168, 1649–1658 (2011).
De Boer, K. et al. APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J. 66, 1053–1065 (2011).
Sears, M. T. et al. NtERF32: a non-NIC2 locus AP2/ERF transcription factor required in jasmonate-inducible nicotine biosynthesis in tobacco. Plant Mol. Biol. 84, 49–66 (2013).
Mason, H. S., DeWald, D. B. & Mullet, J. E. Identification of a methyl jasmonate: responsive domain in the soybean vspB promoter. Plant Cell 5, 241–251 (1993).
Kim, S. R., Choi, J. L., Costa, M. A. & An, G. Identification of G-box sequence as an essential element for methyl jasmonate response of potato proteinase inhibitor II promoter. Plant Physiol. 99, 627–631 (1992).
Guerineau, F., Benjdia, M. & Zhou, D. X. A jasmonate-responsive element within the A. thalianavsp1 promoter. J. Exp. Bot. 54, 1153–1162 (2003).
Xu, B. & Timko, M. Methyl jasmonate induced expression of the tobacco putrescine N-methyltransferase genes requires both G-box and GCC-motif elements. Plant Mol. Biol. 55, 743–761 (2004).
Vom Endt, D., Soares e Silva, M., Kijne, J. W., Pasquali, G. & Memelink, J. Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-hook DNA-binding proteins. Plant Physiol. 144, 1680–1689 (2007).
Zhang, H. et al. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 67, 61–71 (2011).
Xia, Z. et al. RNA-Seq analysis and de novo transcriptome assembly of Hevea brasiliensis. Plant Mol. Biol. 77, 299–308 (2011).
Liu, L. L., Ren, H. M., Chen, L. Q., Wang, Y. & Wu, W. H. A Protein Kinase, calcineurin B-like protein-interacting proteinkinase9, interacts with calcium sensor calcineurin B-like protein3 and regulates potassium homeostasis under low-potassium stress inArabidopsis. Plant Physiol. 161, 266–277 (2012).
Walter, M. et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438 (2004).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Numbers 31260170 and 31060107).
Author information
Authors and Affiliations
Contributions
J.Z. performed most of the experiments; H.H. performed the yeast one-hybrid analysis and part of RT-PCR; H.X. constructed the BIFC vectors and performed transcriptional activity analysis; Y.C. performed the subcellular localization; X.L. performed the BIFC experiment; and X.H. supervised and wrote the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhai, J., Hao, H., Xiao, H. et al. Identification of JAZ-interacting MYC transcription factors involved in latex drainage in Hevea brasiliensis. Sci Rep 8, 909 (2018). https://doi.org/10.1038/s41598-018-19206-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-19206-3
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.