Abstract
Results from live cell imaging of fluorescently tagged Cellulose Synthase (CESA) proteins in Cellulose Synthesis Complexes (CSCs) have enhanced our understanding of cellulose biosynthesis, including the mechanisms of action of cellulose synthesis inhibitors. However, this method has been applied only in Arabidopsis thaliana and Brachypodium distachyon thus far. Results from freeze fracture electron microscopy of protonemal filaments of the moss Funaria hygrometrica indicate that a cellulose synthesis inhibitor, 2,6-dichlorobenzonitrile (DCB), fragments CSCs and clears them from the plasma membrane. This differs from Arabidopsis, in which DCB causes CSC accumulation in the plasma membrane and a different cellulose synthesis inhibitor, isoxaben, clears CSCs from the plasma membrane. In this study, live cell imaging of the moss Physcomitrella patens indicated that DCB and isoxaben have little effect on protonemal growth rates, and that only DCB causes tip rupture. Live cell imaging of mEGFP-PpCESA5 and mEGFP-PpCESA8 showed that DCB and isoxaben substantially reduced CSC movement, but had no measureable effect on CSC density in the plasma membrane. These results suggest that DCB and isoxaben have similar effects on CSC movement in P. patens and Arabidopsis, but have different effects on CSC intracellular trafficking, cell growth and cell integrity in these divergent plant lineages.
Similar content being viewed by others
Introduction
Cellulose is composed of β-1,4-glucan chains that are hydrogen-bonded together to form microfibrils, which are major contributors to the strength of plant cell walls. These microfibrils are synthesized by Cellulose Synthase (CESA) proteins that reside in the plasma membrane within Cellulose Synthase Complexes (CSCs). CSCs both polymerize β-1,4-glucan chains and facilitate their assembly into microfibrils. Mutations in Arabidopsis CESAs result in phenotypes that range from mild dwarfism to lethality, indicating the importance of cellulose in vascular plant development1. Much less is known about the function of cellulose in the development of non-vascular plants such as mosses2.
The study of CESAs and CSCs entered a new era with the development of methods for tagging CESAs with fluorescent proteins (FPs), facilitating live cell imaging of CSC movement behaviors3. These methods have facilitated investigations of CESA intracellular trafficking4,5,6,7, CSC interaction with the cytoskeleton and other proteins8,9,10,11, regulation of CESA and CSC function by endogenous and environmental factors12, and the mechanisms of action of cellulose synthesis inhibitors13,14,15,16,17,18, among other aspects of cellulose biosynthesis. All but one of these investigations have been performed in Arabidopsis, and imaging of CSCs in tip-growing cells has been precluded because FP-CESA fusion proteins fail to accumulate in the plasma membrane of these cell types19. Investigating cellulose synthesis in a nonvascular plant such as the moss Physcomitrella patens would enable us to better understand the evolution of cellulose synthesis and the functions of cellulose in a wider range of developmental processes, including tip growth.
The advantages of P. patens as an experimental organism include a high quality genome sequence20,21 and the capacity for targeted genetic manipulation due to its high rate of homologous recombination22,23. The P. patens plant body is typical of mosses, with two haploid stages: a filamentous protonemal stage, and gametophores consisting of leafy stalks with rhizoids24. The protonemal filaments extend by tip growth in a manner similar to the pollen tubes and root hairs of seed plant species25,26,27. The gametophore leaf cells expand by diffuse growth28 like most cell types in seed plants29.
Seven CESA isoforms have been identified in P. patens30. Mutation analysis has shown that PpCESA5 is required for gametophore development31. ppcesa5 knockout (KO) mutants have strong developmental phenotypes including failure of gametophore buds to sustain meristematic growth and produce leaves31. In addition, a subtle gametophore length phenotype has been reported for one double ppcesa6/7 KO line32. We have recently found that ppcesa8 KO mutants also have a developmental phenotype consisting of reduced cellulose deposition in the midrib stereid cells, which have thickened cell walls33. Because ppcesa5 KO and ppcesa8 KO lines have clear phenotypes, the functionality of mEGFP-PpCESA fusion proteins can be determined by testing transformed lines for complementation of these phenotypes.
One aspect of cellulose biosynthesis that has been clarified through the use of live cell CESA imaging is differences in the mechanisms of action between cellulose biosynthesis inhibitors34. In Arabidopsis, treatment with 2,6-dichlorobenzonitrile (DCB) immobilizes YFP-AtCESA6 in the plasma membrane, whereas treatment with isoxaben causes accumulation of YFP-AtCESA6 in vesicles below the membrane14. Although particle density was not measured, DCB reduced mEGFP-BdCESA particle velocity in Brachypodium distachyon18. In contrast, freeze fracture electron microscopy of protonemal filaments from the moss Funaria hygrometrica indicated that CSCs are lost from the plasma membrane after DCB treatment35. Freeze fracture examination of wheat roots treated for short periods with DCB showed increased CSC density in the plasma membrane of cortical cells36, indicating that this discrepancy is not due to differences in the CSC visualization method. DCB affects growth in widely divergent plants and related phyla, including red37, green38 and brown39 algae, but in most species little is known about its specific effect on CSCs. One possibility is that tip growing cells respond differently to DCB. The effects of DCB on pollen tubes of various plants such as lily, petunia40, and Pinus bungeana41 include distortion of cell walls and changes in cell wall composition40,41. Treatment with DCB also causes tip rupturing of pollen tubes40 and root hairs19, as well as moss protonemal filaments35. Treatment with isoxaben inhibits growth and induces tip swelling in conifer pollen tubes42 and retards growth in Arabidopsis root hairs19.
Here we show that CSC behavior can be analyzed by live cell imaging of mEGFP-PpCESAs in tip growing protonemal filaments of P. patens. Live cell imaging was applied to test the effects of the cellulose synthesis inhibitors DCB and isoxaben on both CSC behavior and protonemal filament growth. mEGFP-tagged PpCESA particles exhibited linear motility at the cell surface that was similar to the behavior of FP-CESA particles observed in Arabidopsis cells. Similar to results in Arabidopsis, treatment with DCB inhibited mEGFP-PpCESA particle motility without changing particle density at the cell surface. Whereas isoxaben treatment also greatly diminished mEGFP-PpCESA particle motility, it did not cause complete loss of particles from the cell surface in contrast to the case in Arabidopsis. Protonemal growth rates were not inhibited by either DCB or isoxaben treatment, but DCB treatment frequently resulted in tip bursting. Together, these data indicate that cellulose synthesis, cell growth control, and the regulation of wall integrity in tip growing protonemal filaments of P. patens share fundamental similarities and differences with these processes in diffusely growing Arabidopsis cells.
Results
Construction and characterization of mEGFP-PpCESA fusion protein expression lines
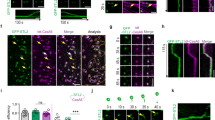
To create FP-CESA fusion protein expression lines for live cell imaging of CESA dynamics, we transformed mEGFP-PpCESA expression vectors into cognate mutant lines with clear visual phenotypes. Transformation of ppcesa5KO-231 with the Ubi::mEGFP-PpCESA5 expression vector produced six stable hygromycin resistant lines in which the gametophore-deficient phenotype was rescued. We chose a single line for further analysis (Fig. 1a) based on screening for fluorescence intensity by spinning-disk confocal microscopy. Because our ppcesa8KO-5B line is hygromycin resistant33, we excised the lox-p flanked hygromycin resistance cassette by transient expression of the CRE protein43. Transformation of a hygromycin susceptible line with Act1::mEGFP-PpCESA8 produced six stable hygromycin resistant lines. We chose one for further analysis based on screening for fluorescence intensity by spinning-disk confocal microscopy. Expression of mEGFP-PpCESA8 partially rescued the cellulose deficient midrib phenotype base on polarization microscopy (Fig. 1b). Quantitative analysis of S4B fluorescence intensity confirmed that the Act1::mEGFP-PpCESA8 line is partially rescued (mean fluorescence intensity = 5,939 ± 146 (SE) Arbitrary Units (AU)) in comparison to the parental ppcesa8KO line (mean fluorescence intensity = 3,971 ± 247 (SE) AU) and the wild type (mean fluorescence intensity = 7,938 ± 247 (SE) AU). The mEGFP-PpCESA8 line differed significantly from both ppcesa8KO (p = 0.00101) and the wild type (p = 0.00101) based on the results of one-way ANOVA with Tukey HSD test. Although complementation was tested for gametophore phenotypes, both PpCESA5 and PpCESA8 are expressed in protonema44. We also tested for cellulose deficiency phenotypes in ppcesa5KO-2 and ppcesa8KO-5B (Fig. S1). No significant differences were detected. However, PpCESA3 and PpCESA8 are partially redundant in secondary cell wall deposition33 and constitutive expression of both PpCESA3 and PpCESA8 can rescue the cesa5KO gametophore phenotype (Scavuzzo-Duggan et al., in review). So, lack of a cellulose deficiency phenotype for the cesa5KO and cesa8KO protonema can be explained by functional redundancy, and thus does not preclude a role for PpCESA3 and PpCESA8 in normal protonemal cell wall deposition.
Expression of mEGFP-PpCESAs rescue the cognate mutant phenotypes. (a) Transformation of the gametophore defective ppcesa5KO-2 line with Ubi::GFP-PpCESA5 restores wild type gametophore development. (b) Transformation of the ppcesa8KO-5B line, which is characterized by weak midrib birefringence due to reduced secondary cell wall deposition, was partially restored to the wild type phenotype by transformation with Act1::GFP-PpCESA8. (c) Quantitative analysis of fluorescence intensity in midribs stained with S4B confirmed partial restoration of the wild type phenotype by expression of Act1::GFP-PpCESA8 the ppcesa8KO-5B line.
After confirming that mEGFP-PpCESA5 and mEGFP-PpCESA8 rescue the phenotypes of ppcesa5 and ppcesa8 knockouts, respectively, we imaged these fusion proteins in protonemal cells using variable angle epifluorescence microscopy (VAEM). mEGFP-PpCESA5 particles were visible (Fig. 2a; Supplementary Movie S1), and moved across the cell surface along short linear paths (Fig. 2b,c) that were not aligned into co-linear arrays, as has been observed for FP-CESA particles that move in alignment with underlying cortical microtubule arrays in Arabidopsis3. We also detected mEGFP-PpCESA8 particles (Fig. 2d; Supplementary Movie S2), which moved across the cell surface along longer paths that were sometimes curved (Fig. 2e,f), but were also only rarely co-linear.
Imaging particle motility for mEGFP-PpCESA5 and mEGFP-PpCESA8. (a,d) Single frames from 10 min time-lapse imaging experiments. (b,e) Average projections of time-lapse images, with lines used to generate kymograph in yellow. (c,f) Kymographs of particle movement along lines in (b) and (e); diagonal lines represent movement over time. Images were acquired with VAEM with 2 s time interval.
Protonemal growth rate is not inhibited by DCB or isoxaben
In preparation for testing the effect of cellulose synthesis inhibitors DCB and isoxaben on mEGFP-PpCESA particle movement, we examined their effect on P. patens protonemal filaments by live cell imaging to determine sensitivity relative to the related species Funaria hygrometrica35. In initial experiments, 6 of 9 protonemal tips treated with 20 µM DCB ruptured within 10 min. In contrast, none of the tips treated with 20 µM isoxaben (n = 15), 10 µM DCB (n = 10), or control medium containing 0.05% ethanol (n = 9) ruptured after treatment (Fig. 3; Supplementary Fig. S2). Even at isoxaben concentrations as high as 100 µM, no tip rupture was observed. To measure the effect of inhibitors on tip growth, we collected image series for 20 min (20 µM DCB) or 15 minutes (20 µM isoxaben and control) and subjected them to kymograph analysis (Supplementary Fig. S3). For tips treated with 20 µM DCB, 13 of 20 ruptured with a mean rupture time of 11.5 ± 1.1 (SE) min. Tip growth rates measured up to the time of rupture were 0.216 ± 0.0325 (SE) µm min−1. Tip growth rates for control and isoxaben treatments were 0.244 ± 0.0283 (SE) µm min−1 (n = 12) and 0.222 ± 0.0300 (SE) µm min−1 (n = 14), respectively, with no significant differences among any of the treatments (p = 0.832). Control and isoxaben treated tips did not rupture. These data indicated that cellulose synthesis inhibitors do not affect the kinetics of protonemal tip growth per se in P. patens, in agreement with data for F. hygrometrica35, but that DCB can affect protonemal cell integrity in actively growing cells.
Protonemal tips rupture after treatment with DCB. Wild type P. patens protonemal filaments grown on solid PPNO3 medium for 7 days were treated with liquid PPNO3 medium with 0.05% ethanol (a,b), 10 µM DCB + 0.05% ethanol (c,d), 20 µM DCB + 0.05% ethanol (e,f), or 20 µM isoxaben + 0.05% ethanol (g,h) and were imaged at 30 sec intervals. Protonemal tips ruptured after treatment with 20 µM DCB (f), but not 20 µM isoxaben (h), or 10 µM DCB (d). The number of ruptured tips/number of imaged tips is shown for each treatment.
PpCESA-containing CSC particle motility, but not density at the cell surface, is reduced after DCB or isoxaben treatment
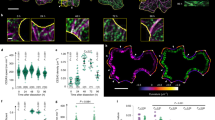
We then measured mEGFP-PpCESA particle density and motility under control conditions and after treatment with DCB or isoxaben. mEGFP-PpCESA5 particle velocities under control conditions averaged 262 ± 80 (SD) nm/min (Fig. 4a,d; Supplementary Movie S3), similar to the ~250–300 nm/min FP-CESA particle velocities measured in Arabidopsis seedlings3,5,14 and slightly faster than the mean of 164 nm/min measured in B. distachyon using a different quantification method18. Treatment of protonemal filaments expressing mEGFP-PpCESA5 with DCB or isoxaben before imaging dramatically decreased mEGFP-PpCESA5 motility (Fig. 4b–d; Supplementary Movies S4, S5). However, we did not measure large changes in mEGFP-PpCESA5 particle density at the cell surface after treatment with DCB and isoxaben, as compared to controls (Fig. 4a–c).
DCB and isoxaben inhibit mEGFP-PpCESA5 particle motility, but do not affect particle density at the cell surface. (a) Control protonemal cell treated with 0.1% ethanol. Mean CSC density = 0.48 ± 0.11 particles/μm2 (SD), n = 8 time-lapses. (b) Protonemal cell treated with 40 μM DCB. Mean CSC density = 0.51 ± 0.09 particles/μm2 (SD), n = 8 time-lapses. (c) Protonemal cell treated with 40 μM isoxaben. Mean CSC density = 0.44 ± 0.11 particles/μm2 (SD), n = 8 time-lapses. Scale bars = 5 μm. (d) Velocity distribution of mEGFP-PpCESA5 particles pooled from 24 time lapses. Mean velocities ± standard deviation in nm/min were: 262 ± 80 (Control); 18 ± 37 (40 μM DCB); and 36 ± 66 (40 μM isoxaben).
We also tested the effects of DCB and isoxaben on mEGFP-PpCESA8 particle velocity, which in control cells averaged 253 ± 79 (SD) nm/min (Fig. 5a,d; Supplementary Movie S6), remarkably similar to the velocities of particles containing mEGFP-PpCESA5. Also similar to the case for mEGFP-PpCESA5, treatment with DCB or isoxaben dramatically decreased mEGFP-PpCESA8 particle velocity compared to controls (Fig. 5b–d; Supplementary Movies S7, S8) with no large changes in mEGFP-PpCESA8 particle density at the cell surface. Although mEGFP-PpCESA8 particle densities were qualitatively lower than mEGFP-PpCESA5 particle density, we could not compare these densities statistically because they were estimated using different methods. In total, these results indicate that under control conditions, mEGFP-PpCESA particles behave similarly to FP-CESA particles in Arabidopsis in moving along linear trajectories at the cell surface at speeds of 250–300 nm/min, but that both DCB and isoxaben inhibit PpCESA particle motility and fail to completely remove FP-CESA particles from the cell surface, as has been reported for isoxaben-treated Arabidopsis cells14 .
DCB and isoxaben inhibit mEGFP-PpCESA8 particle velocity, but do not affect particle density at the cell surface. (a) Control protonemal cell treated with 0.05% ethanol. Mean CSC density = 0.20 ± 0.09 (SD) particles/μm2, n = 11 time-lapses. (b) Protonemal cell treated with 20 μM DCB. Mean CSC density 0.15 ± 0.11 particles/μm2, n = 5 time-lapses. (c) Protonemal cell treated with 20 μM isoxaben. Mean CSC density 0.07 ± 0.09 (particles/μm2), n = 5 time-lapses. Scale bars = 5 μm. (d) Velocity distribution of mEGFP-PpCESA8 particles pooled from 21 time lapses. Mean velocities ± standard deviations in nm/min were: 253 ± 79 (Control); 47 ± 38 (20 μM DCB); and 42 ± 40 (20 μM isoxaben).
Discussion
Whereas live-cell CESA imaging in Arabidopsis has contributed greatly to our understanding of the cell biology of cellulose synthesis, AtCESA fusion proteins have not been observed in the plasma membrane in tip growing cells19. Using live-cell CESA imaging in P. patens, we have shown that PpCESA-containing particles move in the membranes of tip-growing cells with velocities similar to those observed in diffusely growing cells of Arabidopsis3,5,14. Complementation of ppcesa mutant phenotypes by expression of the cognate mEGP-PpCESAs provides support for the normal behavior of the fusion proteins expressed with either the rice Actin1 promoter or maize Ubiquitin promoter.
The cellulose synthesis inhibitors isoxaben and DCB were confirmed to affect mEGFP-PpCESA5 and mEGFP-PpCESA8 motility, since velocities of PpCESAs decreased dramatically upon treatment with either cellulose synthesis inhibitor (Figs 4 and 5). The slowing of FP-PpCESA particle motility by DCB and isoxaben verify the potency of these drugs in P. patens, but their lack of effect on cell surface FP-PpCESA particle density and protonemal growth rates suggest that moss and seed plant cells respond to inhibition of cellulose synthesis by different mechanisms. In seed plants, cellulose synthesis inhibitors and CESA mutations activate a cell wall integrity-sensing pathway, resulting in production of reactive oxygen species, ectopic lignification, and growth inhibition (reviewed in45). It is possible that an inability to sense cell wall damage is responsible for the lack of growth inhibition following treatment with cellulose synthesis inhibitors in P. patens.
Although both DCB and isoxaben inhibited PpCESA motility, only DCB promoted bursting of protonemal tips. Differences in DCB and isoxaben mechanisms of action are manifested as opposing effects on CESA persistence in the plasma membrane in Arabidopsis14. Neither inhibitor altered CESA particle density at the cell surface in P. patens, in contrast to a report that the density of rosette CSCs as visualized by freeze fracture declined in response to DCB treatment in the related species F. hygrometrica35. In Arabidopsis, root hair tip growth is sensitive to DCB46, but not isoxaben19, perhaps because only DCB inhibits Cellulose Synthase-like D proteins (CSLDs), which contribute to cell wall deposition in root hairs and pollen tubes19. Thus, it is possible that CSLD activity is required to prevent bursting in both Arabidopsis root hairs and P. patens protonemal filaments.
Our data suggest that cellulose synthesis inhibitors affect CESA motility, and presumably the patterning and extent of cellulose polymerization, in P. patens protonemal cells, and also that tip growth in P. patens is differentially resilient to the inhibition of cellulose synthesis by these drugs. Differential effects, including the regulation of CSC trafficking and the sensing of cell wall integrity, will serve as fertile ground for future investigations into the unique and common mechanisms by which mosses and seed plants construct and expand their cell walls to control organismal growth and morphology during development.
Materials and Methods
PpCESA KO vector construction
Vectors for expressing mEGFP-PpCESA fusion proteins were constructed using Multisite Gateway Pro according to the manufacturer’s instruction (Invitrogen, Grand Island, NY, USA). A pDONR P1-P5r entry clone containing the coding sequence for mEGFP47 was linked to a pDONR P5-P2 entry clone containing the coding sequence of PpCESA548 and inserted into the pTHUbiGate destination vector, which drives expression with the maize Ubiquitin promoter49. The P1-P5r mEGFP entry clone was linked to a pDONR P5-P2 entry clone containing the coding sequence of PpCESA833 and inserted into the pTHAct1Gate destination vector, which drives expression with the rice Actin1 promoter50. By using different destination vectors, we were able to test two constitutive promoters for use in this application. Both vectors confer hygromycin resistance and target the expression cassette to the intergenic 108 locus22. Vectors were cut with SwaI for transformation into their respective knockout lines.
Culture and transformation of ppcesaKOs
All cultures were maintained in a growth chamber on basal medium supplemented with ammonium tartrate (BCDAT) as described previously51. The hph resistance cassette was removed from a ppcesa8KO-5B line33 by transforming protoplasts with NLS-Cre-Zeo, selecting for 7 d on BCDAT plates containing 50 μg mL−1 zeocin, replica plating the zeocin resistant colonies on BCDAT with and without 15 μg mL−1 hygromycin, and recovering hygromycin-sensitive colonies43.
Ppcesa5KO and ppcesa8KO-lox lines were transformed with mEGFP-PpCESA5 and mEGFP-PpCESA8 expression vectors, respectively, and subjected to two rounds of hygromycin selection as described previously51. Complementation of the ppcesa5KO31 and ppcesa8KO-lox33 morphological phenotypes was confirmed as described previously. Cellulose content of protonema cell walls was determined as described previously33 and the Kruskal-Wallis rank sum with Turkey-Kramer (Nemenyi) post hoc test (http://astatsa.com/KruskalWallisTest/) was used for statistical analysis.
GFP positive transgenic lines were selected using a Zeiss Cell Observer SD spinning disk confocal microscope (Zeiss, Carl-Zeiss-Strasse 22, 73447 Oberkochen) with a 488 nm excitation laser, a 525/50 emission filter, and a 100 × 1.40 NA oil immersion objective.
Tip growth analysis by live cell imaging
For cellulose synthesis inhibitor assays, protonemal filament explants were cultured on PPNO3 solid medium52 in glass bottom petri dishes (P35G-0.17-14-C, Mat Tek, Ashland, MA) for 7 d under continuous light53. DCB and isoxaben (40 mM stock in 100% ethanol) were added to PPNO3 liquid medium52 at a final concentration of 10 or 20 μM (0.05% ethanol); 0.05% ethanol was added to control medium. The dishes were flooded with 100 μL of medium and filament tips were imaged every 30 sec for 20 to 25 min starting immediately after treatment using a Zeiss Axiovert 200 M DICII contrast microscopy (Zeiss) with dimensions set at 516 × 516 with AxioCam software. Image stacks were assembled into kymographs (Supplemenary Fig. S3) using the MultipleKymograph plugin in ImageJ (http://www.embl.de/eamnet/html/body_kymograph.html) and tip growth distance was calculated as described previously53. Means and standard errors were calculated from the combined results of two independent experiments. One-way ANOVA with post-hoc Tukey HSD test (http://astatsa.com/OneWay_Anova_with_TukeyHSD/) was used for statistical analysis.
VAEM imaging of CESA-mEGFP particle trafficking
For VAEM imaging, 5- to 8-day-old plants regenerated from protoplasts were placed on an agar pad in Hoagland’s medium (4 mM KNO3, 2 mM KH2PO4, 1 mM Ca(NO3)2, 89 μM Fe citrate, 300 μM MgSO4, 9.93 μM H3BO3, 220 nM CuSO4, 1.966 μM MnCl2, 231 nM CoCl2, 191 nM ZnSO4, 169 nM KI, 103 nM Na2MoO4), covered by a glass cover slip and sealed with VALAP (1:1:1 parts of Vaseline, lanoline, and paraffin). For inhibitor treatments, 20 or 40 µM of DCB or isoxaben (40 mM stock in ethanol) was added to the Hoagland’s solution in the agar pad and imaging was started within 10 min. Controls were treated with the corresponding concentration of ethanol.
A Nikon Eclipse Ti microscope with a 100 X 1.49 NA TIRF objective and Andor DU-897 EMCCD camera was used to capture images every two seconds to create time-lapse videos of mEGFP-PpCESA5. A Nikon Eclipse Ti microscope with a 100X 1.49 NA TIRF objective and Zyla sCMOS camera (Zyla VSC-01746) was used to capture images every two seconds to create time-lapse videos of mEGFP-PpCESA8. Time lapses were from samples that were independently mounted and treated with inhibitor or solvent (5–8 time lapses per treatment).
Analysis of CESA-mEGFP velocities
Time-lapse files were opened in Fiji image-processing software54. Image contrast was normalized to improve the visibility of particles. CSC velocity was measured by identifying each particle of a size and brightness consistent with Arabidopsis CSCs3 within the first slice of a time-lapse (time 0 seconds), measuring its position, tracking it until it was no longer visible, and measuring its last position. The displacement of the particle and the time necessary for travel were calculated to yield speed measurements for each particle. Particles that did not persist for at least one minute were excluded from analysis, as were particles whose size, speed, or brightness identified them as CESA-containing vesicles rather than CSCs. This process of identifying particles that appeared to be CSCs was repeated at slices corresponding to increments of 2.5 minutes, and identified particles were traced backwards and forwards through the time-lapse so that the beginning and end points would correspond to the first and last appearance of the identified particles. This was done to minimize bias in the experimenter toward particles that moved at “normal” CSC speeds.
Analysis of mEGFP-PpCESA densities
To estimate CSC density in the mEGFP-PpCESA5 protonemal cells, time-lapse files were opened in Fiji and a Region of Interest (ROI) was selected with the freehand tool and its area measured with the measure function. The Particle Detector plugin was used to detect fluorescent CSCs with the following settings: 2 pixel radius, 0 cutoff, 1.9 percentile. The Particle Analysis Point Picker tool was used to select each particle within the ROI to acquire a count. Density was estimated as the number of particles detected by the Particle Detector plugin divided by the area of the ROI.
Signal levels of mEGFP-PpCESA8 particles were too low for the Particle Detector plugin to be able to reliably select CSC particles. Area of the cell was again determined using the freehand tool, but CSC count had to be approximated by the number of measured velocities from within the first slice of each time-lapse. This undercounted the number of CSCs because it ignored any that did not persist for 60s or whose paths could not be tracked during velocity measurements.
Data availability
Additional images generated during the current study are available from the corresponding author on reasonable request.
References
Somerville, C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22, 53–78 (2006).
Roberts, A. W., Roberts, E. M. & Haigler, C. H. Moss cell walls: structure and biosynthesis. Front Plant Sci 3, 166 (2012).
Paredez, A. R., Somerville, C. R. & Ehrhardt, D. W. Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312, 1491–5 (2006).
Crowell, E. F. et al. Pausing of Golgi Bodies on Microtubules Regulates Secretion of Cellulose Synthase Complexes in Arabidopsis. Plant Cell 21, 1141–1154 (2009).
Gutierrez, R., Lindeboom, J. J., Paredez, A. R., Emons, A. M. & Ehrhardt, D. W. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11, 797–806 (2009).
Lei, L. et al. Cellulose Synthase Interactive1 Is Required for Fast Recycling of Cellulose Synthase Complexes to the Plasma Membrane in Arabidopsis. Plant Cell 27, 2926–40 (2015).
Bashline, L., Li, S., Anderson, C. T., Lei, L. & Gu, Y. The endocytosis of cellulose synthase in Arabidopsis is dependent on mu2, a clathrin-mediated endocytosis adaptin. Plant Physiol 163, 150–60 (2013).
Lei, L., Li, S., Du, J., Bashline, L. & Gu, Y. Cellulose synthase INTERACTIVE3 regulates cellulose biosynthesis in both a microtubule-dependent and microtubule-independent manner in Arabidopsis. Plant Cell 25, 4912–23 (2013).
Lei, L., Li, S. & Gu, Y. Cellulose synthase interactive protein 1 (CSI1) mediates the intimate relationship between cellulose microfibrils and cortical microtubules. Plant Signal Behav 7, 714–8 (2012).
Li, S., Lei, L., Somerville, C. R. & Gu, Y. Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc Natl Acad Sci USA 109, 185–90 (2012).
Vain, T. et al. The cellulase KORRIGAN is part of the cellulose synthase complex. Plant Physiol 165, 1521–1532 (2014).
Endler, A. et al. A Mechanism for Sustained Cellulose Synthesis during Salt Stress. Cell 162, 1353–64 (2015).
DeBolt, S. et al. Morlin, an inhibitor of cortical microtubule dynamics and cellulose synthase movement. Proc Natl Acad Sci USA 104, 5854–9 (2007).
DeBolt, S., Gutierrez, R., Ehrhardt, D. W. & Somerville, C. Nonmotile cellulose synthase subunits repeatedly accumulate within localized regions at the plasma membrane in Arabidopsis hypocotyl cells following 2,6-dichlorobenzonitrile treatment. Plant Physiol 145, 334–8 (2007).
Brabham, C. & Debolt, S. Chemical genetics to examine cellulose biosynthesis. Front Plant Sci 3, 309 (2012).
Xia, Y. et al. Acetobixan, an inhibitor of cellulose synthesis identified by microbial bioprospecting. PLoS One 9, e95245 (2014).
Bischoff, V., Cookson, S. J., Wu, S. & Scheible, W. R. Thaxtomin A affects CESA-complex density, expression of cell wall genes, cell wall composition, and causes ectopic lignification in Arabidopsis thaliana seedlings. J Exp Bot 60, 955–65 (2009).
Liu, D. et al. Imaging cellulose synthase motility during primary cell wall synthesis in the grass Brachypodium distachyon. Sci Rep 7, 15111 (2017).
Park, S., Szumlanski, A. L., Gu, F., Guo, F. & Nielsen, E. A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat Cell Biol 13, 973–980 (2011).
Rensing, S. A. et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 (2008).
Zimmer, A. D. et al. Reannotation and extended community resources for the genome of the non-seed plant Physcomitrella patens provide insights into the evolution of plant gene structures and functions. BMC Genomics 14, 498 (2013).
Schaefer, D. G. & Zryd, J. P. Efficient gene targeting in the moss Physcomitrella patens. Plant J 11, 1195–206 (1997).
Reski, R. & Frank, W. Moss (Physcomitrella patens) functional genomics–Gene discovery and tool development, with implications for crop plants and human health. Brief Funct Genomic Proteomic 4, 48–57 (2005).
Cove, D. The moss Physcomitrella patens. Annu Rev Genet 39, 339–358 (2005).
Rounds, C. M. & Bezanilla, M. Growth mechanisms in tip-growing plant cells. Annu Rev Plant Biol 64, 243–65 (2013).
Hepler, P. K., Vidali, L. & Cheung, A. Y. Polarized cell growth in higher plants. Annu Rev Cell Dev Biol 17, 159–187 (2001).
Geitmann, A. & Emons, A. M. The cytoskeleton in plant and fungal cell tip growth. J Microsc 198, 218–45 (2000).
Harrison, C. J., Roeder, A. H., Meyerowitz, E. M. & Langdale, J. A. Local cues and asymmetric cell divisions underpin body plan transitions in the moss Physcomitrella patens. Curr Biol 19, 461–71 (2009).
Geitmann, A. & Ortega, J. K. Mechanics and modeling of plant cell growth. Trends Plant Sci 14, 467–478 (2009).
Roberts, A. W. & Bushoven, J. T. The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Mol Biol 63, 207–219 (2007).
Goss, C. A., Brockmann, D. J., Bushoven, J. T. & Roberts, A. W. A. CELLULOSE SYNTHASE (CESA) gene essential for gametophore morphogenesis in the moss Physcomitrella patens. Planta 235, 1355–1367 (2012).
Wise, H. Z., Saxena, I. M. & Brown, R. M. Jr. Isolation and characterization of the cellulose synthase genes PpCesA6 and PpCesA7 in Physcomitrella patens. Cellulose 18, 371–384 (2011).
Norris, J. H. et al. Functional specialization of cellulose synthase isoforms in a moss shows parallels with seed plants. Plant Physiology 175, 210–222 (2017).
Tateno, M., Brabham, C. & DeBolt, S. Cellulose biosynthesis inhibitors - a multifunctional toolbox. J Exp Bot 67, 533–42 (2016).
Rudolph, U., Gross, H. & Schnepf, E. Investigations of the turnover of the putative cellulose-synthesizing particle ‘rosettes’ within the plasma membrane of Funaria hygrometrica protonema cells. II. Rosette structure and the effects of cycloheximide, actinomycin D, 2,6-dichlorobenzonitrile, biofluor, heat shock and plasmolysis. Protoplasma 148, 57–69 (1989).
Herth, W. Effects of 2,6-DCB on plasma membrane rosettes of wheat root cells. Naturwissenschaften 74, 556–557 (1987).
Orologas, N., Delivopoulos, S. G., Dimopoulou, A. & Tsekos, I. Effects of 2, 6-dichlorobenzonitrile on plasma membrane cellulose synthesizing complexes and cellulose localization in cells of the red alga Erythrocladia subintegra. Phycologia 44, 465–476 (2005).
Mizuta, S. & Brown, R. M. Jr. Effects of 2,6-dichlorobenzonitrile and Tinopal LPW on the structure of the cellulose synthesizing complexes of Vaucheria hamata. Protoplasma 166, 200–207 (1992).
Bisgrove, S. R. & Kropf, D. L. Cell wall deposition during morphogenesis in fucoid algae. Planta 212, 648–58 (2001).
Anderson, J. R., Barnes, W. S. & Bedinger, P. 2,6-Dichlorobenzonitrile, a cellulose biosynthesis inhibitor, affects morphology and structural integrity of petunia and lily pollen tubes. J Plant Physiol 159, 61–67 (2002).
Hao, H., Chen, T., Fan, L., Li, R. & Wang, X. 2, 6-Dichlorobenzonitrile causes multiple effects on pollen tube growth beyond altering cellulose synthesis in Pinus bungeana Zucc. PLoS One 8, e76660 (2013).
Lazzaro, M. D., Donohue, J. M. & Soodavar, F. M. Disruption of cellulose synthesis by isoxaben causes tip swelling and disorganizes cortical microtubules in elongating conifer pollen tubes. Protoplasma 220, 201–207 (2003).
Vidali, L. et al. Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell 22, 1868–1882 (2010).
Tran, M. L. & Roberts, A. W. Cellulose synthase (CESA) gene expression profiling of Physcomitrella patens. Plant Biology 18, 362–368 (2016).
Hamann, T. & Denness, L. Cell wall integrity maintenance in plants: lessons to be learned from yeast? Plant Signal Behav 6, 1706–9 (2011).
Favery, B. et al. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15, 79–89 (2001).
Vidali, L., Rounds, C. M., Hepler, P. K. & Bezanilla, M. Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS One 4, e5744 (2009).
Scavuzzo-Duggan, T. R., Chaves, A. M. & Roberts, A. W. A complementation assay for in vivo protein structure/function analysis in Physcomitrella patens (Funariaceae). App Plant Sci 3, 1500023 (2015).
Anterola, A., Shanle, E., Perroud, P. F. & Quatrano, R. Production of taxa-4(5),11(12)-diene by transgenic Physcomitrella patens. Transgenic Res 18, 655–60 (2009).
Perroud, P. F. & Quatrano, R. S. The role of ARPC4 in tip growth and alignment of the polar axis in filaments of Physcomitrella patens. Cell Motil Cytoskel 63, 162–71 (2006).
Roberts, A. W., Dimos, C., Budziszek, M. J., Goss, C. A. & Lai, V. Knocking out the wall: protocols for gene targeting in Physcomitrella patens. Methods in Molecular Biology 715, 273–290 (2011).
Ashton, N. W., Grimsley, N. H. & Cove, D. J. Analysis of gametophytic development in the moss, Physcomitrella patens, using auxin and cytokinin resistant mutants. Planta 144, 427–435 (1979).
Furt, F., Liu, Y. C., Bibeau, J. P., Tuzel, E. & Vidali, L. Apical myosin XI anticipates F-actin during polarized growth of Physcomitrella patens cells. Plant J 73, 417–28 (2013).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–82 (2012).
Acknowledgements
This research was supported primarily as part of The Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE-SC0001090. The contributions of LV were funded by National Science Foundation award MCB-1253444. We thank Arielle Chaves for technical assistance and Sarah Kiemle for conducting the cellulose assays.
Author information
Authors and Affiliations
Contributions
M.L.T., T.W.M., H.S., and S.-Z.W. carried out live cell imaging experiments. J.H.N. and A.W.R. produced transgenic P. patens lines. M.B., L.V., C.T.A., and A.W.R. supervised the research. M.L.T., T.W.M., C.T.A. and A.W.R. wrote the manuscript with the participation and final approval of all authors.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tran, M.L., McCarthy, T., Sun, H. et al. Direct observation of the effects of cellulose synthesis inhibitors using live cell imaging of Cellulose Synthase (CESA) in Physcomitrella patens. Sci Rep 8, 735 (2018). https://doi.org/10.1038/s41598-017-18994-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-18994-4
This article is cited by
-
Quantitative cell biology of tip growth in moss
Plant Molecular Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.