Abstract
The effects of intensive nitrogen (N) fertilizations on spatial distributions of soil microbes in bioenergy croplands remain unknown. To quantify N fertilization effect on spatial heterogeneity of soil microbial biomass carbon (MBC) and N (MBN), we sampled top mineral horizon soils (0-15 cm) using a spatially explicit design within two 15-m2 plots under three fertilization treatments in two bioenergy croplands in a three-year long fertilization experiment in Middle Tennessee, USA. The three fertilization treatments were no N input (NN), low N input (LN: 84 kg N ha−1 in urea) and high N input (HN: 168 kg N ha−1 in urea). The two crops were switchgrass (SG: Panicum virgatum L.) and gamagrass (GG: Tripsacum dactyloides L.). Results showed that N fertilizations little altered central tendencies of microbial variables but relative to LN, HN significantly increased MBC and MBC:MBN (GG only). HN possessed the greatest within-plot variances except for MBN (GG only). Spatial patterns were generally evident under HN and LN plots and much less so under NN plots. Substantially contrasting spatial variations were also identified between croplands (GG > SG) and among variables (MBN, MBC:MBN > MBC). This study demonstrated that spatial heterogeneity is elevated in microbial biomass of fertilized soils likely by uneven fertilizer application in bioenergy crops.
Similar content being viewed by others
Introduction
Bioenergy crops are important alternative technology for sustainable replacement of fossil fuels1,2. A significant portion (over 30%) of biofuel plant biomass will come from dedicated energy crops such as the perennial switchgrass (Panicum virgatum) and gamagrass (Tripsacum dactyloides L)3,4. Nitrogen (N) fertilizers are widely used to increase yield of various bioenergy crops5,6,7,8,9. In bioenergy crop research field, soil microbial biomass is receiving increasing attentions due to its role in soil fertility and crop yield. Across various soil and plant types, intensive N fertilizations significantly alter soil microbial biomass and activities10,11. However, effects of fertilizations on spatial distributions of soil microbial biomass in bioenergy croplands remain unknown. Understanding effects of fertilization on soil microbial functionality, including spatial structures in various bioenergy croplands may enhance our ability to manipulate nutrient cycling in situ to maintain and improve soil quality, production sustainability and to adapt to climate change.
Intensive inorganic fertilizer inputs substantially restructure spatial heterogeneity of soil biogeochemical and microbial features at a variety of spatial scales12,13,14,15. Long-term cultivation with chemical N fertilizer amendments resulted in weak to moderate spatial heterogeneity of soil total nitrogen and phosphorus in 0–20 cm in the field plot to watershed scales16,17. Effects of mineral fertilizer inputs and the consequent spatial heterogeneity in soil pH exerted key controls on microbial biomass carbon and nitrogen contents as well as their spatial distributions in a long-term field trial of organic agriculture15. Despite lacking such information, indirect evidence supported strong correlation between spatial patterns of soil denitrifiers community with nitrate and other nutrients at scales relevant to land management18. Nevertheless, the altered spatial variation of soil microbial properties and structures is likely to affect the local distribution and abundance of plant species and the performance of individual plants and microorganisms19 and, therefore, to have consequences for both community structure and ecosystem-level processes20,21,22,23.
Although agricultural soils are generally more homogeneous than forests14, they show a substantial level of spatial variability with respect to soil biochemistry. Soil microbial biomass carbon (MBC) and nitrogen (MBN) exhibited moderate spatial dependence24,25,26. Röver and Kaiser25 reported that coefficients of variation of MBC and MBN can reach up to 44%. The spatial distribution of soil MBN demonstrated more hotspots than MBC, implying that MBN might be more sensitive to environmental disturbances24. Spatial structures and variations of bacterial community were found at surface and subsoil horizons at the microscale, and at the centimeter to meter scale27. At the ecosystem scale (>10 m), bacterial community composition and structure were subtly, but significantly, altered by fertilization, with higher alpha diversity in fertilized plots28.
Natural heterogeneity of microbial biomass and activity could be altered likely through soil chemical changes (e.g. pH) caused by fertilizer input15. Spatial patterns and the scale of soil variability differ markedly among edaphically similar sites and these differences are also conditioned likely by intensity and duration of fertilizations13,14,29. On the other hand, bioenergy crop species not only influence biomass yield30,31 but also spatial variations25. Plant trees play a major influence on spatial structures of soil microbial communities32,33,34. Among switchgrass and gamagrass, the latter possessed more significant root biomass and volume35 thus likely favoring nutrients scavenging and microbial activities and contributing to long-term spatial heterogeneity of soil microbial biomass36,37,38,39.
Taken together, previous results suggest that soil microbial features such as MBC and MBN can greatly respond to fertilization in their spatial variability compared with soils without fertilizer input for years in bioenergy croplands. The objective of this study is to investigate effects of N fertilization on spatial distribution of soil MBC, MBN and MBC:MBN in two bioenergy croplands (SG and GG) in a three-year long field experiment at Tennessee State University’s campus farm in Nashville TN, representing a typical bioenergy crop site in Middle Tennessee. Under no tillage or plowing and minor mechanical disturbance, N fertilizer input marked a major management practice in these research plots. We hypothesize that relative to soils that have never been fertilized for years, long-continued N fertilization re-structures spatial patterns of soil MBC, MBN and MBC:MBN at both croplands. The extent of altered spatial heterogeneity varies between variables (MBN > MBC) and crop types (GG > SG). We also explored whether there is significant correlation between soil pH and microbial variables. This study is expected to clarify the fertilization effect on redevelopment of spatial heterogeneity of key soil microbial features in typical bioenergy croplands.
Material and Methods
Site Description and Characteristics
This study was conducted at the Tennessee State University (TSU) Main Campus Agriculture Research and Extension Center (AREC) in Nashville, TN, USA (Lat. 36.12° N, Long. 36.98° W, elevation 127.6 m). Soil at this location is Armour silt loam soil (fine-silty, mixed, thermic Ultic Hapludalfs) with soil pH of 5.97 and organic matter content of 2.4% on average40. A field experiment was established with two crop types and three nitrogen (N) fertilization treatments in 2011 in a randomized block design41. The two crop types include no till cultivation of ‘Highlander’ variety of eastern ‘Alamo’ switchgrass (Panicum virgatum L.) and gamagrass (Tripsacum dactyloides L.). The switchgrass and gammagrass were abbreviated as SG and GG hereafter. The three N fertilization treatments include no N fertilizer input (NN), low N fertilizer input (LN: 84 kg N ha−1 in urea) high N fertilizer input (HN: 168 kg N ha−1 in urea). Each plot has a dimension of 3-m × 6-m and each treatment has four replicate plots.
Soil collections and laboratory analysis
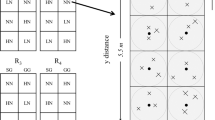
On June 6th 2015, soil cores were collected from 0 to 15 cm depth using soil auger (Thermo Fisher Scientific, Waltham, Massachusetts, USA) from 12 plots (2 crop × 3 N × 2 replicates). That is, two of the four replicated plots were selected in the current study. Within each plot, we identified a sampling area of 2.75-m × 5.5-m rectangle and the southwestern corner point was identified as the origin. Each plot was divided into two-square subplots and within each subplot, four centroids were identified and three cores were collected randomly given random direction and distance relative to each centroid (Fig. 1). When a soil core was collected, we recorded its location in reference to the origin taken as the southwestern corner, i.e. each sampling point had a unique x, y coordinates. Twenty-four cores were collected from each plot yielding 288 soil cores in 12 plots. All soil samples were transported to TSU lab in cooler filled with ice packs and subsequently stored at 4 °C until microbial analysis.
Illustration of a clustered random sampling design within a plot in the three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA. Filled circles represent centroids (n = 8) and each plot consists of eight centroids with one in each sampling region (1.375 × 1.375 m). Xs represent sample locations determined from random directions and distances from a centroid. The extent of an interpolation map was thus determined by the minimum and maximum values at horizontal and vertical axes, and each map can attain its extent less than or equivalent to the study area (2.75 × 5.5 m rectangle).
The visible roots and rocks were removed from soil cores by passing through a 2-mm soil sieve prior to microbial and chemical analysis. For each individual sample, soil gravimetric moisture content was determined by oven drying subsamples for 24 hours at 105 °C. Water extractable soil pH was measured given soil: water = 1:542. The results were summarized in Table S1. Microbial biomass carbon and nitrogen were quantified as described in the following session. Besides, a composited subsample was produced by combining six soil samples of equivalent dry weight for each treatment. The air-dried subsamples were ground to a fine powder and sent to University of North Carolina at Wilmington Center for Marine Science for analysis of soil organic carbon content (SOC) and nitrogen content (TN).
Fresh soil samples (1.0 g) were used to estimate microbial biomass carbon (MBC) and microbial biomass nitrogen (MBN) in each core by chloroform fumigation-K2SO4 extraction and potassium persulfate digestion methods43,44. Briefly, 0.5 M K2SO4 was used to extract soil dissolved organic carbon and nitrogen from fumigated and unfumigated soil samples. Soil extracts were digested with 0.5 M K2S2O4 in oven at 85 °C for 20 hours. The K2SO4-extractable C and N in fumigated and unfumigated samples were determined by Shimadzu TOC-L & TNM-L (Shimadzu Corporation, Kyoto, Japan). MBC or MBN was calculated as the difference in K2SO4-extractable C or N concentration between fumigated and unfumigated soils, divided by 0.45 for C and 0.54 for N, respectively42,45. To minimize the variation likely induced due to unevenly soil mixing, laboratory tests were conducted and specific protocols were created to secure sufficient soil mixing. The variation of each measurement (i.e. coefficient variation) in multiple tests ranged from 2~8% based on our protocol.
Statistical and geospatial analysis
Means, standard errors, and variances were estimated for MBC, MBN and their ratios in each plot. Frequency distributions were produced for each soil variable in each vegetation type after pooling all values of two replicated plots in the three nitrogen treatments. The two-way ANOVA was used to test whether means and coefficient of variance (CV) in MBC, MBN and their ratios differed significantly between fertilization treatments and crop species. To precede the ANOVA, the original data was log transformed if it violated equal variance assumption. The significance level is set at P < 0.05 and the analysis was conducted using R project46.
The Pearson moment correlation coefficients were derived between soil pH, MBC, MBN, and MBC:MBN. Cochran’s C test is used to test the assumption of variance homogeneity. The test statistic is a ratio that relates the largest empirical variance of a particular treatment to the sum of the variances of the remaining treatments. The theoretical distribution with the corresponding critical values can be specified47,48,49. Soil properties that exhibited non-normal distributions were log-transformed to better conform to the normality assumption of the Cochran’s C test14.
The study also derived the sample size requirement (N) in each plot given specified relative errors (γ, 0~100%) in order to evaluate how within-plot variances (i.e. sample size requirements) are altered by N fertilization or crop types at certain relative error.
where CI, \(\bar{X}\), s, n, N, CV and γ denote confidence interval, plot mean, plot standard deviation, sample number (n=24), coefficient of variation, sample size requirement and relative error, respectively. t0.975 = 1.96. The log transformed sample size requirement (N) has a negative linear relationship (i.e. slope = 2) with the log transformed relative error (γ).
In addition to the within-plot variance and derived statistics such as coefficient of variation and sample size requirement, the following geostatistical tools were used to quantify the spatial structure of soil microbial properties within and among plots. The methods were briefly described below and more details can be found in Li et al. (2010).
First, the trend surface analysis (TSA) is the most common regionalized model in which all sample points fit a model that accounts for the linear and non-linear variation of an attribute. The relationships between the soil properties and the x and y coordinates of their measurement location within the sampling plots are estimated with the trend surface model:
The presence of a trend in the data was determined by the significance of any of the parameters β1 to β5, while the β0 term modeled the intercept50,51. Linear gradients in the x or y directions were indicated by significance of the β1 or β2 parameters. A significant β3 term indicated a significant diagonal trend across a plot. Significant β4 and β5 parameters indicated more complex, nonlinear spatial structure such as substantial humps or depressions. Trend surface regressions were estimated using R program46. Model parameters were determined to be significant at a level of P < 0.05.
Second, residuals from the trend surface regressions were saved for subsequent spatial analysis using a Moran’s I index51. The Moran’s I analysis52,53,54 was used to quantify the degree of spatial autocorrelation that existed among all soil cores taken from each plot. The resulting local Moran’s I statistics are in the range from −1 to 1. Positive Moran’s I values indicate similar values (either high or low) are spatially clustered. Negative Moran’s I values indicate neighboring values are dissimilar. Moran’s I values of 0 indicate no spatial autocorrelation, or spatial randomness. A significant autocorrelation is determined if the observed Moran’s I value is beyond the projected 95% confidence interval at certain distance. Correlograms for local Moran indices were estimated for each soil variable in each plot in a range of 0–5.5 meter with 0.25 meter incremental interval.
Third, due to relatively small sample sizes (n = 24) per plot55, we used inverse distance weighting (IDW) interpolation rather than ordinary kriging56. The maps produced by IDW offered direct and visual assessments from which to compare the spatial distributions of the soil properties among the plots. The IDW method derived maps was able to distinguish effects of different land uses on spatial distributions of soil biogeochemical features in South Carolina, USA14. The weights for each observation are inversely proportional to a power of its distance from the location being estimated. Exponents between 1 and 3 are typically used for IDW, with 2 being the most common57. Tests with different IDW exponents indicated that 2 was optimal with these data, as estimated values generated with an exponent of 2.0 showed the best fit with actual data in cross validation tests. ArcGIS 9.0 (ESRI, USA) was used to generate the IDW maps and perform cross validations.
Results
Central tendencies of soil biomass and pH under different treatments
Mean MBC, MBN and MBC:MBN in NN treatment were not significantly different from that in either LN or HN for both SG and GG (Table 1). Mean MBC and MBC:MBN in HN treatment were significantly larger than that in LN treatment, but MBN was not significantly different between LN and HN treatments. These patterns were also reflected by the high frequency of larger MBC values and similar frequency of MBN among different N treatments (Fig. 2). The distribution of MBC:MBN showed higher frequency in lower values in general, but the frequency of higher values were particularly large for HN than LN for GG (Fig. 2). The average water extractable soil pH in different plots ranged between 5.91 and 6.12 (see Supplemental dataset), and showed neither significant differences between treatments nor significant correlations with any of microbial variables (Table S2).
Frequency histograms of soil MBC, MBN concentrations and MBC:MBN under three fertilization treatments (i.e. NN, LN and HN) in SG (panels a~c) and GG (panels d~f) croplands in a three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA. The number on the x-axis (i.e. 0.03, 0.08 in panel a) represents a range of (0, 0.03) and (0.03, 0.08), respectively. The abbreviations are referred to Table 1.
Within-plot variance, within-plot CV and sample size requirement
The largest within-plot variances appeared consistently in one of the HN plots for all three variables in two croplands except for MBN in NN plot in GG (Table 2). Within-plot variances of soil MBC and MBN varied largely among different plots in SG but showed consistently high in GG. Cochran’s C test showed significantly different within-plot variances of MBC and MBN in SG but not in GG (Table 2).
The within-plot CVs ranged from 15~48% for all variables and both croplands (Fig. 3). There is no significant difference of CV between two croplands (P > 0.05). The CVs under NN was significantly lower than that under LN or HN or both for three variables only in SG (P < 0.05). The CVs also differed significantly between LN and HN in GG (Fig. 3). The ranges of CVs for MBC were narrower than that for MBN and MBC:MBN. The largest CVs appeared to be in one of plots in HN treatment in SG, but none in GG (Fig. 3). As for the number of plots in twelve that produced CVs of more than 30% for MBN, MBC:MBN and MBC, respectively, they were six, six and one in SG, and five, four and none in GG. When the threshold of CV set at 40%, the numbers are two, one and none in SG, and three, one and none in GG.
Within-plot CVs of MBC (mgC/gsoil), MBN (mgN/gsoil) concentrations, and MBC:MBN under three N fertilization (i.e. NN, LN and HN) in (a) SG and (b) GG cropland soils in a three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA. The dashed lines represent a CV of 30% and 40%. The abbreviations are referred to Table 1. Different lowercase letters denote significant difference in CV between fertilization treatments for each variable at P < 0.05.
The plotted lines of sample size requirements were more widely separated for MBC than for MBN and MBC:MBN in both croplands (Fig. 4). Given the same relative desired error, much less sample size were required for MBC than for MBN and MBC:MBN in both SG and GG. The largest sample size requirements for all variables appeared to be at a HN plot in SG (Fig. 4a~c), but it is not true in GG (Fig. 4d~f). With such variability for soil MBN, even five samples per plot still will produce a relative error of the mean greater than ±30% (Fig. 4b,e), a sobering result given the level of interest in precise estimates of microbial dynamics. We think it not well appreciated that in either unfertilized or fertilized soils of gamagrass, the relative error of the estimate for MBN to be expected is >±50% if three samples are taken to estimate the mean (Fig. 4).
The relationship between log transformed sample size requirements and desired relative errors. Panels a~f denote the linear regression lines for soil MBC, MBN concentrations and MBC:MBN under three N fertilization treatments (i.e. NN, LN and HN) in SG and GG cropland soils in a three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA. The log scale was applied on both axes. The abbreviations are referred to Table 1.
Spatial heterogeneity under nitrogen fertilization and different crop species
Results of the trend surface analyses (Table 3) indicated that significant linear trends (i.e. diagonal direction) were identified in HN plots only (i.e. MBC at both SG and GG). More linear and nonlinear trends were identified in both LN and HN plots than that in NN plots, for MBC:MBN (SG and GG) and for MBN (GG only). The linear and nonlinear trends were identified for MBN in NN plot in SG only.
Correlograms showed more significant autocorrelations of three variables in LN and HN than NN in either plot or treatment level, only except the equal number of significant autocorrelations of MBC in plots of NN and LN in SG (Fig. 5). For MBC, significant autocorrelations were present in each plot of LN treatment in GG (Fig. 5) and the lagging distances were either positive or negative ranging from 0.5 m to 2.0 m (Table 4). For MBN, only one significant autocorrelation was identified in one plot of NN in SG and none were present in NN plots in GG, whereas, such significant autocorrelations were present much more frequently in one plot of LN in GG (Table 4; Fig. 6). The lagging distances were either positive or negative for these significant autocorrelations and ranged from 0.75 m to 3.75 m (Table 4). For MBC:MBN, no significant autocorrelations were identified in one plot of NN in both croplands (Fig. 7) and one to three significant autocorrelations were present for other plots (Table 4).
Moran’s I correlograms for soil MBC concentration under three N fertilization treatments (i.e. NN, LN and HN) in SG and GG cropland soils in a three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA. Filled circles denote Moran’s I values that exhibited significant positive or negative autocorrelation. Obs: observations; LCL: low confident limit; and UCL: Upper confidence limit. Other abbreviations are referred to Table 1.
Moran’s I correlograms for soil MBN concentration under three N fertilization treatments (i.e. NN, LN and HN) in SG and GG cropland soils in a three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA. Filled circles denote Moran’s I values that exhibited significant positive or negative autocorrelation. Obs: observations; LCL: low confident limit; and UCL: Upper conficence limit. Other abbreviations are referred to Table 1.
Moran’s I correlograms for soil MBC:MBN under three N fertilization treatments (i.e. NN, LN and HN) in SG and GG cropland soils in a three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA. Filled circles denote Moran’s I values that exhibited significant positive or negative autocorrelation. Obs: observations; LCL: low confident limit; and UCL: Upper confidence limit. Other abbreviations are referred to Table 1.
The IDW maps of within-plot patterns of MBC in LN or HN treatments, exhibited rather higher heterogeneity than those in NN treatments in switchgrass or gamagrass soils (Figs 8 and 9). There appeared to have greater within-plot heterogeneity of MBN in LN or HN treatments in SG cropland (Fig. 8), but less so in GG cropland (Fig. 9). It turned out to possess more hotspots of MBC:MBN than that of MBC and MBN across N treatments and croplands (Figs 8 and 9). Last, within-plot heterogeneity of all variables tended to be greater in GG cropland than that in SG cropland.
Spatial distribution of soil MBC and MBN concentrations and MBC:MBN under three N fertilization treatments (i.e. NN, LN and HN) in SG cropland soil in a three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA. The interpolation maps were produced by inverse distance weighting (IDW) method. The abbreviations are referred to Table 1.
Spatial distribution of soil MBC, MBN concentrations and MBC:MBN under three N fertilization treatments (i.e. NN, LN and HN) in GG cropland soil in a three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA. The interpolation maps were produced by inverse distance weighting (IDW) method. The abbreviations are referred to Table 1.
Discussion
N fertilization elevated spatial heterogeneity of soil microbial biomass
Similar to other important agricultural practices (i.e. plowing, mechanical disturbance), N fertilization is generally regarded to potentially homogenize the spatial distribution of soil chemical features in long-term cultivated lands particularly when compared with forests14. Despite very few studies examining effects of N fertilization on soil microbial features27,32,58, it is presumable that the high responsiveness of soil microbial features with disturbance59,60 could potentially override the general prediction of homogenization under N fertilizations. Based on a three-year fertilization experiment in two bioenergy croplands, this study revealed moderate spatial heterogeneity of soil microbial biomass C and N, and indeed, their spatial variations were generally elevated with low or high amount of N fertilizer input. In spite of great plot-plot variations within each fertilization treatment, different geostatistical approaches generally supported more linear and nonlinear surface trends and fine-scale spatial heterogeneity in the fertilized soils.
Possible explanations for the elevated spatial heterogeneity with N fertilizations may lie in the less complexity of management practice in the bioenergy croplands and the essentially high responsiveness of microbial properties in soils. First, the bioenergy croplands were subjected to continuous fertilization management over years but no plowing and less so in mechanical disturbance61,62. This unique management practice in the bioenergy croplands is distinct from the common practices implemented in conventional croplands such as wheat, corn and rice in many regions of world. Under common practices, the year- to decade-long plowing plus mechanical movement acted as the most significant driver that can physically and thoroughly mix and blend soil resources leading to homogenization63. Due to the cumulative effects induced by common practices other than N fertilization, spatial heterogeneity at various scales may be largely masked and turn to be subtle over long-term time period. Second, soil microbial communities and activities are highly responsive to soil nutrient availability such as nitrate and ammonium10,64, while N fertilizer inputs supply readily available nutrients and exert immediate influence on microbes over months to years65,66. This instant effect was also found evident at soil macroaggregate scale67. Therefore, the manual spread of N fertilizers in the field will likely lead to irregularity of nutrient deposit and clusters, and consequently favor the formation of hotspots in soil microbial communities68.
However, a more evident linear surface trend (i.e. diagonal direction) of MBN was identified in soils with no fertilizer input than fertilized soils, which likely attributed to the low sensitivity of this approach applied in the field plots at a scale of meter or broader69,70. Evidence showed the spatial autocorrelation of microbial properties in soil was well described at a scale of centimeter69, two magnitude of scale lower than meter. Consistent with the scale of centimeter, the study identified a unprecedentedly large number of significant autocorrelations, i.e. elevated fine-scale heterogeneity with fertilization in the same scenario. For instance, there are five significant Moran’s I values in one GG plot (i.e. P2 in LN) with the lagging distance ranging from 0.75 to 3.75 meter.
Altered spatial heterogeneity with fertilization varied with crop types
Relative to SG, GG showed greater spatial variations in all three microbial variables such as more detectable linear and nonlinear surface trends, significant autocorrelations and hotspots. This is particularly true revealed by IDW maps that showed widespread appearance of hotspots in both MBC and MBN in GG, which is less evident in SG. The different spatial variations between the two crop species may be largely attributed to the root systems of two plants with contrasting characteristics. Roots are a key plant organs involved in resource competition and stand establishment70. Both GG than SG are warm-season grasses with a thicker and deeper rooting system. Their extensive root channels increased macropore flow in soil and consequently higher water infiltration rates71. They also exhibited enormous tolerance to low soil pH, aluminum toxic condition and high soil strength35,72,73,74. It is well known that plant root and microbes interact closely as mutualist71. The features of root morphology and physiology rendered plants capable of extending their access to large volume of soils for water, nutrients and resources thus favoring for soil microbial growth71. However, the strong effect of rhizosphere on relocating microbial niche may also differ between SG and GG because, though rarely compared quantitatively directly between the two plants, evidence pointed to the role of fine roots and its functions for switchgrass75 which is distinct from the more frequently reported much larger coarse roots and functions for gamagrass71. Though both SG and GG plantations resulted in strong clustering effects on spatial distribution of soil microbial communities, the larger size of roots for GG may play a key role in restructuring the larger patches of soil microbial biomass in soils, which is evident in GG than in SG.
Altered spatial heterogeneity with fertilization varied among microbial variables
Across three fertilization treatments and two croplands, MBC showed relatively narrower within-plot variance and spatial heterogeneity than MBN or MBC:MBN. Also, MBC:MBN mimicked the spatial dependence of MBN, rather than MBC. In addition, the extent of elevated spatial heterogeneity due to fertilization was more pronounced for MBN or MBC:MBN than for MBC. These results collectively corroborated that MBN was a highly responsive variable in spatial dependence as compared with MBC revealed in several previous studies24,25,26. Due to nutrient poor conditions in these bioenergy croplands, the competitions for nutrients (e.g. N) between plant roots and microbes may be more intense given the widespread N limitations in terrestrial ecosystems76,77. On the other hand, the microbes with varying stoichiometry (i.e. C:N:P), that is, intrinsic N demand for growth and enzymatic kinetics78,79, may regulate their movement, colonization and growth given the soil indigenous N availability. This spatial assemblage of these microorganisms may be further complicated when N fertilizer granule manually applied in the field altered the distribution, diffusion and accessibility of readily available N forms, leading to more scattered hotspots of N island and more varied conditions of N hunger or limitation80. It is also likely that a more varied microbial biomass C:N may be driven by both altered physiological and compositional features in soil microbial community under N fertilizations81.
Multiple drivers in restructuring spatial heterogeneity of microbial biomass
Despite the key role of soil pH on microbial growth and activity82 and bacterial community structure and diversity across environmental gradients83,84, no significant correlation was identified between soil pH and microbial biomass in our study (p-value > 0.05). The relatively uniform soil pH (5.7~6.3) across our study plots suggested the structured spatial heterogeneity of microbial biomass were likely driven by other edaphic factors (e.g. water availability), and management practices (e.g. fertilization means). Given the soil sampling conducted in dry summer season and a range of 15~18% gravimetric water content across plots, it is believed that the fertilization itself may overweigh the influence of other factors. In fact, in contrast to conventional cropland management with intensive plowing and mechanical disturbance, our research plots have not been plowed since it was established. Furthermore, fertilizers have been manually applied to soil by different people in our study plots. In addition, the relatively more pronounced spatial heterogeneity in GG than in SG indicates the influence of crop species on the spatial variations of soil microbial biomass. Our preliminary tests showed strong correlations and consistent spatial patterns between soil organic C (SOC), MBC and MBN (p-value < 0.05) in a few of our study plots. It suggests the possibly key control of SOC on MBC and MBN and highlights the keen need in the future to examine their relationships between two crops and under different N fertilization treatments. Therefore, a suite of interrelated edaphic, biochemical and management factors acted as major drivers during the redevelopment of spatial heterogeneity in fertilized soils of bioenergy croplands.
Conclusions
Our study demonstrates that nitrogen fertilizer and the types of bioenergy crop type not only altered soil microbial properties’ central tendencies but also their spatial heterogeneities. In general, N fertilizations elevated the spatial heterogeneity of soil microbial biomass C and N as well as their ratio in both bioenergy croplands. Lacking the commonly applied agricultural practices such as plowing and mechanical disturbance, this study supported that in combination with edaphic and biochemical factors, intensive and uneven fertilizations tended to restructure the spatial heterogeneity of microbial properties, rather than to homogenize it. Substantially contrasting spatial variations were also identified between two bioenergy croplands (GG > SG) and among variables (MBN, MBC:MBN > MBC). Future researchers should better match sample sizes with the heterogeneity of soil microbial property (i.e. MBN) particularly in gamagrass cropland.
References
Tulbure, M. G., Wimberly, M. C. & Owens, V. N. Response of switchgrass yield to future climate change. Environmental Research Letters 7, doi:Artn 04590310.1088/1748-9326/7/4/045903 (2012).
Monti, A., Barbanti, L., Zatta, A. & Zegada-Lizarazu, W. The contribution of switchgrass in reducing GHG emissions. Global Change Biology Bioenergy 4, 420–434, https://doi.org/10.1111/j.1757-1707.2011.01142.x (2012).
Gelfand, I. et al. Sustainable bioenergy production from marginal lands in the US Midwest. Nature 493, 514–517, http://www.nature.com/nature/journal/v493/n7433/abs/nature11811.html#supplementary-information (2013).
Kering, M. K., Butler, T. J., Biermacher, J. T., Mosali, J. & Guretzky, J. A. Effect of Potassium and Nitrogen Fertilizer on Switchgrass Productivity and Nutrient Removal Rates under Two Harvest Systems on a Low Potassium Soil. Bioenerg Res 6, 329–335, https://doi.org/10.1007/s12155-012-9261-8 (2012).
Behrman, K. D., Kiniry, J. R., Winchell, M., Juenger, T. E. & Keitt, T. H. Spatial forecasting of switchgrass productivity under current and future climate change scenarios. Ecological Applications 23, 73–85 (2013).
Robertson, G. P., Hamilton, S. K., Del Grosso, S. J. & Parton, W. J. The biogeochemistry of bioenergy landscapes: carbon, nitrogen, and water considerations. Ecological Applications 21, 1055–1067, https://doi.org/10.1890/09-0456.1 (2011).
Jung, J. Y. & Lal, R. Impacts of nitrogen fertilization on biomass production of switchgrass (Panicum Virgatum L.) and changes in soil organic carbon in Ohio. Geoderma 166, 145–152, https://doi.org/10.1016/j.geoderma.2011.07.023 (2011).
Smith, C. M. et al. Reduced Nitrogen Losses after Conversion of Row Crop Agriculture to Perennial Biofuel Crops. Journal of Environmental Quality 42, 219–228, https://doi.org/10.2134/jeq. 2012.0210 (2013).
Kiniry, J. R. et al. Perennial Biomass Grasses and the Mason-Dixon Line: Comparative Productivity across Latitudes in the Southern Great Plains. Bioenerg Res 6, 276–291, https://doi.org/10.1007/s12155-012-9254-7 (2013).
Jian, S. et al. Soil extracellular enzyme activities, soil carbon and nitrogen storage under nitrogen fertilization: A meta-analysis. Soil Biology and Biochemistry 101, 32–43, https://doi.org/10.1016/j.soilbio.2016.07.003 (2016).
Chen, J. et al. Costimulation of soil glycosidase activity and soil respiration by nitrogen addition. Global Change Biology, n/a-n/a, https://doi.org/10.1111/gcb.13402 (2016).
Guo, L. B. B., Wang, M. B. & Gifford, R. M. The change of soil carbon stocks and fine root dynamics after land use change from a native pasture to a pine plantation. Plant and Soil 299, 251–262, https://doi.org/10.1007/s11104-007-9381-7 (2007).
Fraterrigo, J. M., Turner, M. G., Pearson, S. M. & Dixon, P. Effects of past land use on spatial heterogeneity of soil nutrients in southern appalachian forests. Ecological Monographs 75, 215–230 (2005).
Li, J. W., Richter, D. D., Mendoza, A. & Heine, P. Effects of land-use history on soil spatial heterogeneity of macro- and trace elements in the Southern Piedmont USA. Geoderma 156, 60–73, https://doi.org/10.1016/j.geoderma.2010.01.008 (2010).
Heinze, S., Raupp, J. & Joergensen, R. G. Effects of fertilizer and spatial heterogeneity in soil pH on microbial biomass indices in a long-term field trial of organic agriculture. Plant and Soil 328, 203–215, https://doi.org/10.1007/s11104-009-0102-2 (2010).
Wang, Y. Q., Zhang, X. C. & Huang, C. Q. Spatial variability of soil total nitrogen and soil total phosphorus under different land uses in a small watershed on the Loess Plateau, China. Geoderma 150, 141–149, https://doi.org/10.1016/j.geoderma.2009.01.021 (2009).
Zhang, A., Jiang, L., Qi, Q., Li, X. & Pi, L. Spatial heterogeneity of surface soil nutrients in small scale in the black soil region of Northeast China. 1–4, https://doi.org/10.1109/Agro-Geoinformatics.2014.6910654 (2014).
Enwall, K., Throbäck, I. N., Stenberg, M., Söderström, M. & Hallin, S. Soil Resources Influence Spatial Patterns of Denitrifying Communities at Scales Compatible with Land Management. Applied and Environmental Microbiology 76, 2243–2250, https://doi.org/10.1128/aem.02197-09 (2010).
Abbott, K. C. et al. Spatial Heterogeneity in Soil Microbes Alters Outcomes of Plant Competition. PLoS ONE 10, e0125788, https://doi.org/10.1371/journal.pone.0125788 (2015).
Tilman, D. Plant Strategies and the Dynamics and Structure of Plant Communities. Monographs in Population Biology. Princeton University Press. 360 pp. (1988).
Robertson, G. P. & Gross, K. L. Assessing the heterogeneity of belowground resources: quantifying pattern and scale. In: Caldwell, M.M., Pearcy, R.W. (Eds.), Exploitation of Environmental Heterogeneity by Plants. Academic Press, San Diego, CA (1994).
Schlesinger, W. H., Raikes, J. A., Hartley, A. E. & Cross, A. E. On the spatial pattern of soil nutrients in desert ecosystems. Ecology 77, 364–374 (1996).
Van Veen, J. A. & Kuikman, P. J. Soil structural aspects of decomposition of organic matter by micro-organisms. Biogeochemistry 11, 213–233, https://doi.org/10.1007/bf00004497 (1990).
Liu, S. et al. Spatial variability of soil microbial biomass carbon, nitrogen and phosphorus in a hilly red soil landscape in subtropical China. Soil Science & Plant Nutrition 56, 693–704, https://doi.org/10.1111/j.1747-0765.2010.00510.x (2010).
Röver, M. & Kaiser, E.-A. Spatial heterogeneity within the plough layer: low and moderate variability of soil properties. Soil Biology and Biochemistry 31, 175–187, https://doi.org/10.1016/S0038-0717(97)00272-1 (1999).
Wang, L. X., Mou, P. P., Huang, J. H. & Wang, J. Spatial heterogeneity of soil nitrogen in a subtropical forest in China. Plant and Soil 295, 137–150, https://doi.org/10.1007/s11104-007-9271-z (2007).
Nunan, N., Wu, K., Young, I. M., Crawford, J. W. & Ritz, K. In situ Spatial Patterns of Soil Bacterial Populations, Mapped at Multiple Scales, in an Arable Soil. Microbial Ecology 44, 296–305 (2002).
O’Brien, S. L. et al. Spatial scale drives patterns in soil bacterial diversity. Environmental Microbiology 18, 2039–2051, https://doi.org/10.1111/1462-2920.13231 (2016).
Basaran, M., Erpul, G., Tercan, A. E. & Canga, M. R. The effects of land use changes on some soil properties in Indagi Mountain Pass - Cankiri, Turkey. Environmental Monitoring and Assessment 136, 101–119, https://doi.org/10.1007/s10661-007-9668-4 (2008).
Song, Y. et al. The Interplay Between Bioenergy Grass Production and Water Resources in the United States of America. Environmental Science & Technology 50, 3010–3019, https://doi.org/10.1021/acs.est.5b05239 (2016).
Song, Y., Jain, A. K., Landuyt, W., Kheshgi, H. S. & Khanna, M. Estimates of Biomass Yield for Perennial Bioenergy Grasses in the USA. Bioenerg Res 8, 688–715, https://doi.org/10.1007/s12155-014-9546-1 (2015).
Saetre, P. & Bååth, E. Spatial variation and patterns of soil microbial community structure in a mixed spruce–birch stand. Soil Biology and Biochemistry 32, 909–917, https://doi.org/10.1016/S0038-0717(99)00215-1 (2000).
Wang, L. X., D’Odorico, P., Manzoni, S., Porporato, A. & Macko, S. Soil carbon and nitrogen dynamics in southern African savannas: the effect of vegetation-induced patch-scale heterogeneities and large scale rainfall gradients. Climatic Change 94, 63–76, https://doi.org/10.1007/s10584-009-9548-8 (2009).
Kuramae, E., Gamper, H., van Veen, J. & Kowalchuk, G. Soil and plant factors driving the community of soil-borne microorganisms across chronosequences of secondary succession of chalk grasslands with a neutral pH. FEMS Microbiology Ecology 77, 285–294, https://doi.org/10.1111/j.1574-6941.2011.01110.x (2011).
Clark, R. B. et al. Eastern gamagrass (Tripsacum dactyloides) root penetration into and chemical properties of claypan soils. Plant and Soil 200, 33–45, https://doi.org/10.1023/A:1004256100631 (1998).
Rhoades, C. C. Single-tree influences on soil properties in agroforestry: Lessons from natural forest and savanna ecosystems. Agroforestry Systems 35, 71–94 (1997).
Dockersmith, I. C., Giardina, C. P. & Sanford, R. L. Persistence of tree related patterns in soil nutrients following slash-and-burn disturbance in the tropics. Plant and Soil 209, 137–156 (1999).
Gross, K. L., Pregitzer, K. S. & Burton, A. J. Spatial Variation in Nitrogen Availability in 3 Successional Plant-Communities. Journal of Ecology 83, 357–367 (1995).
Wang, L. X., Okin, G. S., Caylor, K. K. & Macko, S. A. Spatial heterogeneity and sources of soil carbon in southern African savannas. Geoderma 149, 402–408, https://doi.org/10.1016/j.geoderma.2008.12.014 (2009).
Yu, C. L. et al. Responses of corn physiology and yield to six agricultural practices over three years in middle Tennessee. Sci Rep-Uk 6, Artn 27504 https://doi.org/10.1038/Srep27504 (2016).
Dzantor, E. K., Adeleke, E., Kankarla, V., Ogunmayowa, O. & Hui, D. Using Coal Fly Ash in Agriculture: Combination of Fly Ash and Poultry Litter as Soil Amendments for Bioenergy Feedstock Production. Coal Combustion and Gasification Products 7, 33–39, https://doi.org/10.4177/CCGP-D-15-00002.1 (2015).
Brookes, P. C., Landman, A., Pruden, G. & Jenkinson, D. S. Chloroform fumigation and the release of soil nitrogen: A rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biology and Biochemistry 17, 837–842, https://doi.org/10.1016/0038-0717(85)90144-0 (1985).
Brookes, P. C., Landman, A., Pruden, G. & Jenkinson, D. S. Chloroform Fumigation and the Release of Soil-Nitrogen - a Rapid Direct Extraction Method to Measure Microbial Biomass Nitrogen in Soil. Soil Biology & Biochemistry 17, 837–842 (1985).
Vance, E. D., Brookes, P. C. & Jenkinson, D. S. An extraction method for measuring soil microbial biomass C. Soil Biology and Biochemistry 19, 703–707 (1987).
Wu, J., Joergensen, R. G., Pommerening, B., Chaussod, R. & Brookes, P. C. Measurement of soil microbial biomass C by fumigation-extraction—an automated procedure. Soil Biology and Biochemistry 22, 1167–1169, https://doi.org/10.1016/0038-0717(90)90046-3 (1990).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vinnna, Austria ISBN 3-900051-07-0, URL http://www.R-project.org/ (2015).
Cochran, W. G. Testing a Linear Relation among Variances. Biometrics 7, 17–32 (1951).
Underwood, A. J. Experiments in Ecology: Their Logical Design and Interpretation Using Analysis of Variance. Cambridge University Press (1997).
Cochran, W. G. The distribution of the largest of a set of estimated variances as a fraction of their total. Annals of Eugenics 11, 47–52 (1941).
Gittins, R. Trend-Surface Analysis of Ecological Data. Journal of Ecology 56, 845 (1968).
Legendre, P. & Legendre, L. Numerical ecology. Elserier Science, Amsterdam, the Netherlands. (1998).
Legendre, P. & Fortin, M. J. Spatial Pattern and Ecological Analysis. Vegetatio 80, 107–138 (1989).
Cressie, N. Statistics for Spatial Data. John Wiley & Sons, New York (1993).
Moran, P. A. P. Notes on continuous stochastic phenomena. Biometrika 37, 17–23 (1950).
Webster, R. & Oliver, M. A. Sample adequately to estimate variograms of soil properties. Journal of Soil Science 43, 177–192, https://doi.org/10.1111/j.1365-2389.1992.tb00128.x (1992).
Isaaks, E. H. & Srivastava., R. M. Introduction to applied geostatistics. Oxford University Press, New York. (1989).
Gotway, C. A., Ferguson, R. B., Hergert, G. W. & Peterson, T. A. Comparison of kriging and inverse-distance methods for mapping soil parameters. Soil Science Society of America Journal 60, 1237–1247 (1996).
Oliver, M. A. & Webster, R. Semi-Variograms for Modeling the Spatial Pattern of Landform and Soil Properties. Earth Surface Processes and Landforms 11, 491–504 (1986).
Allison, S. D. & Martiny, J. B. H. Resistance, resilience, and redundancy in microbial communities. Proceedings of the National Academy of Sciences of the United States of America 105, 11512–11519, https://doi.org/10.1073/pnas.0801925105 (2008).
Liang, C. & Balser, T. C. Warming and nitrogen deposition lessen microbial residue contribution to soil carbon pool. Nat Commun 3, 1222 (2012).
Le, X., Hui, D. & Dzantor, E. Characterizing Rhizodegradation of the Insecticide Bifenthrin in Two Soil Types. Journal of Environmental Protection 2, 940–946 (2011).
Anderson, E. K. et al. Impacts of management practices on bioenergy feedstock yield and economic feasibility on Conservation Reserve Program grasslands. GCB Bioenergy 8, 1178–1190, https://doi.org/10.1111/gcbb.12328 (2016).
Robertson, G. P., Crum, J. R. & Ellis, B. G. The Spatial Variability of Soil Resources Following Long-Term Disturbance. Oecologia 96, 451–456 (1993).
Chen, J. et al. Costimulation of soil glycosidase activity and soil respiration by nitrogen addition. Global Change Biology 23, 1328–1337, https://doi.org/10.1111/gcb.13402 (2017).
Jangid, K. et al. Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biology & Biochemistry 40, 2843–2853, https://doi.org/10.1016/j.soilbio.2008.07.030 (2008).
Yu, P. Y., Zhu, F., Su, S. F., Wang, Z. Y. & Yan, W. D. Effects of nitrogen addition on red soil microbes in the Cinnamomum camphora plantation. Huan Jing Ke Xue 34, 3231–3237 (2013).
Xie, H. T. et al. Long-term manure amendments enhance neutral sugar accumulation in bulk soil and particulate organic matter in a Mollisol. Soil Biology & Biochemistry 78, 45–53, https://doi.org/10.1016/j.soilbio.2014.07.009 (2014).
Kuzyakov, Y. & Blagodatskaya, E. Microbial hotspots and hot moments in soil: Concept & review. Soil Biology & Biochemistry 83, 184–199, https://doi.org/10.1016/j.soilbio.2015.01.025 (2015).
Bourceret, A., C. Leyval, et al. Mapping the Centimeter-Scale Spatial Variability of PAHs and Microbial Populations in the Rhizosphere of Two Plants. PLOS ONE 10(11), e0142851 (2015).
Rovira, A. D. Root excretions in relation to the rhizosphere effect: iv. Influence of plant species, age of plant, light, temperature, and calcium nutrition on exudation. Plant and Soil 11, 53–64 (1959).
Perrygo1, C. L., Shirmohammadi, A., Ritchie, J. C. & Rawls, W. J. Effect of eastern gamagrass on infiltration rate and soil physical and hydraulic properties. Pp. 265-268 in: DavidBosch & Kevin King (eds.), Preferential Flow Water: Movement and Chemical Transport in the Environment, Proc. 2nd Intl. Symp. (3–5 January 2001, Honolulu, Hawaii, USA), St. Joseph, Michigan: ASAE. 701P0006 (2001).
Lux, A. & Rost, T. L. Plant root research: the past, the present and the future. Annals of Botany 110, 201–204, https://doi.org/10.1093/aob/mcs156 (2012).
Foy, C. D. Tolerance of eastern gamagrass to excess aluminum in acid soil and nutrient solution. Journal of Plant Nutrition 20, 1119–1136, https://doi.org/10.1080/01904169709365322 (1997).
Gilker, R. E., Weil, R. R., Krizek, D. T. & Momen, B. Eastern gamagrass root penetration in adverse subsoil conditions. Soil Science Society of America Journal 66, 931–938 (2002).
Tufekcioglu, A., Raich, J. W., Isenhart, T. M. & Schultz, R. C. Fine root dynamics, coarse root biomass, root distribution, and soil respiration in a multispecies riparian buffer in Central Iowa, USA. Agroforestry Systems 44, 163–174, https://doi.org/10.1023/a:1006221921806 (1998).
Luo, Y. et al. Progressive nitrogen limitation of ecosystem responses to rising atmospheric carbon dioxide. Bioscience 54, 731–739 (2004).
Vitousek, P. M. & Howarth, R. W. Nitrogen Limitation on Land and in the Sea - How Can It Occur. Biogeochemistry 13, 87–115 (1991).
Sinsabaugh, R. L., Hill, B. H. & Shah, J. J. F. Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature 462, 795–U117, https://doi.org/10.1038/nature08632 (2009).
Sinsabaugh, R. L. et al. Stoichiometry of soil enzyme activity at global scale. Ecology Letters 11, 1252–1264, https://doi.org/10.1111/j.1461-0248.2008.01245.x (2008).
Smith, J. l. & Paul, E. a. in: Bollag J.M., Stotz ky G. (eds.): SoilBiochemistry, vol. 6. Marcel Dekker, inc., New York: 357–396. (1990).
Allison, S. D., Hanson, C. A. & Treseder, K. K. Nitrogen fertilization reduces diversity and alters community structure of active fungi in boreal ecosystems. Soil Biology and Biochemistry 39, 1878–1887, https://doi.org/10.1016/j.soilbio.2007.02.001 (2007).
Rousk, J., Brookes, P. C. & Bååth, E. Contrasting Soil pH Effects on Fungal and Bacterial Growth Suggest Functional Redundancy in Carbon Mineralization. Applied and Environmental Microbiology 75, 1589–1596, https://doi.org/10.1128/aem.02775-08 (2009).
Kim, M. et al. Highly Heterogeneous Soil Bacterial Communities around Terra Nova Bay of Northern Victoria Land, Antarctica. PLoS ONE 10, e0119966, https://doi.org/10.1371/journal.pone.0119966 (2015).
Fierer, N. & Jackson, R. B. The diversity and biogeography of soil bacterial communities. Proceedings of the National Academy of Sciences of the United States of America 103, 626–631, https://doi.org/10.1073/pnas.0507535103 (2006).
Acknowledgements
This research received financial support from the USDA Evans-Allen grant awarded to JL (No. 1005761), China Scholarship Council ([2015]3069) to CG, USDA National Institute of Food and Agriculture (2010–38821) to DEK. We thank assistance received from staff members at the TSU’s Main Campus Agriculture Research and Extension Center (AREC) in Nashville TN.
Author information
Authors and Affiliations
Contributions
J.W.L. conceived the project. J.W.L and C.L.G. contributed equally to the work. C.L.G. and S.Y.J. conducted the laboratory and data analysis. J.W.L., S.Y.J, Q.D., and C.L.Y. conducted field soil collections. K.E.D. managed the field site. J.W.L. led the manuscript preparation. All contributed to data analysis, interpretation, and writing the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, J., Guo, C., Jian, S. et al. Nitrogen Fertilization Elevated Spatial Heterogeneity of Soil Microbial Biomass Carbon and Nitrogen in Switchgrass and Gamagrass Croplands. Sci Rep 8, 1734 (2018). https://doi.org/10.1038/s41598-017-18486-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-18486-5
This article is cited by
-
Nitrogen Fertilization Restructured Spatial Patterns of Soil Organic Carbon and Total Nitrogen in Switchgrass and Gamagrass Croplands in Tennessee USA
Scientific Reports (2020)
-
Effects of nitrogen fertilization and bioenergy crop species on central tendency and spatial heterogeneity of soil glycosidase activities
Scientific Reports (2020)
-
Nitrogen-inputs regulate microbial functional and genetic resistance and resilience to drying–rewetting cycles, with implications for crop yields
Plant and Soil (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.