Abstract
TFs are important proteins regulating plant responses during environmental stresses. These insults typically induce changes in cellular redox tone driven in part by promoting the production of reactive nitrogen species (RNS). The main source of these RNS is nitric oxide (NO), which serves as a signalling molecule, eliciting defence and resistance responses. To understand how these signalling molecules regulate key biological processes, we performed a large scale S-nitrosocysteine (CySNO)-mediated RNA-seq analysis. The DEGs were analysed to identify potential regulatory TFs. We found a total of 673 (up- and down-regulated) TFs representing a broad range of TF families. GO-enrichment and MapMan analysis suggests that more than 98% of TFs were mapped to the Arabidopsis thaliana genome and classified into pathways like hormone signalling, protein degradation, development, biotic and abiotic stress, etc. A functional analysis of three randomly selected TFs, DDF1, RAP2.6, and AtMYB48 identified a regulatory role in plant growth and immunity. Loss-of-function mutations within DDF1 and RAP2.6 showed compromised basal defence and effector triggered immunity, suggesting their positive role in two major plant defence systems. Together, these results imply an important data representing NO-responsive TFs that will help in exploring the core mechanisms involved in biological processes in plants.
Similar content being viewed by others
Introduction
Nitric oxide (NO), a small, gaseous, redox-active molecule, has gained attention due to its regulatory role in animal and plant physiology. Therefore, considerable research has been conducted for exploring its role in different biological processes1,2. Despite the attention this small molecule gained in animals for its myriad roles, the first report of NO generation in a biological system came from plants3 where it is reported to play a key role in physiological functions like growth, development, environmental interactions, defence4, stomatal regulation, and senescence5. Furthermore, NO’s role in mitigating abiotic stresses such as drought, cold and salt stress has also been studied6,7,8,1,9.
Despite the diverse role of NO in plant biology and the completion of various plant genome projects, the main source of NO production in higher plants is still unknown. In mammals, NO synthesis is associated with oxidative mechanisms involving NO synthase (NOS), which consists of three well-established isoforms: endothelial (eNOS), neuronal (nNOS), and inducible (iNOS)10. In contrast, seven possible routes have been proposed for NO production in plants6. The oxidative mechanisms include NO synthesis from L-arginine (L-Arg), polyamines, or hydroxyl amines while the reductive routes are dependent upon nitrite being reduced by nitrate reductase. A related protein isolated from the alga Ostreococcus tauri is 45% similar to human NOS enzyme. This enzyme was found to have NOS activity in vitro with an equally similar Km for NADPH oxidation11. However, no such enzyme has been isolated from higher plants.
Contrary to classical signal transduction, which mostly relies on the interaction of macromolecular shapes, NO and related reactive nitrogen species (RNS) act through chemical reactions with specific targets in various proteins, resulting in covalent modifications12. The amino acid cysteine (Cys) of different proteins sometimes consists a low pKa sulfahydryl group that significantly enhances their susceptibility to redox-based post-translational modifications13. Among these redox-based post-translational modifications, NO-mediated S-nitrosylation is perhaps the most studied one13; a phenomenon in which an NO moiety is covalently attached to a solvent-exposed cysteine residue to form S-nitrosothiols (SNOs). Protein S-nitrosylation is reported to be important in cellular processes for regulating enzyme activity, protein-protein interaction, and protein localization14. Genes that are involved in key physiological and biochemical processes have been reported to be S-nitrosylated, including NPR114, AtSABP315, and the auxin receptor TIR116. Recently, Hu, et al.17 showed that the proteome of Arabidopsis GSNOR knockout mutant atgsnor1-3 (which reported to have significantly higher level of SNOs)18 contains 926 and 1195 S-nitrosylated proteins and peptides, respectively.
NO’s role in transcriptional control has also been reported in some studies. Following abiotic stress or pathogen ingress, cellular RNS levels increase, resulting in swift transcriptional re-programming. Microarray and RNA-seq analysis have made it easy to identify genome-wide differentially expressed genes (DEGs) in response to a particular stimulus. A number of NO-mediated transcriptomic studies have been performed so far. In a study, involving Arabidopsis roots and leaves, GSNO-mediated transcriptome analysis showed the differential expression of 3263 genes19. Similarly, microarray analysis of Arabidopsis roots treated with different concentrations of SNP, resulted in differential expression of 422 genes including 342 up- and 80 down-regulated genes20.
Transcription factors (TFs) are important regulatory proteins that control the on/off switch of genes. They contain a DNA-binding domain that binds with specific sequences in the promoter of different genes, thereby activating or repressing their expression to regulate various physiological functions. Therefore, higher plants have a fine-tuned, complex regulatory system consisting of transcriptional activators and repressors that manage the expression of genes involved in plant development and defence21. Changes in cellular NO levels can regulate the expression of important genes, including TFs such as HY5, MYB, and Trx22. This indicates a regulatory role for NO during physiological and molecular process via the transcriptional as well as translational control of different genes. Identification of NO-responsive TFs will help establish the regulatory role of NO in plant tolerance to abiotic stress and disease. In this study, we used high-throughput transcriptome analysis to identify 673 TFs in Arabidopsis leaves that showed differential expression in response to 1 mM CySNO.
In a previous study we performed RNA-Seq mediated transcriptome analysis of Arabidopsis leaves infiltrated with 1 mM CySNO generating an average of 91 million reads. CySNO infiltration induced changes in the expression of 6436 (2988 down-regulated and 3448 up-regulated) genes after 6 h23. This study is focused on the differentially expressed genes coding different transcription factors. Here we used both in silico and in vivo analysis to highlight the importance of TFs in plant responses to nitrosative stress.
We examined three knockout mutant lines representing three TFs ddf1, rap2.6 and atmyb48 for their possible role in plant growth, development and defence. The DDF1 (dwarf and delayed flowering) is characterized by an AP2 domain. Overexpression of this gene results in dwarfism and delayed flowering and increase resistance to cold, drought and heat stress in Arabidopsis24. RAP2.6 an ERF TF family member is reportedly involved in enhanced resistance towards beet cyst nematode Heterodera schachtii in Arabidopsis roots through callose deposition. In addition, overexpression of this gene showed induced expression of JA-responsive genes25. According to TAIR annotation, AtMYB48 is involved in cell differentiation, response to salicylic acid and DNA-templated transcription factor activity. We have also used atgsnor1-3 mutant as susceptible control due to its established role in plant immunity through SNO homeostasis in the cell26.
Results
Data mining and analysis
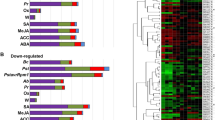
A total of 673 TFs that changed significantly in expression profile in response to exogenously applied 1 mM CySNO were identified by RNA-seq mediated transcriptome analysis at a cutoff Q-value of 0.05. A Heatmap generated from the expression values of controls and CySNO treated leaves (n = 3) revealed hierarchical clustering of all of the TFs genes with P-values < 0.05 (Fig. 1a). Of the TFs, 385 genes were up-regulated, while 288 were down-regulated in response to CySNO (Fig. 1b). An average of 23.1% TFs showed 10-fold change in their expression while 94.5% showed at least a 2-fold change, suggesting significant transcriptional changes in response to cellular NO accumulation (Supplementary Table S1). An MDS plot revealed no dispersion among control samples, however, some dispersion was found among CySNO treated samples, which may be due to the presence of TFs with very low basal expression levels (Fig. S1). A list of the top ten up- and down-regulated genes, along with their annotations, is given (Supplementary Tables S2, S3). All of the raw sequences and gene expression data were submitted to the public repository Gene Expression Omnibus (GEO) and Short Read Archive at NCBI (http://www.ncbi.nlm.nih.gov/) and are available under the accession numbers GSE81361 and SRP074890, respectively.
Clustering of NO-responsive TFs. (a) Heatmap showing the expression patterns of transcriptome-wide, differentially expressed transcription factor genes in response to CySNO. (b) A total of 673 TFs were identified at a Q-value < 0.05 in the transcriptome. Among these 385 were up-regulated and 288 down-regulated. The red-green colour key indicates changes in expression level (≥log2-fold change).
GO-Enrichment analysis of NO-induced TFs
All CySNO-responsive TFs were analysed for their respective gene ontology (GO) terms and fold enrichment through the GO consortium and PANTHER classification system, using Arabidopsis thaliana as a reference genome. Out of 673 total TFs, 664 were successfully found in the reference Arabidopsis genome. GO-terms for biological processes, molecular functions and cellular components were determined. A number of TFs were involved in biological processes showing different fold enrichment (FE) (Fig. S2A; Supplementary Table S4). These included localization (3 FE), developmental processes (5.30 FE), responses to external stimuli (2.50 FE), biological regulation (9.60 FE) and biogenesis (4.30 FE); the most common categories were metabolic processes (41.90 FE) and cellular processes (30.60 FE) (Fig. S2A). Further analysis within the metabolic processes category indicated TFs involved in nitrogen metabolism (21.20 FE), biosynthetic processes (18.40 FE), catabolic processes (3.10 FE), and primary metabolic processes (54.50 FE). Detailed analysis within the cellular processes category indicated that highest FE (85.20) was observed for TFs involved in regulation of the cell cycle, followed by cell communication (11.10 FE) (Fig. S2A). Similarly, TFs associated with binding activity (75.40 FE) and catalytic activity (19.60 FE) were the dominant categories in terms of FE in GO terms for molecular functions. Further analysis within the catalytic activity category revealed key catalytic activities including transferase (46.30 FE), hydrolase (31.70 FE), ligase (7.30 FE), and enzyme regulation (7.30 FE) (Fig. S2B). Within the binding activity category, nucleic acid binding (68.80 FE), protein binding (17.60 FE), and chromatin binding (13.20 FE) were the major categories (Fig. S2B). GO analysis for cellular components revealed TF genes localized to membranes (5.20 FE), macromolecular complexes (13.90 FE), organelles (35.80 FE), and other cellular parts (45.10 FE). The highest FE for TFs related to organelles were observed for those localized in the nucleus (93.80 FE, Fig. S2C).
NO responsive major transcription factor families
All of the CySNO-responsive TFs were analysed to identify major TFs families using MapMan. A number of well-known, key transcription factor families were induced in response to NO accumulation. The basic helix-loop-helix (bHLH) was the dominant TF family, having 53 TFs (Fig. 2). bHLH proteins are a superfamily of TFs that bind to DNA as a dimer and are characterized by the presence of a ~ 60 amino acid bHLH domain. This domain is well-characterized in mammals27; in plants, however, it is not well understood. AP2 (APETALA2) and EREBPs (ethylene-responsive element binding protein) was the second major TF family, with 44 TFs, that showed differential expression to CySNO (Fig. 2). AP2- EREBP are the archetypic members of this family, found only in plants28. It is a large, multi-gene family and is the key regulator of many developmental processes and responses to various environmental and biotic stimuli28.
MapMan analysis of NO-responsive TFs involved in different pathways. All the transcription factors that showed differential expression to CySNO were analysed for functional classification into different pathways using TAIR 10 (Aug 2012) as a reference genome database in MapMan. This figure is a schematic representation of gene regulation through different transcription factors and major transcription factor families that showed differential expression in response to CySNO. The red-blue key indicates up- and down-regulated genes, respectively. The illustrations were made using Microsoft PowerPoint (2013) and CorelDRAW X7.
We also identified 36 MYB and 16 MYB-related TFs in the CySNO-responsive transcriptome analysis (Fig. 2). MYB family genes have been reported in a number of plant and animal species including Arabidopsis, maize, populous, soybean and brassica for their diverse roles reviewed in29. MYB has been implicated for its role in ABA signalling and interaction with other TFs in plants29. The WRKY TF family is one of the largest TF families found in plants. Members of this family are characterized by presence of at least one conserved WRKY domain. We found that out of 74 total WRKY TFs found in Arabidopsis, 33 of these showed differential expression in response to CySNO (Fig. 2). The C2H2 Zinc-Finger TF family is another TF family found in plants that regulates genes involved in growth and development, as well as both biotic and abiotic stress conditions30. Of 211 C2H2 TF genes in Arabidopsis, 31 showed differential expression in response to CySNO (Fig. 2). Auxin/indole-3-acetic acid (Aux/IAA) proteins are short-lived TFs that are responsible for auxin-dependent transcriptional regulation. We found 12 Aux/IAA family members that showed differentially expression in response to NO-donor (Fig. 2).
Similarly, other transcription factor families showed differential expression in response to CySNO (Fig. 2) including: bZIP (basic region/leucine zipper motif), which regulates light and defence signalling responses in plants31; ARF (Auxin responsive factor family); C2C2-Gata, reportedly involved in regulation of light or circadian rhythms32; C3H; HB (home box), a 60 amino acid peptide initially discovered in Drosophila that regulates embryogenesis and pathogenesis33; the SET-domain, recognized as having methyl-transferase activity34; the NAC domain, which is involved in transcriptional re-programming during plant biotic and abiotic stress responses35; MADS box, involved in plant developmental processes36; Trihelix, involved in the regulation of light, pathogen attack, and salt responses in plants37; GRAS, a key player of GA signalling, thus regulating growth and development38; and PHOR1, a U-box component of GA signalling with a role in proteasome degradation39. However, a large number of TF genes (74) were mapped as unspecified (Fig. 2).
Transcription factor genes putatively involved in cellular processes and defence
Almost 30% (201/673) of the CySNO-responsive TFs showed putative involvement in biotic and abiotic stress responses (Fig. 3). Out of these, 95.5% (193/201) were related to biotic stress. A heatmap showing the expression of TFs involved in defence is also given (Supplementary Fig. S3). These include a variety of TFs like ERF, bZIP, WRKY, MYB and DOF (the detail of these TFs is given in the “major transcription factors” section). Among these, twenty-five TFs were mapped to biotic-stress-mediated hormone signalling, seven were mapped to proteolysis, and four were putatively involved in biotic-stress-mediated secondary metabolite production (Fig. 3). Similarly, among other cellular processes, thirty-one TFs were putatively involved in development, seven in protein degradation, three in DNA synthesis, and a single TF each in cell division and cell cycle, DNA repair, RNA synthesis, and transport (Fig. 3).
An average of 30% TFs were involved in defence. Functional classification of TFs involved in different cellular process and defence. Out of all NO-induced TFs, 193 were involved in defence and 31 in development. The red-blue colour key indicates up- and down-regulated genes, respectively. The bottom-right section of the figure represents ubiquitin proteasome system (UPS) that acts in ATP-dependent manner with Regulatory Particle Triple-A ATPase 3 (RPT3). Proteins are targeted to the proteasome by tagging with small ubiquitin protein (red balls) through a 3 step processes. First E1 ubiquitin-activating enzyme is covalently linked with an ubiquitin protein which is then transferred to a E2 ubiquitin-conjugating enzyme. This activates the E3 ubiquitin ligase complex having RING U-box and HECT (homologous to E6-assoicated protein C terminus) domain proteins that mediates transfer of ubiquitin to lysine residue in the substrate thus marking them for proteasomal degradation. About 7 different NO-responsive TFs were mapped to the E3 ubiquitin-ligase complex suggesting NO role in UPS.
TFs putatively involved in biotic stress signalling
We identified seven TFs that were putatively involved in signalling during stress conditions using MapMan (Fig. 3). Among these, 6 were up-regulated while only one was down-regulated in response to CySNO. These include AT1G35160 (GF14PHI), involved in brassinosteroid signalling; At2g32250 (FRS2), involved in light signalling; At3g59220 (PRN); At4g09570 (CPK4), involved in ABA signalling; and At5g60890 (AtMYB34), involved in the tryptophan pathway, according to TAIR (https://www.arabidopsis.org/) annotations. The single down-regulated TF involved in signalling showed a more than 9-fold change in expression level and is negative regulator of the GA signalling pathway (Fig. 3).
TFs putatively involved in growth and development
The role of TFs in regulating plant growth and development has already been studied in detail40. We identified thirty-one (17 up- and 14 down-regulated) TFs involved in growth and development that showed differential expression to CySNO (Fig. 3). These include members of the GRAS, NAC, AP2/EREP, and MADS TF families. The role of these TF families in growth and development has been already reported36. The highest-fold change was in At1g52890 (NAC TF), which showed a more than 150-fold change in expression after CySNO. The expression of this gene is induced by drought, high salinity, and ABA41. We also found a SQUAMOSA promoter-binding protein-like (SPL) TF (At1g76580) that is involved in key biological processes like plant phase transition, flower and fruit development, GA signalling, sporogenesis, plant architecture, and response to copper and fungal-released toxins42. Among down-regulated TFs was Origin Recognition Complex subunit 2 (AtORC2), which is involved in initiation of DNA replication during the cell cycle.
TFs related to protein degradation
Swift degradation of particular target proteins by proteasome-dependent pathways is an important part of many cellular regulatory processes/mechanisms. We identified seven (4 up- and 3 down-regulated) TFs putatively involved in protein degradation (Fig. 3). This indicates a regulatory role of NO in protein degradation. Among the up-regulated TFs in Arabidopsis, MATH-BTB domain proteins (AtBPM1) have a direct interaction with targets of proteasomal degradation. Another TF, ADO2 (At2g18915), whose expression was changed by two fold, encodes a member of the F-box protein family (including 2 proteins in Arabidopsis, ZTL and FKF1) that regulates circadian rhythms (Supplementary Table S1). Similarly, among down-regulated TFs, we identified a jumonji TF that has a putative function in Zn binding (Supplementary Table S1). These TFs are thus good candidates for NO-regulated metal biology.
RAP2.6 negatively regulates shoot length while DDF1 negatively regulates root length under nitrosative stress
To identify a possible role of NO-induced TFs in growth and development, the selected knockout mutant lines ddf1, rap2.6, and atmyb48 were screened for their growth performance under oxidative and nitrosative stress induced by 2 mM H2O2 and 1 mM CySNO respectively. Under control conditions, there was no significant difference in cotyledon development frequency (CDF) among all mutant lines compared to wild type (WT) (Fig. 4d); however, under nitrosative stress conditions, all mutant lines showed a significant reduction in CDF compared to WT (Fig. 4d). Similarly, under oxidative stress condition, all mutant lines except ddf1 showed a reduction in CDF (Fig. 4d). Furthermore, under control and oxidative stress conditions, shoot length measurements revealed no significant difference between mutant lines and WT, except atgsnor1-3 (Fig. 4a,b). The ddf1 line showed significantly higher root length compared to WT under control and nitrosative stress conditions (Fig. 4a,c); however, under oxidative stress conditions, only atmyb48 showed a significant reduction in root length compared to wild type (Fig. 4c).
ddf1, rap2.6 and atmyb48 response to oxidative and nitrosative stress. (a) Phenotypes of the indicated genotypes under control, oxidative (H2O2) and nitrosative (CySNO) stress conditions. (b) Shoot length and (c) root length and (d) cotyledon development frequency (CDF) compared to WT control. All the data points represent the mean of three replicates with 8 plants per replicate. The experiment was repeated three times with similar results. Error bars represents standard error (±SE). Significant differences are shown by asterisks (Student’s t-test with 95 and 99% confidence levels).
Nitric oxide responsive TFs regulate ROS-RNS cross-talk
Bulk of literature reports extensive crosstalk between reactive oxygen and reactive nitrogen species in living cells; the ultimate aim of which is to maintain a critical balance between these two redox-pools for smooth functioning. The transcription factor knock out mutant plants rap2.6, ddf1 and myb48 showed an interesting response when grown on oxidative and nitrosative stress media (Fig. 4). When grown on basal medium all the mutant plants showed significantly higher expression of the NADPH oxidase 1 (NOX1) as compared to Col-0 indicating that these mutants would accumulate more ROS. However, as expected the mutant plants also showed significantly higher expression levels of genes encoding the three catalase enzymes (CAT1, CAT2 and CAT3) and the gene encoding the superoxide dismutase enzyme (APX1) on basal media (Fig. 5). The mutant plants also showed a significantly higher expression of NIA1 the enzyme responsible for the production of nitric oxide via the reductive pathway (Fig. 6). This indicates an increase in the production of RNS in order to counterbalance the increasing cellular ROS.
Transcript accumulation of key marker genes involved in oxidative stress. The expression of selected genes involved in regulation of oxidative stress regulation were analyzed through quantitative real-time PCR. Each data point is the mean of three replicates and error bars represent standard error (±SE).
Furthermore, NOX1 expression in rap2.6, and myb48 dramatically increased on MS medium containing H2O2 whereas, it diminished in the ddf1 mutant (Fig. 6). However, on CySNO medium the expression of this gene decreased significantly in rap2.6 and ddf1 whereas, increased in myb48. Similarly, the expression of NIA1 increased significantly in all the mutant plants grown on CySNO medium as compared to those grown on H2O2 and basal medium whereas, NIA2 expression showed a varied response indicating redundancy in the response of these two isoforms of the nitrate reductase enzyme (Fig. 6). Expression of GSNOR1 (which controls global levels of S-Nitrosothiols in plants) was significantly higher in mutant plants grown on H2O2 medium but lower in mutant plants grown on CySNO medium (Fig. 6). Similarly, significantly higher expression levels of APX1 were observed in plants grown on H2O2 medium but lower in plants grown on CySNO medium whereas, APX2 showed a varied response (Fig. 5).
Reactive oxygen and nitrogen species have been known to regulated drought tolerance in plants by regulating ABA biosynthetic and signaling pathways and via ABA mediated stomatal closure. Nitrate reductase-mediated NO generation is required for ABA-induced stomatal closure43. The rap2.6, ddf1 and myb48 plants showed significantly higher basal expression level of ABI2 as compared to WT plants but the expressions of ABA2 were significantly lower than WT plants. However, rap2.6 and myb48 showed higher expression of ABI2 when grown on media containing H2O2 but lower expression on media containing CySNO. Inversely, ddf1 showed lower expression of ABI2 on H2O2 medium but higher expression on CySNO medium (Fig. 6). These results indicate that NO-responsive TFs play an important role in regulating the cross talk between reactive oxygen and nitrogen species.
RAP2.6 and DDF1 positively regulates basal disease resistance
The basal defence system uses pathogen-associated molecular patterns (PAMPs) that serve to limit the growth of virulent pathogens within the susceptible host. To examine if AtMYB48, RAP2.6, and DDF1 are required for basal defence, we inoculated plants with Pst DC3000, which is virulent on the Arabidopsis Col-0 accession. Both rap2.6 and ddf1 showed increased susceptibility as shown by significantly higher pathogen growth compared to WT, while atmyb48 showed resistance towards the virulent Pst DC3000 (Fig. 7a). Thus, mutations in RAP2.6 and DDF1 compromise basal defence response while those in AtMYB48 induce it. These results were further supported by real time PCR analysis of PR1 and PR2 genes. rap2.6 and ddf1 plants showed significantly lower expression of these genes after 12 and 24 hours of inoculation with Pst DC3000 (Fig. 7b). Whereas, myb48 showed significantly higher expression of PR1 and PR2 following Pst DC3000 inoculation (Fig. 7b) indicating that MYB48 negatively regulates basal defence system in Arabidopsis.
AtMYB48 negatively regulates basal defence and effector triggered immunity (ETI) while DDF1 and RAP2.6 positively regulate them. Pathogen growth from leaves pressure infiltrated with (a) virulent Pst DC3000 (b) PR gene expression in response to virulent Pst DC3000 inoculation. (c) Pathogen growth in response to Pst DC3000 (avrB) at 5 × 105 colony forming units (CFU) mL−1 (unless stated otherwise) after 0 and 4 days of inoculation (d) PR gene expression after attempted Pst DC3000 (avrB) inoculation. Error bars represent ± SE (n = 3). Significant differences are represented by asterisks (student t-test with 95% confidence interval).
AtMYB48 negatively regulates effector-triggered immunity
We examined the impact of AtMYB48, RAP2.6 and DDF1 mutations during effector-triggered immunity (ETI; R gene-mediated resistance). The major Arabidopsis R genes encode NBS-LRR (nucleotide binding sites -lucine rich repeats) proteins44. NB-LRR proteins have the capability to recognize pathogens from diverse kingdoms and activate defence responses called ETI. We determined if AtMYB48, RAP2.6 and DDF1 are required for ETI by examining the growth of pathogenic Pst DC3000 expressing avrB within the inoculated leaves. WT plants demonstrated ETI against Pst DC3000 (avrB) due to the presence of the CC-NBS-LRR protein RPM145. The mutant lines rap2.6 and ddf1 showed increased pathogen growth compared to WT (Fig. 7c) with a concomitant lower expression of PR1 and PR2 genes (Fig. 7d). Conversely, atmyb48 exhibited resistance to Pst DC3000 (avrB) compared to WT with significantly higher expression levels of PR1 and PR2 genes within the first 12 hours of inoculation (Fig. 7d). This indicates that AtMYB48 negatively regulates ETI, while RAP2.6 and DDF1 positively regulate it.
qRT-PCR validation
To further validate the CySNO-mediated transcriptional changes in TF genes, we analysed the transcript accumulation of selected representative genes after 6 h of 1 mM CySNO infiltration in Arabidopsis leaves, using qRT-PCR. These genes were involved in different key biological process such as growth and development, abiotic stress, protein degradation, signalling, secondary metabolite production, and plant defence (Fig. 8a–f and Supplementary Fig. S5). Fold change was calculated from qRT-PCR data and correlated with RNA-seq results using Microsoft Excel (version 2016; https://www.office.com/). Our correlation coefficient of 0.951 indicates a high, significant correlation between the RNA-seq and qRT-PCR analyses (Fig. 8a–f).
Validation of RNA-seq data by qRT-PCR analysis. A total of 22 genes involved in different biological processes that showed differential response to CySNO in RNA-seq analysis were selected for confirmation through qRT-PCR. These genes include those involved in (a) growth and development, (b) abiotic stress, (c) protein degradation, (d) signalling, (e) secondary metabolite production, and (f) plant defence. The correlation coefficient (R) of 0.951 indicates that qRT-PCR results are significantly correlated with RNA-seq analysis. Each data point is the mean of three replicates and error bars represents standard error (±SE).
Discussion
Nitric oxide (NO) is a major signalling molecule that regulates a plethora of biological process in both plants and animals. After its initial identification in mammalian systems1, scientists started to hunt for its presence in plants. Since its first report in a plant system, extensive research has been conducted on different aspects of NO biology. Changes in NO-mediated cellular redox status can affect the expression of important genes like peroxidases and catalases, receptor proteins16, protein kinases, and transcription factors like MYB, Trx and HY522. CySNO is a well-known and widely used NO source due to its low molecular weight and high diffusivity into plant tissues compared to other NO donors. Reports suggest that many NO-modulated genes are regulatory proteins or transcription factors (TFs)22. Therefore, studying global changes in the expression of TFs will help clarify the underlying mechanisms involved in the regulation of key biological processes.
A large-scale CySNO-mediated RNA-seq analysis, covering almost 176 million reads, was performed to describe transcriptional changes in response to nitrosative stress. The details of the differentially expressed genes (DEGs) and their analysis has already been reported46. A total of 98.6% (664) of the TFs were successfully mapped to the Arabidopsis genome. The remaining seven TFs may be a potential source of novel TFs in Arabidopsis.
GO analysis indicated TFs putatively involved in biological process regulation such as cell communication, cell cycle regulation, developmental processes, biogenesis, biosynthetic processes, and nitrogen metabolism (Fig. 2a). These results are consistent with a number of published reports claiming the involvement of NO in regulating whole plant growth, primary root development, hypocotyls, shoot, germination and leaf senescence [reviewed in6]. Similarly, GO annotation for Molecular Function indicated TFs putatively involved in two major categories, binding activity (75.40 FE) and catalytic activity (19.60 FE) (Fig. S2B). The large number of TFs classified to the binding category is because of their ability to bind with DNA at specific sequences in the promoter region22. Crosstalk between NO and transcription factor binding sites (TFBSs) has also been studied, and eight families of TFBSs, including WRKY and ocs element-like sequences, were found at least 15% more often in the promoter regions of NO-responsive genes compared to 28447 control genes in Arabidopsis22. TFs classified to catalytic activity confirm the putative involvement of NO-responsive TFs in the regulation of enzyme activity. Changes in the expression level of important enzymes in response to CySNO were also evident in our previous study involving RNA-seq and qRT-PCR analyses. These included disulfide isomerases, mono- and di-hydro ascorbate reductase, glutathione peroxidases, oxido-reductases, catalases (CAT2 and CAT3), carboxy methyltransferase, MAP kinases, MAPK phosphatases etc46.
In this study, we found many important NO-induced transcription factor families, such as bHLH, AP2-EREBP, MYB, WRKY, C2H2, Aux/IAA, bZIP, and DOF (Fig. 2). All these TFs regulate key biological processes like light signalling; abiotic and biotic stress responses; ABA, Auxin, and GA signalling; salt response; pathogenesis; etc. Some of these TFs are already known for their regulatory role under abiotic and/or biotic stress, such as NAC35 MYB, WRKY, C2H2 Zinc-finger reviewed in29 etc. A promoter analysis of NO-responsive genes showed many TF binding sites22. These include WRKY, GBOX, DOFF, HEAT, MADS, MYB, and CAAT, indicating extensive crosstalk between NO and these TFs. The remaining 74 unspecified TFs may be a good source for uncovering the function of these uncharacterised genes.
The production of NO in response to biotic stress like pathogenesis is well studied. This results in changes in cellular redox tone, which is highly toxic for plants. Plants have a finely tuned system to convert highly reactive and toxic NO into a mobile, non-toxic, and easily available form GSNO. There is an intricate balance between NO production and its conversion to GSNO, catalysed by a key enzyme, GSNOR. Mutations in this gene result in compromised basal defence and R-gene mediated resistance26. Our results showed almost 30% of total NO-responsive TFs were related to stress tolerance, among which 95.5% were related to biotic stress (Fig. 3). These include members of the ERF, bZIP, WRKY, MYB and DOF families, among others (Fig. 4, Supplementary Fig. S3). As an example, the bZIP TF is reportedly involved in the regulation of abiotic and biotic stress47. A class of bZIP-binding elements associated with defence is formed by octopoine synthase (OCS). These OCS-like elements are imperative for the expression of specific pathogenesis-related genes such as PR1 in Arabidopsis48. This was further supported by the finding that the Arabidopsis TGA/ocs element-binding factor interacts with NPR1, the main player of the SA-mediated defence signalling pathway49.
Experiments were also conducted to confirm the regulatory role of TFs in growth, development, and plant defence. Our results suggested a significant increase in root length for mutant line ddf1 under control and nitrosative stress conditions compared to WT. Literature reports reveal that overexpression of DDF1 results in a dwarf phenotype24. This suggests that DDF1 negatively regulates plant growth, which is also evident from our results (Fig. 4a,b). Furthermore, atmyb48 had a significant reduction in root length under H2O2-mediated oxidative stress compared to wild type, suggesting a positive regulatory role of AtMYB48 in root development (Fig. 4a,c). This might be because most MYB TFs are involved in abiotic stress tolerance, particularly to oxidative stresses such as drought, salinity, and osmotic stress. Reports suggest that MYB TFs such as AtMYB2, AtMYB74, and AtMYB102 are up-regulated in response to drought and various other abiotic stresses50.
We also investigated role of these TFs in basal defence and ETI by pathogen inoculation with Pst DC3000 (virulent) and Pst DC3000 (avrB) respectively. The mutant lines ddf1 and rap2.6 showed compromised basal defence and ETI and showed significantly higher pathogen growth compared to WT, indicating a positive regulatory role of these genes in basal and ETI (Fig. 7a,c). These results are in accordance with previous results that showed a regulatory role of rap2.6 in response to type III secretion system of virulent and avirulent strains of P. syringae and to nematode pathogens25. Reports suggested that RAP2.6 has been involved in responses to various biotic and abiotic stress conditions, suggesting its role in the regulation of these stresses51.
We also validated our RNA-seq results by selecting representative TF genes involved in key biological processes through qRT-PCR (Fig. 8). The high coefficient value (0.95) indicates high fidelity to the RNA-seq results46. NO-induced TFs may bind to enhancer proteins, enabling the recruitment of RNA polymerase for transcription initiation (Supplementary Fig. S5). Alternatively, TFs may also bind to repressor proteins, blocking RNA polymerase attachment and hence inhibiting transcription. Thus, depending upon the type, TFs may activate or repress transcription52. We identified genes involved in various biological processes with an RNA-seq mediated transcriptome of Arabidopsis leaf in response to 1 mM CySNO and validated them through qRT-PCR (Fig. 8, Supplementary Fig. S5), suggesting that TFs plays key role in the mechanistic control of diverse cellular processes.
Collectively, this information regarding NO-induced transcription factors will help in portraying a clearer picture of NO-mediated transcriptional control of different regulatory genes. The biological data have confirmed significant crosstalk between oxidative and nitrosative stress conditions, in addition to plant defence.
Methods
Plant material, growth and treatment
Seeds of Wild Type (WT) Arabidopsis thaliana (accession Col-0) were obtained from the Nottingham Arabidopsis Stock Centre (NASC) (http://arabidopsis.info/) and grown under long day conditions (16 h of light and 8 h of dark) at 23 ± 2 °C. The plants were infiltrated with 1 mM CySNO on the abaxial side of leaves at rosette leaf stage (4-week-old plants, unless stated otherwise). The 1 mM CySNO concentration is a standard dose for studying the effect of nitrosative stress on the phenotype and the expression of a plethora of genes, of Arabidopsis thaliana. The same dose have been used in different published reports e.g.53,54. The control plants were infiltrated with buffer. Leaf samples were collected after 6 h of infiltration (after screening for optimum effect over time; supplementary Fig. S4).
RNA and cDNA library construction
An RNeasy® Plant Mini Kit (Qiagen) was used to extract RNA from leaf samples (3 replicates) using the standard protocol suggested by manufacturer. RNA quality and integrity were analysed (Agilent® 2100 Bio-analyzer, Agilent) and RNA was treated with DNase 1 to remove gDNA contamination. For sequencing purposes, mRNA was synthesized from 2 µg of total RNA by an already-described method46. A TruSeqTM RNA library prep kit (Illumina) was used to generate RNA libraries. Single-stranded cDNA was synthesized using hexamer priming of mRNA to generate double stranded cDNA libraries, which were then quantified using a KAPA library quantification kit (Illumina) and sequenced through a HiSeq-2500 sequencer (Illumina).
Cufflinks and Cuffdiff analysis for identifying Differentially Expressed Genes (DEGs)
Raw sequence reads (low quality reads with adaptors included) were further processed to identify high quality reads. The threshold level for high quality reads was Q20 > 40%; and the reads less than the threshold level or having more than 10% ambiguous bases were removed. The reads were further processed for high quality using a customized analysis program developed by Theragen ETEX (Korea). The high-quality reads obtained were compared to the reference Arabidopsis thaliana genome using Ensemble.
The reads were then aligned with the reference genome through TopHat55, using default values. After aligning the reads, we inferred the abundance of the transcripts using Cufflinks package v2.2.156. Cufflinks assembles the fragments and calculates the transcript abundance based on the number of reads. The gene and transcript expression levels were calculated for control and treated conditions and were tested for significant differences using Cuffdiff v.2.2.156 to identify differentially expressed genes (DEGs). The genes having significant differential expression (Q < 0.05) were selected. A Heatmap showing the hierarchical clustering of differentially expressed genes was generated using R (https://www.r-project.org/).
Identification, annotation, and Gene Ontology analysis of transcription factors
The selected DEGs were analysed for annotation and GO terms using the NCBI (www.ncbi.nlm.nih.gov) and Gene Ontology Consortium databases (http://geneontology.org/). All the DEGs were then mapped against the Arabidopsis “Ath_AGI_LOCUS_TAIR10_Aug2012.m02” database in MapMan57 to identify transcription factors (TFs). MapMan identifies TFs based on its comparison with the reference genome. The TFs thus obtained were validated manually using the annotated list of all the DEGs and were analysed for any duplicated values using conditional formatting in Microsoft EXCEL. A Heatmap representing the hierarchical clustering of expression differences between treated and control samples using FPKM values was generated using R (version 3.3.1). Dispersion in the data was analysed using a Multi-Dimensional Scatter (MDS) plot using R (version 3.3.1). Further, the identified TFs were analysed for functional classification through the PANTHER classification system using Arabidopsis thaliana as reference genome.
Classification of transcription factors into families and pathways using MapMan analysis
RNA-seq data, usually comprising of several thousand genes and transcripts, is challenging to analyse efficiently. To study the involvement of NO-responsive transcription factors in different pathways and cellular processes, we analysed all TFs with their expression values using MapMan version 3.6.0RC1. MapMan is an omics data analysis software that allows visualization of omics data using a hierarchical BIN-based ontology system46.
To study in detail the involvement of NO-responsive TFs, we mapped all the TFs (Q < 0.05) against the already-mapped Arabidopsis database (TAIR 10 Aug 2012) in MapMan. The analysed pathways and cellular processes were then collectively mapped using their BIN and sub-BIN numbers on custom-made images to present in concise and succinct form.
Oxidative and nitrosative stress assay
To examine the role of TFs in oxidative and nitrosative stress tolerance, we selected three different TFs one each from different categories including those involved in defence (RAP2.6; At1g43160 an ERF TF as majority of defence related TFs were related to ERF family), growth and development (DDF1; At1g12610) and secondary metabolite production (MYB48, At3g46130 involved in flavonol biosynthesis) based on their annotation and fold change (all the selected candidates having more than 25 fold change) for studying downstream functions through functional genomics. The mutant lines ddf1, rap2.6, and atmyb48 were genotyped at the rosette leaf stage in order to identify homozygous lines. Arabidopsis WT and all other lines used in the study were of the Col-0 genetic background. Plants were tested for different growth parameters under nitrosative and oxidative stress conditions. All the seeds were surface sterilized and germinated on half-strength MS medium in triplicate, containing 2 mM H2O2 (for oxidative stress) or 1 mM S-nitrocysteine (CySNO) (for nitrosative stress), as described previously58. Data on cotyledon development frequency (CDF), shoot length, and root length were recorded 2 weeks after treatment started. The term CDF was used to mention the number of developed, green seedlings as in case of nitrosative stress seeds of mutants that lack NO scavenging mechanism (e.g atgsnor 1-3) may germinate but cannot survive.
Pathogenicity assessment
The virulent Pseudomonas syringae pv. tomato strain DC3000 and avirulent Pst DC3000 expressing the avrB effector were grown and maintained, as described previously26. Briefly, the bacterial strains were grown on LB (Luria-Bertani)-agar media with appropriate antibiotics for selection (rifampicin for virulent Pst DC3000 and kanamycin and rifampicin for Pst DC3000 (avrB). Following this, 5 mL of liquid culture was made using a single colony in LB broth and incubated at 28 °C overnight. Both bacterial strains were then harvested by centrifugation at 13,000 rpm for 1 min in 10 mM MgCl2, and syringe-infiltrated into the abaxial side of leaves at rosette leaf stage at a concentration of 5 × 105 CFU mL−1. Control plants were only infiltrated with 10 mM MgCl2. Leaf samples (1 cm each) from plants inoculated either with Pst DC3000 or Pst DC3000 (avrB) were collected at two and four days post inoculation (DPI) and crushed in 1 mL of sterile 10 mM MgCl2. The homogenate was diluted by a factor of 10 and spread on LB-agar plates containing appropriate antibiotics. Plates were incubated at 28 °C for 48 h and colonies were counted.
Validation of RNA-seq data through qRT-PCR analysis
To validate the expression level of TF genes that showed differential expression in response to CySNO in RNA-seq mediated transcriptome analysis, we selected representative genes involved in key processes like growth and development, signalling, defence, protein degradation etc. for quantitative real time PCR (qRT-PCR) analysis. A detailed list of the genes and primers is given in supplementary Table S5. For qRT-PCR analysis, RNA was extracted using Trizol (Ambion, Life Technologies USA). The RNA concentration was measured using nanoQ (OPTIZEN Korea), and 2 µg of total RNA with good integrity and purity was used to synthesize cDNA using a DiaStarTM RT Kit (SolGent, Korea). To measure transcript accumulation, a two-step qRT-PCR reaction was performed using the Illumina (USA) EcoTM real time PCR system using a 2× Quantispeed SYBR Kit (PhileKorea) according to the manufacturer’s recommended protocol.
References
Delledonne, M., Xia, Y., Dixon, R. A. & Lamb, C. Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588 (1998).
Guo, F.-Q., Okamoto, M. & Crawford, N. M. Identification of a Plant Nitric Oxide Synthase Gene Involved in Hormonal Signaling. Science 302, 100–103, https://doi.org/10.1126/science.1086770 (2003).
Klepper, L. Nitric oxide (NO) and nitrogen dioxide (NO2) emissions from herbicide-treated soybean plants. Atmospheric Environment (1967) 13, 537–542, https://doi.org/10.1016/0004-6981(79)90148-3 (1979).
Durner, J. & Klessig, D. F. Nitric oxide as a signal in plants. Current Opinion in Plant Biology 2, 369–374, https://doi.org/10.1016/S1369-5266(99)00007-2 (1999).
Bright, J., Desikan, R., Hancock, J. T., Weir, I. S. & Neill, S. J. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. The Plant Journal 45, 113–122, https://doi.org/10.1111/j.1365-313X.2005.02615.x (2006).
Yu, M., Lamattina, L., Spoel, S. H. & Loake, G. J. Nitric oxide function in plant biology: a redox cue in deconvolution. New Phytol 202, 1142–1156, https://doi.org/10.1111/nph.12739 (2014).
Neill, S. J., Desikan, R. & Hancock, J. T. Nitric Oxide Signalling in Plants. New Phytol 159, 11–35, https://doi.org/10.2307/1514348 (2003).
Abat, J. K. & Deswal, R. Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: Change in S-nitrosylation of Rubisco is responsible for the inactivation of its carboxylase activity. Proteomics 9, 4368–4380, https://doi.org/10.1002/pmic.200800985 (2009).
Zhao, L. et al. Nitric Oxide Functions as a Signal in Salt Resistance in the Calluses from Two Ecotypes of Reed. Plant Physiol 134, 849–857, https://doi.org/10.1104/pp.103.030023 (2004).
Alderton, W. K., Cooper, C. E. & Knowles, R. G. Nitric oxide synthases: structure, function and inhibition. Biochem J 357, 593–615 (2001).
Foresi, N. et al. Characterization of a Nitric Oxide Synthase from the Plant Kingdom: NO Generation from the Green Alga Ostreococcus tauri Is Light Irradiance and Growth Phase Dependent. The Plant Cell 22, 3816–3830, https://doi.org/10.1105/tpc.109.073510 (2010).
Nathan, C. Specificity of a third kind: reactive oxygen and nitrogen intermediates in cell signaling. Journal of Clinical Investigation 111, 769–778, https://doi.org/10.1172/JCI200318174 (2003).
Spadaro, D. et al. The redox switch: dynamic regulation of protein function by cysteine modifications. Physiol Plantarum 138, 360–371, https://doi.org/10.1111/j.1399-3054.2009.01307.x (2010).
Tada, Y. et al. Plant Immunity Requires Conformational Charges of NPR1 via S-Nitrosylation and Thioredoxins. Science (New York, N.Y.) 321, 952–956, https://doi.org/10.1126/science.1156970 (2008).
Wang, Y.-Q. et al. S-Nitrosylation of AtSABP3 Antagonizes the Expression of Plant Immunity. J Biol Chem 284, 2131–2137, https://doi.org/10.1074/jbc.M806782200 (2009).
Terrile, M. C. et al. Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J 70, 492–500, https://doi.org/10.1111/j.1365-313X.2011.04885.x (2012).
Hu, J. L. et al. Site-Specific Nitrosoproteomic Identification of Endogenously S-Nitrosylated Proteins in Arabidopsis. Plant Physiol 167, 1731–1746, https://doi.org/10.1104/pp.15.00026 (2015).
Feechan, A. et al. A central role for S-nitrosothiols in plant disease resistance. Proceedings of the National Academy of Sciences of the United States of America 102, 8054–8059, https://doi.org/10.1073/pnas.0501456102 (2005).
Begara-Morales, J. C. et al. Differential transcriptomic analysis by RNA-Seq of GSNO-responsive genes between Arabidopsis roots and leaves. Plant Cell Physiol, pcu044 (2014).
Parani, M. et al. Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnol J 2, 359–366 (2004).
Eulgem, T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10, 71–78, https://doi.org/10.1016/j.tplants.2004.12.006 (2005).
Palmieri, M. C. et al. Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: a bioinformatics approach. J Exp Bot 59, 177–186, https://doi.org/10.1093/jxb/erm345 (2008).
Hussain, A. et al. Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Front Plant Sci 7, https://doi.org/10.3389/fpls.2016.00975 (2016).
Kang, H. G. et al. Overexpression of FTL1/DDF1, an AP2 transcription factor, enhances tolerance to cold, drought, and heat stresses in Arabidopsis thaliana. Plant Sci 180, 634–641, https://doi.org/10.1016/j.plantsci.2011.01.002 (2011).
Ali, M. A., Abbas, A., Kreil, D. P. & Bohlmann, H. Overexpression of the transcription factor RAP2.6 leads to enhanced callose deposition in syncytia and enhanced resistance against the beet cyst nematode Heterodera schachtii in Arabidopsis roots. Bmc Plant Biol 13, doi:Artn 4710.1186/1471-2229-13-47 (2013).
Yun, B. W. et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–U161, https://doi.org/10.1038/nature10427 (2011).
Ledent, V. & Vervoort, M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res 11, 754–770, https://doi.org/10.1101/gr.177001 (2001).
Riechmann, J. L. & Meyerowitz, E. M. The AP2/EREBP family of plant transcription factors. Biol Chem 379, 633–646 (1998).
Ambawat, S., Sharma, P., Yadav, N. R. & Yadav, R. C. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Pla 19, 307–321, https://doi.org/10.1007/s12298-013-0179-1 (2013).
Liu, Q., Wang, Z., Xu, X., Zhang, H. & Li, C. Genome-Wide Analysis of C2H2 Zinc-Finger Family Transcription Factors and Their Responses to Abiotic Stresses in Poplar (Populus trichocarpa). Plos One 10, e0134753, https://doi.org/10.1371/journal.pone.0134753 (2015).
Jakoby, M. et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci 7, 106–111, https://doi.org/10.1016/S1360-1385(01)02223-3 (2002).
Borello, U., Ceccarelli, E. & Giuliano, G. Constitutive, light-responsive and circadian clock-responsive factors compete for the different l box elements in plant light-regulated promoters. Plant J 4, 611–619 (1993).
Mark, M., Rijli, F. M. & Chambon, P. Homeobox genes in embryogenesis and pathogenesis. Pediatr Res 42, 421–429, https://doi.org/10.1203/00006450-199710000-00001 (1997).
Ng, D. W. K. et al. Plant SET domain-containing proteins: Structure, function and regulation. Bba-Gene Struct Expr 1769, 316–329, https://doi.org/10.1016/j.bbaexp.2007.04.003 (2007).
Nuruzzaman, M., Sharoni, A. M. & Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol 4, https://doi.org/10.3389/fmicb.2013.00248 (2013).
Parenicova, L. et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15, 1538–1551, https://doi.org/10.1105/tpc.011544 (2003).
Kaplan-Levy, R. N., Brewer, P. B., Quon, T. & Smyth, D. R. The trihelix family of transcription factors - light, stress and development. Trends Plant Sci 17, 163–171, https://doi.org/10.1016/j.tplants.2011.12.002 (2012).
Hirsch, S. & Oldroyd, G. E. D. GRAS-domain transcription factors that regulate plant development. Plant Signaling & Behavior 4, 698–700 (2009).
Monte, E., Amador, V., Russo, E., Martinez-Garcia, J. & Prat, S. PHOR1: A U-Box GA signaling component with a role in proteasome degradation? J Plant Growth Regul 22, 152–162, https://doi.org/10.1007/s00344-003-0029-4 (2003).
Ramachandran, S., Hiratsuka, K. & Chua, N. H. Transcription factors in plant growth and development. Curr Opin Genet Dev 4, 642–646 (1994).
Takasaki, H. et al. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J 84, 1114–1123, https://doi.org/10.1111/tpj.13067 (2015).
Chen, X. et al. SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J Integr Plant Biol 52, 946–951, https://doi.org/10.1111/j.1744-7909.2010.00987.x (2010).
Desikan, R., Griffiths, R., Hancock, J. & Neill, S. A new role for an old enzyme: Nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 99, 16314–16318, https://doi.org/10.1073/pnas.252461999 (2002).
Jones, J. D. G. & Dangl, J. L. The plant immune system. Nature 444, 323–329, https://doi.org/10.1038/nature05286 (2006).
Grant, S. R., Fisher, E. J., Chang, J. H., Mole, B. M. & Dangl, J. L. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol 60, 425–449, https://doi.org/10.1146/annurev.micro.60.080805.142251 (2006).
Hussain, A. et al. Nitric Oxide Mediated Transcriptome Profiling Reveals Activation of Multiple Regulatory Pathways in Arabidopsis thaliana. Front Plant Sci 7, https://doi.org/10.3389/fpls.2016.00975 (2016).
Hobo, T., Kowyama, Y. & Hattori, T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. P Natl Acad Sci USA 96, 15348–15353, https://doi.org/10.1073/pnas.96.26.15348 (1999).
Lebel, E. et al. Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J 16, 223–233, https://doi.org/10.1046/j.1365-13x.1998.00288.x (1998).
Zhou, J. M. et al. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant Microbe In 13, 191–202, https://doi.org/10.1094/Mpmi.2000.13.2.191 (2000).
Denekamp, M. & Smeekens, S. C. Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol 132, 1415–1423, https://doi.org/10.1104/pp.102.019273 (2003).
Chen, W. et al. Expression Profile Matrix of Arabidopsis Transcription Factor Genes Suggests Their Putative Functions in Response to Environmental Stresses. The Plant Cell Online 14, 559–574, https://doi.org/10.1105/tpc.010410 (2002).
Lee, T. I. & Young, R. A. Transcription of eukaryotic protein-coding genes. Annu Rev Genet 34, 77–137, https://doi.org/10.1146/annurev.genet.34.1.77 (2000).
Martinez-Ruiz, A. & Lamas, S. Detection and proteomic identification of S-nitrosylated proteins in endothelial cells. Archives of Biochemistry and Biophysics 423, 192–199, https://doi.org/10.1016/j.abb.2003.12.006 (2004).
Lam, Y. W. et al. Comprehensive identification and modified-site mapping of S-nitrosylated targets in prostate epithelial cells. Plos One 5, e9075, https://doi.org/10.1371/journal.pone.0009075 (2010).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111, https://doi.org/10.1093/bioinformatics/btp120 (2009).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–U174, https://doi.org/10.1038/nbt.1621 (2010).
Thimm, O. et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37, 914–939, https://doi.org/10.1111/j.1365-313X.2004.02016.x (2004).
Imran, Q. et al. Nitric oxide responsive heavy metal-associated gene AtHMAD1 contributes to development and disease resistance in Arabidopsis thaliana. Front Plant Sci 7, https://doi.org/10.3389/fpls.2016.01712 (2016).
Acknowledgements
We would like to thank Mr. Rolly Nkulu Kabange for providing help in creating some illustrations used in custom-made images. This work was supported by a grant from the Next-Generation BioGreen 21 Program (SSAC, Grant No. PJ01110201), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Contributions
Q.M.I., A.H., and B.-W.Y. designed the study, Q.M.I. and N.F. performed experiments, S.U.L. and B.-G.M., analyzed the data, Q.M.I., A.H., wrote the manuscript, Q.M.I. designed and made the illustrations. G.J.L. and B.-W.Y. supervised the study and critically reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Imran, Q.M., Hussain, A., Lee, SU. et al. Transcriptome profile of NO-induced Arabidopsis transcription factor genes suggests their putative regulatory role in multiple biological processes. Sci Rep 8, 771 (2018). https://doi.org/10.1038/s41598-017-18850-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-18850-5
This article is cited by
-
Revisiting the Critical Role of ROS and RNS in Plant Defense
Journal of Plant Growth Regulation (2023)
-
Comprehensive analysis of the MYB transcription factor gene family in Morus alba
BMC Plant Biology (2022)
-
Nitric Oxide Improved Growth and Yield in Soybean (Glycine max) by Mediating Physiological, Anatomical, and Transcriptional Modifications
Journal of Plant Growth Regulation (2022)
-
SWATH-MS based quantitive proteomics reveal regulatory metabolism and networks of androdioecy breeding system in Osmanthus fragrans
BMC Plant Biology (2021)
-
A prognostic model based on seven immune-related genes predicts the overall survival of patients with hepatocellular carcinoma
BioData Mining (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.