Abstract
Species of Pomacea, commonly known as apple snails, are native to South America, and have become widely distributed agricultural and environmental pests in southern China since their introduction in the 1980s. However, only since 2010 have researchers recognized that at least two species, P. canaliculata and P. maculata, are present in China. Although impacts of apple snails have been extensively documented, confusion still persists regarding current distributions and origin of the species in China. To resolve this confusion, we used phylogenetic and phylogeographic methods to analyze 1464 mitochondrial COI sequences, including 349 new sequences from samples collected in southern China and 1115 publicly available sequences from snails collected in the native and introduced ranges. Pomacea canaliculata was found at all sampled localities, while P. maculata was found at only five sampled localities in the Sichuan basin and Zhejiang province. Our data indicate that Chinese populations of P. canaliculata share an Argentinian origin, consistent with multiple introductions of this species elsewhere in Asia. In addition, just a single lineage of P. maculata is established in China, which shares with populations in Brazil.
Similar content being viewed by others
Introduction
Apple snails (Ampullariidae), are freshwater gastropods native to South America1, and several species in the genus Pomacea have been introduced and become established in many parts of the world including other Asian countries, North America, islands of the Pacific, and Europe2,3. They have a voracious appetite4,5, reproduce rapidly6, are resistant to desiccation during dry down periods7, and act as vectors of zoonotic diseases8, all of which have made them serious agricultural9, environmental1, and potential human health pests10.
With its highly diverse biogeography, topography, and climate, China offers numerous opportunities for a range of invasive species, and those that have been introduced have impacted China significantly11. Apple snails were initially introduced to the mainland of China, from Taiwan to Zhongshan city, Guangdong province, in 1981 for aquaculture12,13. The first ten years after introduction saw a rapid expansion of the range of apple snails in China, with a boom in aquaculture and economic interests as the main driver12. In the early 1980s, apple snails were introduced to at least 18 cities in11 provinces/municipalities, including Guangdong, Guangxi, Fujian, Zhejiang, Jiangxi, Jiangsu, Shanghai, Anhui, Hubei in the south, and Beijing and Liaoning in the north. In the mid-1980s, another 12 cities reported introduction of apple snails, including the southern provinces of Zhejiang, Yunnan, Sichuan, Chongqing and Jiangxi, and the northern provinces of Gansu and Tianjin12. Intentional introductions declined sharply after the 1990s, because of the poor market benefits and realization of significant crop damage caused by the snails. The spread also slowed, with limited expansion due to unintentional human transport and natural diffusion12.
Pomacea canaliculata (Lamarck, 1822) and P. maculata Perry, 1810 are the two most common and highly invasive apple snail species14. However, many other alien apple snail species were difficult to differentiate from P. canaliculata and P. maculata, which were frequently misidentified as these two species3,15,16. Additionally, for a long time, P. canaliculata was presumed to be the only alien apple snail species in Asia and was listed as one of 100 of the world’s worst invasive alien species17. However, Hayes et al., using a combination of morphological and DNA sequence data recognized four species of Pomacea as having been introduced into Asia3. Subsequently, Hayes et al. provided clear anatomical and biogeographic data for delineating between these two previously conflated species14.
In Asia, P. canaliculata was introduced to Asia more than once from multiple locations in Argentina, while P. maculata was introduced to Asia from Brazil and Argentina independently3. However, only 5 samples of P. canaliculata was recorded in China by Hayes et al.3. Subsequently, Song et al. and Lv et al. reported that both species, P. canaliculata and P. maculata, were established in China13,18. However, after the distribution pattern of apple snails sampled in 2006 and 2007 in China by Lv et al.13, there are no tracking updates of their spread until now. In this study, we combined phylogenetic and phylogeographic analyses of mtDNA COI sequences of P. canaliculata and P. maculata collected from across their range in China to fully document their origin and current distributions in China.
Material and Methods
Sample collection and DNA extraction
August 2014 – July 2015, we surveyed 34 localities in 14 provinces in mainland China in which apple snails might occur, collecting 44 adults and 305 egg masses from 31 locations in 12 provinces (Table 1). Subsamples of foot tissues and eggs, from each sampled population, were preserved in 100% ethanol and stored at −20 °C prior to extraction of DNA.
Genomic DNA was extracted from approximately 10 mg of foot tissue or a single egg from each clutch using the DNeasy Blood and Tissue Extraction Kit (QIAGEN) following the manufacturers’ protocol, with final elution of 200 µL. Eggs from each clutch were separated using a 10% sodium hydroxide solution19.
Amplification and sequencing
A portion of the mitochondrial gene cytochrome c oxidase subunit I (COI) was amplified using the primers LCO1490 and HCO219820 in 25 µL reactions containing 0.625 U TaKaRa Ex Taq, 1 × Ex Taq Buffer, 5 mM dNTP mixture, 10 µM of each primer and 1 µL of genomic DNA. Cycle conditions consisted of an initial denaturation for 3 min at 95 °C, followed by 35 cycles of 30 s at 95 °C, 30 s at 50 °C, and 1 min at 72 °C, followed by a final extension step of 72 °C for 8 min and 10 min at 4 °C. Amplicons were visualized and checked for specificity via gel electrophoresis and single product amplicons were sent to Sunny Biotechnology (Shanghai, China) for sequencing in both directions. All sequences were checked for errors and edited manually in Chromas 1.021. We finally obtained 349 COI sequences of 657 bp. Species were prior distinguished through phylogenetic analyses, and then all sequences were deposited in GenBank (Table 1).
COI datasets
We added 607 COI sequences from Hayes et al. to the 349 COI sequences generated in this study3. The sequences from Hayes et al. included sequences from the native and introduced ranges, with 466 sequences of P. canaliculata, 18 from China (five from the mainland and 13 from Taiwan), and 141 sequences of P. maculata3.
We downloaded another 151 sequences from GenBank from other studies, excluding those from Hayes et al.3. However, given the widespread issues with misidentification of Pomacea species, we filtered COI sequences from GenBank using the following criteria: (1) sequences were published after 2007 when it became possible to distinguish P. maculata sequences from those of P. canaliculata2; (2) sequences were verified as being correctly identified through phylogenetic systematic approaches (see below). After filtering, we discarded 33 sequences and added 118 to our matrix, including 52 sequences of P. canaliculata, and 66 sequences of P. maculata (Supplementary Table S1). We also added 390 sequences from Lv et al. Appendix S1, including 389 from P. canaliculata and one sequence of P. maculata (Table 1 of Lv et al.)13.
Phylogenetic analyses
The total matrix consisted of 1464 sequences that varied in length from 503 bp13 to 657 bp (this study). We added COI sequences from P. lineata (FJ710310)22 and P. paludosa (EU528477)3 to serve as outgroups. All sequences were assembled and aligned in ClustalW implemented in MEGA 6.023. The best sequence substitution model (GTR + I + G) for the data set was selected using the AIC in jModelTest ver. 2.1.724. Phylogenetic relationships among all COI sequences was reconstructed under Maximum Likelihood implemented in MEGA 6.0 with node support assessed using 1000 bootstrap replicates23,25.
Haplotype distribution and network analyses
Clades containing both P. maculata and P. canaliculata were identified from the phylogenetic analyses based on COI sequences, and these sequences were used to created haplotype networks in TCS 1.21 for each species26. The parsimony connection limit for haplotype network reconstruction was set to 95% for all analyses. We also mapped haplotype distributions in China using ArcGIS 10.2.
Prior to analyses unique haplotypes for each species were identified in DnaSP 5.127. Because the sequence lengths were different, prior to haplotype analysis three datasets consisting of only P. canaliculata and P. maculata haplotypes were constructed. (1) sequences from our study only (Dataset 1), 2) sequences from our study, plus those of Hayes et al., and those filtered from GenBank (Dataset 2), and 3) dataset 2 plus Lv et al. sequences (Dataset 3)3,13. Datasets were created by trimming all sequences to the shortest length for each species. Dataset 1 was 657 bp for both species, Dataset 2 was 558 bp for P. canaliculata and 577 bp P. maculata, and Dataset 3 was 503 bp for both.
Mismatch distribution analyses
Introduction scenarios and signals of historical population expansion was examined with mismatch analyses28,29. Theoretically, a mismatch distribution analyses for populations after bottlenecks followed by sudden expansions should generate well-separated peak patterns for each population, with each unique introduction source generating a separate peak3.
Patterns of genetic variation for P. canaliculata and P. maculata in mainland China were examined based on mismatch distribution analyses. We conducted the mismatch distribution analyses by comparing the number of pairwise differences at all sites of the COI sequences using DnaSP 5.1.
Data accessibility
All sequences were submitted in GenBank under accession numbers KP310264-KP310445, KP310474, KP310480-KP310496, KR020942-KR021020, KR021027, KR021034-KR021040, KT852706-KT852762, and KT852782-KT852786.
Results
Phylogenetic systematics
Apple snails were found at 31 of the 34 sites surveyed. Phylogenetic analyses recovered all sequences from these newly collected samples and all others on mainland China in two well supported, monophyletic clades. Of the 1464 COI sequences, 1226 were recovered in a clade identified as P. canaliculata, and the remaining 238 sequences were P. maculata (Fig. S1).
Haplotype distribution in China
There were no appreciable differences in the results derived from the three different datasets of different lengths, as such we only report the results from Dataset 3, which contained all sequences trimmed to 503 bp. From this dataset, the 1226 P. canaliculata sequences produced 58 unique haplotypes (PcH1- PcH58), while there were only 37 unique haplotypes from P. maculata (PmH1-PmH37; Table 2).
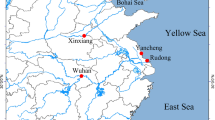
Seven P. canaliculata haplotypes representing 319 sequences (PcH1~PcH7) and two P. maculata haplotypes representing 30 sequences (PmH1and PmH2) were recovered. Pomacea canaliculata was found at all 31 sites, and P. maculata haplotypes were recorded from only five populations in the Sichuan province, Chongqing municipality, and Zhejiang province (Fig. 1). Twenty-two (71%) of the populations contained multiple haplotypes, and nine only had a single haplotype (four with only PcH1 and five with PcH2; Fig. 1).
Geographical distribution and frequency of P. canaliculata (PcH) and P. maculata (PmH) haplotypes in China. The map was created in ArcGIS 10.2 software (ESRI Inc., Redlands, CA, USA). URL http://www.esri.com/software/arcgis/arcgis-for-desktop. Red circles indicate survey sites with snails sampled during this study, and colors of the associated pie charts represent haplotype frequencies at each site. Stars indicate localities sampled by Lv et al.13 and triangles represent localities from Song et al.18.
Among the P. canaliculata haplotypes, PcH2 was the most widely distributed geographically (24 sites), accounting for 40% (n = 129) of the snails. Snails with this haplotype were mainly distributed along the eastern and southern coastal regions and at the northern edge of the range (Fig. 1). The second most widely distributed haplotype, PcH1, was detected in 21 sites, but made up 50% of the snails, which were primarily collected in the rural interior (Fig. 1). The remaining haplotypes, PcH3, PcH4, and PcH5 came from 5–14 sequences each, and were less widely distributed, found in two populations of Zhejiang province, two populations of Zhejiang and Jiangxi provinces, and four populations of Zhejiang, Guangdong, Guangxi, and Chongqing provinces, respectively. Two haplotypes, PcH6 and PcH7, were represented by a single sequence each and were found in only one population each, in Jiangsu and Hainan provinces, respectively (Fig. 1). Two haplotypes were recovered from the 30 P. maculata sequences, with PmH1 represented by 29 sequences occurring in five populations of Sichuan, Chongqing, and Zhejiang provinces. The other haplotype, PmH2, was represented by only one sequence in Zhejiang province (Fig. 1). Together, Dataset 3 (all sequences) produced 25 P. canaliculata haplotypes and 3 P. maculata haplotypes from China.
Haplotype networks and phylogenetic analyses
Under a 95% parsimony limit, haplotype analyses produced three independent networks for P. canaliculata and seven separate networks for P. maculata. Only two of the P. canaliculata networks and one of the P. maculata network included haplotypes from mainland China (Fig. 2a,b, and c). Networks for both P. canaliculata and P. maculata corresponded to well supported (BS values ≥ 95%) clades in the ML tree.
COI haplotype networks reconstructed using 95% connection limit both P. canaliculata and P. maculata. (a) Network A and (b) Network B represent for P. canaliculata, respectively. (c) Network C represents for P. maculata. They are the three networks containing haplotypes present in mainland China. The colors indicate haplotypes from different countries. For each haplotype, the size of the circle is proportional to the observed frequencies. PcH and PmH represent haplotypes for P. canaliculata and P. maculata, respectively.
For P. canaliculata, the 24 haplotypes from China occurred in two of the networks (Network A and Network B), with 209 sequences representing six haplotypes (PcH2 and PcH43~ PcH47) in Network A and 509 sequences representing 18 haplotypes (PcH1, PcH3~PcH7, and PcH48~ PcH58) in Network B (Fig. 2a,b). The third network contained eight haplotypes, including six unique to Argentina, one unique to Uruguay, and one shared by Taiwan and Japan (Fig. S2).
For Network A, PcH2 was the only shared haplotype out of the six detected haplotypes in China. It shared among populations found in Argentina (native) and non-native ranges, including China, the Philippines, Japan, Korea, Vietnam, Myanmar, USA, and Papua New Guinea (non-native) (Fig. 2a). The remaining 25 unique haplotypes and one shared haplotype (Argentina, Japan and the Philippines) were one to five steps away from PcH2, creating a star-like structure which indicated that PcH2 was an founding haplotypes (Fig. 2a). Among 23 haplotypes in Network B, PcH13 was the only one found in Argentina and 18 haplotypes were found in China (Fig. 2b). Five haplotypes (PcH1, PcH3~PcH6) detected in China were shared by snails found in Asian countries; one haplotype unique to Taiwan and the remaining 12 haplotypes were unique to mainland China (Fig. 2b).
For P. maculata, one (Network C) of seven networks contained haplotypes detected in China (Fig. 2c). The other six networks contained 15 haplotypes representing 39 sequences sampled from their native range in Brazil (Fig. S3). In Network C, PmH1 was the only haplotypes shared by snails found in Brazil and China, and also other non-native countries, including Japan, Vietnam, Thailand, Cambodia, Singapore, USA, Spain, and Belgium. PmH3 and PmH5 were the shared haplotypes found in Argentina (native range) and non-native ranges from USA, Thailand, Japan or Korea (Fig. 2c).
Mismatch distribution
The mismatch distribution for Chinese P. canaliculata sequences produced two distinct and well separated peaks, which exhibited high frequencies of number of nucleotide differences, with the intermediate peak representing single rare introduced samples (Fig. 3). However, the mismatch distribution for Chinese P. maculata sequences produced a single major peak (Fig. 3).
Mismatch distributions of P. canaliculata (blue) and P. maculata (red) sequences from China. The major peaks of samples in both distributions correspond to the haplotype groups recovered with network analysis (Fig. 2).
Discussion
China is the world’s fourth-largest country in terms of landmass, and its highly diverse topography and climate provides numerous opportunities for non-native species to find suitable habitats, establish, and potentially become invasive pests11. There are 560 confirmed invasive alien species in China, resulting in an estimated annual economic loss of more than US$18.9 billion30,31. The surge in economic growth following the implementation of the landmark Reform and Opening in 1978 was a milestone in China’s national policy and economic development, but resulted in the 1980s and 1990s in the introduction and spread of large numbers of invasive species11. Apple snails were one of the pests introduced and spread rapidly during this period.
The agricultural and environmental impacts, and associated economics costs of introduced species have led to a rising interest in studies on their ability to disperse, colonize, and establish in novel habitats32,33. Apple snails have colonized a wide range of aquatic systems in China, including rivers, paddies, pools, and ponds. The irrigated rice and wild rice (Zizania latifolia) ecosystems in southern areas provide an ideal environment for the dispersal and growth of the snails. Although the species of apple snails introduced to Asia and their origins have been elucidated3, their current distributions and origins in China have been less well understood.
Generally, ancestral populations possess higher levels of gene diversity than more recently established populations, which often display low diversity and few haplotypes34. The low haplotype diversity may be attributed either to the founder effect, such that invasive populations experience bottlenecks and genetic drift19,35, or to the bridge-head effect, in which the introduction of alien organisms to a non-native location may not be directly from the native range, but from a successful invasive population elsewhere36.
Both our and previous studies confirmed a much lower haplotype diversity of P. maculata in populations of China than in their native countries Argentina and Brazil with a statistic ratio of 3: 343,13,18. However, unlike non-native P. canaliculata populations in Hawaii with a single haplotype represented by sequences from 89 snails37, snails in China possess higher haplotype diversity than in native populations. We found 25 haplotypes from Chinese populations of P. canaliculata, thus five more than that from both Argentina and Uruguay (Table 2). It was indicated that apple snail populations had admixed in the course of invasion in China18. These admixed populations support the conclusion by Hayes et al. of multiple source introductions initially out of South America3. Such introduction scenarios increase genetic diversity of introduced populations over that of a single source introduction, thus possibly facilitating the establishment despite a bottleneck38. However, there were 14 haplotypes from Chinese populations of P. canaliculata reported by Lv et al., and not recovered in any other study or shared with any other countries. Since unique mutations were carefully checked and ambiguous bases were confirmed by Lv et al.13, the most parsimonious explanation is that these unique haplotypes come from unsampled populations in the native range. Two explanations are possible for the discrepancy in haplotype diversity between the previous study and this one for P. canaliculata: (1) the larger sample size of Lv et al. and the less extensive sampling in other countries for this study13, and/or 2) apple snails in China may have lost haplotypes as following a bottleneck39. However, further genetic analysis is needed to clarify this in any situation.
Haplotype diversity and distributions revealed consistent patterns with which revealed by Hayes et al. that Chinese populations of P. canaliculata shared an Argentinian origin with other introduced apple snails in Asia and experienced multiple introductions3. Different from P. canaliculata, Hayes et al. also indicated two introduction lineages of non-native P. maculata from Brazil and Argentina independently3. However, just single lineage of P. maculata from Brazil was introduced into and established in China. The presence of diverse shared haplotypes among different populations from different countries indicated a complicated pattern of introduction into China and other non-native countries.
According to early accounts, driven by the commercial benefits of aquaculture, apple snails were introduced to national wide including cities in both southern and northern China, like Beijing, Tianjin, and Liaoning province12. Our study of the current distribution of apple snails in China revealed that apple snails have established natural populations in most of southern China but none in north area. We found natural populations of apple snails in north area of Zhejiang provinces (longitude 30.31°N) and south area of Jiangsu provinces (longitude 31.23°N). Comparing with the sampling sites in previous studies13,18,40, our data indicated that apple snails tended to expand into northern China. In addtion,We discovered a new P. maculata population in Zhejiang province, which is ~1876 km far from the reported P. maculata populations in Sichuan and Chongqing basin, indicating an invisible or unobtrusive spread of apple snails.
It is recorded that apple snails were first introduced into Asia via Taiwan in 1979, and then introduced to other Asian countries, including Japan and the Philippines9,41,42. Subsequently, the prevalence of snails-farming and frequent agriculture contacts among our neighbor countries made a round introduction of apple snails and speed the wide spread of apple snails in Asia43,44. Nevertheless the native origins of invasive apple snails were explicated, such complicated pattern in introduced ranges was probably result from extensive influence by human activities. Human factors were also the most likely driver for the fast spread of apple snails in China. Our study for understanding the origin and distribution of apple snails is important for early detection and control of these invasive snails to slow the rate of new invasions in China.
References
Hayes, K. A. et al. Insight from an integrated view of the biology of apple snails (Caenogastropoda: Ampullariidae). Malacologia 58, 245–302 (2015).
Rawlings, T. A., Hayes, K. A., Cowie, R. H. & Collins, T. M. The identity, distribution, and impacts of non–native apple snails in the continental United States. BMC Evol. Biol. 7, 1–14 (2007).
Hayes, K. A., Joshi, R. C., Thiengo, S. C. & Cowie, R. H. Out of South America: multiple origins of non-native apple snails in Asia. Divers. Distrib. 14, 701–712 (2008).
Carlsson, N. O. L., Bronmark, C. & Hansson, L. A. Invading herbivory: the golden apple snail alters ecosystem functioning in Asian wetlands. Ecology 85, 1575–1580 (2004).
Qiu, J. W., Chan, M. T., Kwong, K. L. & Sun, J. Consumption, survival and growth in the invasive freshwater snail Pomacea canaliculata: does food freshness matter? J Mollus. Stud. 77, 189–195 (2011).
Barnes, M. A., Fordham, R. K., Burks, R. L. & Hand, J. J. Fecundity of the exotic apple snail, Pomacea insularum. J N. Am. Benthol. Soc. 27, 738–745 (2008).
Havel, J. E., Bruckerhoff, L. A., Funkhouser, M. A. & Gemberling, A. R. Resistance to desiccation in aquatic invasive snails and implications for their overland dispersal. Hydrobiologia 741, 89–100 (2014).
Kim, Y. S. & Choi, K. C. 215 snail and slug, markers of epithelial mesenchymal transition, appeared to be altered by alkyl-phenols, bisphenol a and nonyl-phenol, in ovarian cancer cells expressing estrogen receptors. Reprod. Fert. Develop. 27, 198 (2014).
Mochida, O. Spread of freshwater Pomacea snails (Pilidae, Mollusca) from Argentina to Asia. Micronesica Suppl. 3, 51–62 (1991).
Lv, S. et al. Invasive snails and an emerging infectious disease: results from the first national survey on Angiostrongylus cantonensis in China. PLoS Neglect. Trop. D. 3, e368 (2009).
Wan, F. H. & Yang, N. W. Invasion and management of agricultural alien insects in China. Annu. Rev. Entomol. 61, 77–98 (2016).
Yang, Y. et al. Historical invasion, expansion process and harm investigation of Pomacea canaliculata in China. Chinese Agri. Sci. Bull. 26, 245–250 (2010).
Lv, S. et al. Phylogenetic evidence for multiple and secondary introductions of invasive snails: Pomacea species in the People’s Republic of China. Divers. Distrib. 19, 147–156 (2013).
Hayes, K. A., Cowie, R. H., Thiengo, S. C. & Strong, E. E. Comparing apples with apples: clarifying the identities of two highly invasive Neotropical Ampullariidae (Caenogastropoda). Zool. J Linn. Soc. 166, 723–753 (2012).
Cazzaniga, N. J. Old species and new concepts in the taxonomy of Pomacea (Gastropoda: Ampullariidae). Biocell 26, 71–81 (2002).
Cowie, R. H., Hayes K. A. & Thiengo, S. C. What are apple snails? Confused taxonomy and some preliminary resolution in Global advances in ecology and management of golden apple snails (eds Joshi, R. C. & Sebastian, L. S.) 3–24 (Philippine Rice Research Institute, Philippines, 2006).
Lowe, S., Browne, M., Boudjelas, S. & DePoorter, M. 100 of the world’s worst invasive alien species, a selection from the global invasive species database. Published by The Invasive Species Specialist Group (ISSG) a specialist group of the Species Survival Commission (SSC) of theWorld Conservation Union (IUCN), 12 pp. First published as special lift-out in Aliens 12 December 2000. Updated andreprinted version: November 2004.
Song, H. M. et al. Sequencing cytochrome oxidase subunit I of mitochondrial DNA and the taxonomic status of apple snails. Chinese J. Zool. 45, 1–7 (2010).
Matsukura, K., Okuda, M., Cazzaniga, N. J. & Wada, T. Genetic exchange between two freshwater apple snails, Pomacea canaliculata and Pomacea maculata invading East and Southeast Asia. Biol. Invasions 15, 2039–2048 (2013).
Folmer, O., Black, M., Hoeh, W., Lutz, R. & Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 3, 294–299 (1994).
Goodstadt, L. & Ponting, C. P. CHROMA: consensus-based colouring of multiple alignments for publication. Bioinformatics 17, 845–846 (2001).
Hayes, K. A., Cowie, R. H. & Thiengo, S. C. A global phylogeny of apple snails: Gondwanan origin, genetic relationships, and the influence of outgroup choice (Caenogastropoda: Ampullariidae). Biol. J Linn. Soc. 98, 61–76 (2009).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9, 772 (2012).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Clement, M., Posada, D. & Crandall, K. A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659 (2000).
Librado, P. & Rozas, J. DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009).
Rogers, A. & Harpending, H. Population growth curves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569 (1992).
Harpending, H. Signature of ancient population growth in a low resolution mitochondrial DNA mismatch distribution. Hum. Biol. 66, 591–600 (1994).
Xu, H. G. et al. Aninventory of invasive alien species in China. NeoBiota 15, 1–26 (2012).
Ding, H., Li, M. Y. & Xu, H. G. Assessing economic costs of invasive exotic species in China in Alien Species Invasion, Biosafety and Genetic Resources (eds Xu, H. G., Wang, J. M., Qiang, S. & Wang, C. Y.) 78–128 (Beijing: Science, (2004).
Cote, J., Fogarty, S., Weinersmith, K., Brodin, T. & Sih, A. Personality traits and dispersal tendency in the invasive mosquitofish (Gambusia affinis). Proc. Biol. Sci. 277, 1571–1579 (2010).
Sabour, B. et al. Sargassum muticum (Yendo) Fensholt (Fucales, Phaeophyta) in Morocco, an invasive marine species new to the Atlantic coast ofAfrica. Aquat. Invaions. 8, 97–102 (2013).
Wu, Y., Mcpheron, B. A., Wu, J. J. & Li, Z. H. Genetic relationship of the melon fly, Bactrocera cucurbitae, (Diptera: Tephritidae) inferred from mitochondrial DNA. Insect Sci. 19, 195–204 (2012).
Shirk, R. Y., Hamrick, J. L., Zhang, C. & Qiang, S. Patterns of genetic diversity reveal multiple introductions and recurrent founder effects during range expansion in invasive populations of Geranium carolinianum (Geraniaceae). Heredity 112, 497–507 (2014).
Lombaert, E. et al. Bridgehead effect in the worldwide invasion of the biocontrol harlequin ladybird. PLoS One 5, e9743 (2010).
Tran, C. T., Hayes, K. A. & Cowie, R. H. Lack of mitochondrial DNA diversity in invasive apple snails (Ampullariidae) in Hawaii. Malacologia 50, 351–357 (2008).
Simon, A. et al. Invasive cyprinid fish in Europe originate from the single introduction of an admixed source population followed by a complex pattern of spread. PLoS One 6, e18560 (2011).
Nei, M., Maruyama, T. & Chakraborty, R. The bottleneck effect and genetic variability in populations. Evolution 29, 1–10 (1975).
Lv, S. et al. The emergence of angiostrongyliasis in the People’s Republic of China: the interplay between invasive snails, climate change and transmission dynamics. Freshwater Biol. 56, 717–734 (2011).
Naylor, R. Invasions in agriculture: assessing the cost of the golden apple snail in Asia. Ambio 25, 443–448 (1996).
Joshi, R. C. & Sebastian, L. S. Global advances in the ecology and management of golden apple snails. (Muñoz Nueva Ecija: Philippine Rice Research Institute, Philippines, 2006).
Cowie, R. H. Apple snails (Ampullariidae) as agricultural pests: their biology, impacts and management. In: G. M. Barker ed.. Molluscs as crop pests (pp. 145–192. CAB–International, Wallingford, 2002).
Yusa, Y., Sugiura, N. & Wada, T. Predatory potential of freshwater animals on an invasive agricultural pest, the apple snail Pomacea canaliculata (Gastropoda: Ampullariidae), in southern Japan. Biol. Invasions 8, 137–147 (2006).
Acknowledgements
We thank Dr. Robert H. Cowie (University of Hawaii) and Dr. Kenneth A. Hayes (Howard University) for their constructive comments and reviewing language in this manuscript. We thank Yalin Bian, Guangfu Liu, Yinbo Li, Dr. Zihong Ye, Dr. Yafen Zhang for their assistance in sample collection. This work was supported by a grant from Zhejiang Natural Science Foundation (No. LQ15C140002) and Yucai Project of Zhejiang Association for Science and Technology Foundation (No. 2017YCGC006).
Author information
Authors and Affiliations
Contributions
X.-P. Y. and Q.-Q.Y. conceived and designed this project. S.-W.L., Q.-Q.Y. and C.H. performed the molecular experiments. Q.-Q.Y., S.-W.L. and X.-P.Y. wrote the paper. All authors review the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, QQ., Liu, SW., He, C. et al. Distribution and the origin of invasive apple snails, Pomacea canaliculata and P. maculata (Gastropoda: Ampullariidae) in China. Sci Rep 8, 1185 (2018). https://doi.org/10.1038/s41598-017-19000-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-19000-7
This article is cited by
-
A smartphone-based crowd-sourced real-time surveillance platform (apple snail inspector) for the invasive snails: a design and development study
Parasites & Vectors (2024)
-
Comparison of development and overwintering rates and feeding efficiency on rice seedlings among two invasive freshwater apple snails and their hybrid
Hydrobiologia (2024)
-
Global distribution of the invasive apple snail Pomacea canaliculata: analyzing possible shifts in climatic niche between native and invaded ranges and future spread
Aquatic Sciences (2024)
-
Niche conservatism and geographical range expansion of Pomacea canaliculata and Pomacea maculata in non-native United States and China
Biological Invasions (2023)
-
First report of the invasive snail Pomacea canaliculata in Kenya
CABI Agriculture and Bioscience (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.