Abstract
It has been repeatedly demonstrated that the centromere-specific histone H3 (CENH3), a key component of the centromere, shows considerable variability between species within taxa. We determined the molecular structure and phylogenetic relationships of CENH3 in 11 Secale species and subspecies that possess distinct pollination systems and are adapted to a wide range of abiotic and biotic stresses. The rye (Secale cereale) genome encodes two paralogous CENH3 genes, which differ in intron-exon structure and are transcribed into two main forms of the protein, αCENH3 and βCENH3. These two forms differ in size and amino acid substitutions. In contrast to the reported differences in CENH3 structure between species within other taxa, the main forms of this protein in Secale species and subspecies have a nearly identical structure except some nonsynonymous substitutions. The CENH3 proteins are strictly controlled by genetic factors responsible for purifying selection. A comparison between Hordeum, Secale and Triticum species demonstrates that the structure of CENH3 in the subtribes Hordeinae and Triticinae evolved at different rates. The assumption that reticulate evolution served as a factor stabilizing the structure and evolutionary rate of CENH3 and that this factor was more powerful within Secale and Triticum than in Hordeum, is discussed.
Similar content being viewed by others
Introduction
The pivotal role in the proper chromosome segregation during meiosis and mitosis lies with centromeres. In most species the centromere identity is defined by the presence of the centromere-specific variant of histone H3 known in plants as centromere-specific histone H3 variant CENH3 (for review, see1,2). Any error in transcription, translation, modification or import can affect the assembly of intact CENH3 chromatin, which would result in the loss of CENH3 from the centromeres and hence in the centromere identity (reviewed in3). In contrast to the conserved structure of canonical histone Н3, CENH3 shows considerable variability across species4,5. Different domains of this molecule evolved differently. An extended N-terminal tail (NTT) and loop 1 of the histone fold domain (HFD) putatively interact with centromeric DNA6 and show signatures of positive selection in some animal and plant species7,8, while the part of the HFD domain outside loop 1 is generally conserved8,9,10.
Most of the diploid plant species (Arabidopsis thaliana, maize and rice), in which the structure and copy number of CENH3 have been determined, have this gene as a single copy8,11,12. However, some species in the Triticeae tribe have CENH3 in two variants. They are tetraploid and diploid wheat (Triticum) species13, diploid barley (Hordeum) species14 and Aegilops species13. The levels of expression of these two CENH3 variants and the efficiency of their incorporation at centromeres vary across different tissues as demonstrated for barley15 and between wild and cultivated tetraploid wheats, which is considered as a signature of adaptive evolution13.
Rye (Secale) is a small but important genus of the Triticeae tribe adapted to a wider range of environmental and climatic conditions than wheat or barley16. Cultivated, weedy and wild species in Secale have different pollination systems (self-incompatible, allogamous vs self-compatible, autogamous) and life-cycle durations (perennials vs annuals). Sencer & Hawkes17 classified this genus as consisting of three biological species: the outcrossing perennial S. strictum Presl., the outcrossing annual S. cereale L., and the autogamous annual S. sylvestre Host. This classification received further support from morphometrical data18 and molecular analysis19. Traditional rye varieties are panmictic populations displaying high levels of heterozygosity and heterogeneity20, which might have resulted from outcrossing pollination and facilitated interspecies hybridization. Because the CENH3 proteins and genes encoding them in Secale species have yet to be known, it is intriguing to explore the molecular structure and the evolutionary dynamics of this central component of centromere specification and function.
We have identified and characterized CENH3 variants in Secale species and subspecies, the intron-exon structure of the CENH3 genes and their phylogenetic relationships in Secale and closely related genera, Triticum and Hordeum, in Triticeae. We found that CENH3 sequences in Secale species and subspecies have a nearly identical structure except some nonsynonymous substitutions. This implies that the general view about rapidly evolving CENH3s is not universal – at least, it does not apply to the genus Secale. A comparison of Hordeum, Secale and Triticum species demonstrated that the CENH3 structure in the subtribes Hordeinae and Triticinae (the latter including Triticum and Secale species21) evolved at different rates. We hypothesize that past remote hybridization events (reticulate evolution) served as a factor stabilizing the structure of the CENH3 genes and proteins and that this factor was more powerful within Secale and Triticum than it was in the other cereals taxa, including Hordeum.
Results
Identification and characterization of the CENH3 forms in Secale
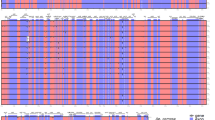
We searched the NCBI SRA database for CENH3 of S. cereale and found partial sequences with homology to αCENH3 of H. vulgare and βCENH3 of T. urartu. Based on these, PCR primers were designed and used for amplifying complete CENH3 transcripts of rye. After cloning of PCR products and sequencing of randomly selected clones the presence of two main forms of CENH3 (called αScCENH3 and βScCENH3) were revealed in rye. The αScCENH3 sequence is 501 bp in length and the deduced protein is made up of 166 amino acids. In S. cereale, βCENH3 is distinct from αCENH3 in that the former has several deletions in the N-terminal tail (NTT) and the insertion of three nucleotides, АСС, which encode the amino acid threonine (framed in Fig. 1), in the histone fold domain (HFD). Thus, βScCENH3 has an overall length of 456 bp and encodes a protein made up by 151 amino acids. Most of the amino acid sequences of the NTT in αCENH3 and βCENH3 do not align well with each other (Fig. 1). The average nucleotide identity between αCENH3 and βCENH3 is 81–83%. In the HFD the main differences are concentrated in the α1-helix and loop 1, that is, in the centromere-targeting domain (CATD).
Multiple alignment of the amino acid sequences of CENH3 proteins. αCENH3-v1 and βCENH3-v1 from rye accessions are aligned with CENH3 proteins in Triticum, Aegilops, Hordeum, and Oryza sativa accessions: αCENH3 of T. aestivum (JF969285.1), T. urartu (KM507181.1), A. tauschii (KM507183.1), A. speltoides (KM507182.1), H. vulgare (JF419328.1), H. bulbosum (GU245882.1), O. sativa (AY438639.1) and βCENH3 of T. urartu (KM507184.1), A. tauschii (KM507186.1), A. speltoides (KM507185.1), H. vulgare (JF419329.1), H. bulbosum (JF419330.1). For convenience, alpha and beta forms are grouped into two separate blocks. Separate HFD regions are singled out according to27. The amino acid threonine that occurs in the βCENH3 HFD, but not in the αCENH3 HFD, is framed. Amino acid residues identical in all species are shaded in dark gray.
In addition to the clones with 501-bp long sequences (αScCENH3-v1), which were the most frequently occurring in the pool of the αCENH3 clones randomly selected for sequencing, we found clones with shorter inserts, 492 bp in length (Supplementary Fig. S1), with 94% nucleotide identity to αScCENH3-v1 and also with 99% nucleotide identity to one of the CENH3 sequences identified previously in the genome of T. aestivum (JF969287.1) and to αCENH3 in the genome of T. urartu (KM507181.1). Beside different lengths, they also contain different amino acids at the same positions in different αScCENH3 clones and thus probably reflect individual sequence differences. The shorter variant should be designated as minor, αScCENH3-v2, because the percentage of these clones in the pool is low. The highest frequency of αScCENH3-v2 is 18%, which is in the annual S. cereale ssp. cereale (S. cereale throughout) cv. Otello.
βScCENH3, too, occurs in two variants. The two βScCENH3 variants differ by 14 amino acid substitutions, of which nine are nonsynonymous and three of these are found in loop 1 and α2-helix (Supplementary Fig. S1). βCENH3-v1 is 456 bp in length and has 95% nucleotide identity to T. urartu (KM507184.1). βCENH3-v2 has 95% nucleotide identity to βCENH3-v1, 99% nucleotide identity with the βCENH3 of A. tauschii (KM507186.1) and is 6 bp longer than βCENH3-v1.
Ten additional Secale species and subspecies that possess distinct pollination systems and are adapted to a wide range of abiotic and biotic stresses were included in this study. αCENH3-v2 was only in five Secale accessions and βCENH3-v2 only in four (Table 1). The frequency of αCENH3-v2 is 8–10% in the perennial self-pollinated S. strictum ssp. africanum (S. africanum throughout) and the annual cross-pollinated S. cereale ssp. dighoricum (S. dighoricum throughout) (Table 1). βCENH3-v2 is present in cross-pollinated subspecies: the annual S. cereale ssp. afghanicum (S. afghanicum throughout), in which it makes up 60% of the clones sequenced, and the perennial S. strictum ssp. strictum (S. strictum throughout) with 45%. However, it is important to take into account the fact that the non-observation of any rare variant in PCR products for cDNA does not necessarily mean that this variant is not present in the genome. Thus, rye species and subspecies possess their own genus-specific alpha and beta forms of CENH3, as well as variants of these forms, which are also present in Triticum and Aegilops species (Table 1).
Transcripts with the characteristics of the main forms of CENH3 were found in all the 11 rye species and subspecies analyzed (the rye CENH3 forms given in Fig. 1 are actually αCENH3-v1 and βCENH3-v1). The nucleotide identity of αCENH3 and βCENH3 sequences between Secale species and subspecies is 98–100%. Deletions in the NTT of βCENH3 in the rye species and subspecies are noted for having fixed lengths, these lengths being exactly the same as those of deletions in T. urartu and other donors of the hexaploid wheat genome, A. tauschii and A. speltoides. Surprisingly, the structure of this region in the rye species is closer to that in T. aestivum than to that of αCENH3 in T. aestivum progenitors. A high level of similarity in the nucleotide sequences of the CENH3 genes between allopolyploid wheats and various rye species and subspecies, which is 96–97% for αCENH3, is reflected by a high level of similarity in their amino acid sequences. A comparison of the αCENH3 sequences in four S. cereale cultivars (Otello, Black Winter, Imperial and Korotkostebelny 69) with their counterpart in T. aestivum cv. Chinese Spring (JF969285.1) revealed as few as six amino acid substitutions in the NTT and one in the HFD. In contrast to the close similarities in CENH3 sequences between the rye and wheat species, the structures of CENH3 have considerable differences between the barley species H. vulgare and H. bulbosum (JF419329.1 and JF419330.1). Compared to H. bulbosum αHbCENH3, H. vulgare αHvCENH3 contains 10 amino acid substitutions in the HFD, largely in loop 1, and three additional amino acids in the NTT. The differences are especially high between the beta forms of CENH3. βHvCENH3 is distinct from βHbCENH3 in that it has 30 nonsynonymous amino acid substitutions throughout the molecule and four additional amino acids. Compared to the beta forms of rye and wheat CENH3, those of barley have longer deletions in the NTT, and this accounts for the differences in the size of this domain: 108 bp in H. bulbosum, 111 bp in H. vulgare, and 165 bp in S. cereale. The mean pairwise distance between the αCENH3 paralogs of S. cereale and H. vulgare is 0.122 at nucleotide level and 0.269 at amino acid level; that between S. cereale and H. bulbosum, 0.097 and 0.221, correspondingly; and that between S. cereale and T. aestivum, 0.033 and 0.043, correspondingly. This comparison shows that both main forms of CENH3 have a surprisingly high structural similarity between Secale, Triticum and Aegilops species, but it is different in barley.
Phylogeny of rye CENH3
With the Neighbor Joining (NJ) algorithm, a phylogenetic tree was constructed for the amino acid sequences of CENH3 in 11 accessions of rye, the closest rye relatives within Triticeae (wheat, barley and Aegilops species), and some monocotyledonous species as well as for the sequence of canonical histone H3 of O. sativa as an outgroup (Fig. 2). The first node is where two major clusters arise from, one with alpha forms of CENH3 and another with beta forms. In all Secale accessions analyzed, αCENH3-v1s fall in the same domain within the cluster. However, the second variant of rye αCENH3, αCENH3-v2, is in another domain, together with the wheat and Aegilops accessions. Of the other Triticeae species, the closest to rye αCENH3 was T. aestivum CENH3 (93–97% nucleotide identity, p (pair-wise distance between orthologs) = 0.040). For other cereal species, A. sativa and O. sativa, the corresponding values varied from 78% to 73% and from 0.365 to 0.469.
Both βCENH3 variants form the second major cluster, together with beta forms in the Aegilops species and T. urartu. The alpha and beta forms of the barley species are in the major tree clusters together with their rye, wheat and Aegilops counterparts. The CENH3 sequence of O. sativa forms a separate branch and is in the same major cluster as the alpha forms of Triticeae species.
Divergence of rye CENH3s
A comparison of NTT and HFD sequences done using the McDonald—Kreitman test22 in the subspecies within S. cereale and S. strictum revealed a lack of “fixed divergence”, as McDonald and Kreitman put it. We aligned the sequences of all subspecies of each of the given species in one data set and estimated Ka/Ks ratios (also denoted as ω). For both domains, the ratio of nonsynonymous (Ka) to synonymous substitutions (Ks) in the alpha and beta forms between species is significantly less than 1 (Table 2), which appears to be a signature of stabilizing selections. To identify potential sites under positive selection, we estimated ω between subspecies. The results of the pair-wise comparisons of subspecies include a few ω >1 instances (one such comparison is given in Supplementary Table S1). Although the difference between the ω value and 1 was not statistically significant in any of these instances, it suggests that within species divergence has signatures of positive selection. Noteworthy, the ω value was higher in the NTT than in the HFD (Table 2), suggesting that the NTT has evolved faster than the HFD. Similarly, the ω values for the beta forms of CENH3 are in all cases higher than those for the alpha forms.
Even though the full-length NTT and HFD sequences reveal signatures of stabilizing selection, it is still possible that some of the sites within these domains have been under other modes of selection. Supplementary Table S2 summarizes the characteristics of these variable codons, which occur in at least several accessions, that is, their variability is not accounted for by random effects or by sequencing errors. Codon 34 containing only nonsynonymous substitutions in the αNTT of the annual subspecies S. dighoricum, S. cereale ssp. ancestrale (S. ancestrale throughout) and the perennial subspecies S. anatolicum is under diversifying selection. Codons 22 and 48 in the αNTT and codons 92 and 136 in the αHFD, as well as just one substitution at codon 51 in the βNTT as being under negative selection. Synonymous substitutions occur at these codons in all the S. cereale and S. strictum subspecies. Noteworthy, all the accessions with codons under diversifying or negative selection are cross-pollinated. Variable codons that are not undergoing selection occur most frequently in the most ancient annual self-pollinated S. sylvestre. Thus, diversifying selection operates at a very few sites of the N-terminal tail of αCENH3 and the HFD of βCENH3 of cross-pollinated species and adds little to the structural diversity of the existing forms of CENH3 proteins.
Divergence of CENH3 histone fold domains in Triticeae
Considering an important functional role of the HFD, which is crucial for nucleosome assembly and targeting of CENH3 to centromeres23, we extended the analysis of its structure to Triticum, the genus most closely related to Secale, and species progenitors to polyploid wheat species (Table 1).
Two types of CENH3s had previously been identified in diploid and tetraploid wheat species13. The HFD sequences of Triticum, Aegilops and Secale accessions are shown on Fig. 3. In Triticum and Aegilops, the C-terminal part of ßCENH3 is distinct from that of αCENH3 in that (a) some of its positions are polymorphic, which leads to synonymous and nonsynonymous amino acid substitutions, and (b) it has the asparagine-encoding trinucleotide АAС inserted near the top of loop 1.
In diploid ancestors, the main HFD forms appear in two variants, having specific amino acids at particular positions, one in the αHFD (not shown) and six in the βHFD (asterisked in Fig. 3). The amino acid sequences of the HFDs of allopolyploid wheats display no synonymous substitutions. In rye αHFD sequences have the highest level of similarity in both between v-1, v-2 variants and between species. The specific amino acids that make the rye species distinct from all Triticum accessions occupy only six positions: one in the αHFD (in loop N) and five in the βHFD, four of which are in the САТD (Fig. 3). Importantly, four out of five amino acids occur in βHFD-v1, suggesting that was the preferred beta form variant during Secale evolution. Comparisons for βHFD-v2 revealed signatures of positive selection in the genomes of allopolyploid wheat species (Table 3). However, according to Fisher’s exact test, ω values >1 in these cases failed to reach significance because the Ka/Ks ratio was just slightly higher between than within species. All the ω values for ßCENH3 sequences in the H. vulgare and H. bulbosum genomes are similar to their counterparts in wheat and rye species; however, the absolute values of Ka and Ks are several times higher. Noteworthy, the differences are more salient for the cultivated barley H. vulgare (Table 3).
Alternative splicing of ScCENH3s
Alignment of the cDNA sequences of αScCENH3 and βScCENH3 with publically available genomic rye sequences24 confirmed that the open reading frame of αScCENH3 comprises 7 exons and 6 introns, while that of βScCENH3, 4 exons and 3 introns (Fig. 4А). Exons 2 and 3 of αScCENH3 are very short, 25 bp and 39 bp, respectively; exon 4 of βScCENH3 is 52 bp in length. In addition to the main ScCENH3 forms, we obtained several variants of transcripts which will be referred to as alternative splicing (AS) isoforms throughout. The N-terminal domain of the αScCENH3 gene is the source of most AS products. The following isoforms were identified: (1) ScCENH3-AS1 flanked by splice sites and containing a 21-bp deletion in exon 1 from position 67 to position 87; (2) ScCENH3-AS2 flanked by splice sites and containing a 66-bp deletion, which removes exon 2 (104–129 nt) and exon 3 (130–169 nt) entirely; (3) ScCENH3-AS3, with retention of a 43-bp fragment of intron 1 after position 103; and 4) ScCENH3-AS4, with retention of a 97-bp fragment of intron 2 after position 129. These isoforms are consistent with the main types of alternative splicing25: ScCENH3-AS1 appears due to the proximity of two splice sites; ScCENH3-AS2, due to mutually exclusive exons; ScCENH3-AS3 and ScCENH3-AS4, due to intron retention.
Intron-exon structure of CENH3 genes in S.cereale. (A) Schematic of splicing isoforms. Exons are enumerated and are depicted as light gray rectangles; introns are depicted as black lines connecting exons; the ranges on top of exons indicate exon boundaries. Introns are not to scale. Deletions are depicted as dashed rectangles. Right angled triangles point to retained intron fragments, with their sizes as indicated below. Putatively functional forms with conserved HFD structure are asterisked. Exons with reading frame shifts due to intron retention events are depicted as dark gray rectangles. (B) Percentage of frequency of occurrence of splicing isoforms in rye accessions. AS1-AS4 isoforms expressed as a percentage of the total αCENH3 NTT clones. AS5 isoform expressed as a percentage of the total βCENH3 HFD clones.
Only the most frequent isoform AS4 occurs in all Secale accessions. For example, isoform AS2 was found in annual S. segetale and rye cultivars Otello and Imperial (2.8% of all αScCENH3 transcripts, Fig. 4B), but not in perennial species. Noteworthy, although the 21-bp and 66-bp deletions shorten the N-terminal domain, they do not cause a shift in the open reading frame – nor do they modify the amino acid sequence of the HFD of αCENH3. The 21-bp deletion contains CAG, a false acceptor site.
βScCENH3 was found to contain only one AS isoform (retention of intron 1). It has an unusual localization in the HFD (Fig. 4A). The 15-bp insertion is before loop 1 and has therefore no effect on the βHFD CATD structure. Thus, the protein sequences of AS isoforms have the same CATD structure as the main ScCENH3 forms and can potentially participate in the formation of centromeric nucleosomes, thus increasing the diversity of rye CENH3 proteins.
Discussion
We have identified two distinct forms of CENH3 transcripts, αCENH3 and βCENH3, in 11 rye species and subspecies. This finding indicates that the rye genome contains at least two copies of the CENH3 gene. The closest relatives of rye–wheat, Aegilops and barley–had previously also been found to have two main forms of CENH313,14. In barley, these copies are encoded by two different chromosomes14. Thus, the presence of these two forms in Triticeae appears to be one of the features of this tribe. Both copies avoided elimination from the rye genome, even though elimination by massive local deletions is often imminent after duplications26. This strongly suggests that both CENH3 paralogs are required for proper centromere function in Triticeae. On the other hand, functional inactivation of βCENH3 did not result in an obvious somatic phenotype in barley27.
Alpha CENH3s cluster with CENH3 of О. sativa, a precursor of the Triticeae species, encoding only one variant of CENH3, therefore it is likely that the alpha forms of Triticeae species appeared earlier than the beta forms. The Ka and Ks values as well as their ratios between the beta forms of CENH3 are higher than the corresponding values for the alpha forms (Table 2, Supplementary Table S1), as if the beta forms were under relaxed selective constraints, which result in elevated rates of evolution. A similar tendency is observed in the genus Triticum, in which the αCENH3 HFDs are under negative (stabilizing) selection and the βCENH3 HFDs are undergoing positive (adaptive) evolution13.
The amino acid sequences of CENH3 in the rye species and subspecies display a surprisingly high level of similarity, despite the differences that they have in morphology, life-cycle duration and pollination systems as well as environmental and growing conditions. The highest number of amino acid differences was observed in the βCENH3 of the most ancient species, S. silvestre, but only two of them were found to be nonsynonymous substitutions (Supplementary Table S2). Low ω values obtained from most of the pair-wise comparisons (Table 2, Supplementary Table S1) suggest that Secale CENH3 appears to have evolved under strong purifying selection. Triticum also exhibits the evolutionary conservatism of the CENH3 structure (Table 3), which is even more pronounced in polyploid wheat species. The complete structure of the alpha and beta forms of CENH3 was determined in the cultivated H. vulgare and its close wild relative H. bulbosum 14. A comparison of these two species reveals the heterogeneity of CENH3 structure, which contrasts with the homogeneity of this protein in Secale and is even higher than the genus-specific differences between S. cereale and T. aestivum.
Comparison of the dynamics of evolutionary changes occurring in the CENH3s of Hordeum, Secale and Triticum leads to the assumption that the structure of this protein evolved at different rates in these genera. Considerable differences between the Hordeum species suggest that this genus is the only one among the three that have rapidly evolving CENH3 genes. What factors could possibly account for different rates of structural changes in CENH3 within closely related genera of a tribe? According to by far the most extensive molecular marker-based study, most Hordeum species are not older than 2 MY28. The rye/wheat split time is estimated at approximately 3.5–4 MYA29 and 3–9 MYA30. Later on, the genome donors of hexaploid wheat split off between 2.1–2.9 MYA29 and the age of the Secale crown group is estimated at 1.7 MY31. Thus, the existing estimates of split times for Hordeum, Secale and Triticum species suggest that age alone is unlikely to be a factor that accounts for such strong differences in the rates of evolutionary changes in CENH3 structure between the barley species on the one hand and the Secale/Triticum complex on the other.
Hybridization and associated introgression of genetic material are powerful evolutionary factors, and remote hybridization played an important role in plant speciation. Compared to Hordeum, the genus Secale consists of much fewer taxa, most of which are cross-pollinated. Most rye species and subspecies cross readily with each other and with cultivated rye and produce vigorous hybrids with completely normal meiosis and high pollen fertility32, suggesting the absence of reproductive barriers33. Indeed, a genome-wide comparative analysis showed that the rye genome represents a concatenation of genomic segments of different evolutionary origin and is likely to have been shaped by introgressive hybridization or reticulate evolution23. In support based on the work by Escobar and the co-workers34 Hordeum species follow a tree-like pattern of evolution, while Secale, Triticum, Aegilops are more reticulated than any other clade. Thus, our data favor the assumption that the process of genome formation for Secale was accompanied by ancestral hybridization events. It appears that such reticulate evolution served as a factor stabilizing the structure of the CENH3 genes and proteins, and this factor was more powerful within Secale and Triticum than it was in the other taxa, including Hordeum.
Three splicing isoforms were found among the rye subspecies that do not disrupt the CATD structure: the 21- and 66-bp deletions in the αNTT were largely found in the annual species, and the 15-bp insertion, in the βHFD (Fig. 4). Alternative splicing at the C-terminus in rye does not affect the structure of the DNA-binding domain, but can influence CENH3 binding to other kinetochore proteins, for example, CENP-C, which, in addition to being a DNA-binding protein, can bind to the C-terminal tail of CENP-A35.
Thus, the factors responsible for CENH3 diversity in the rye species are (1) the occurrence of CENH3 in two forms, αCENH3 and βCENH3, with two variants of each of these forms, and (2) the products of alternative splicing, which are presumably driven by positive selection36. In wheat, 95% of alternative transcripts from a particular gene exhibited different expression profiles, as was revealed by a hierarchical clustering of 30,232 transcripts37. It appears that AS isoforms have complementary functions, thereby enhancing the adaptation-related potential of proteins. This finding is indicative of an evolutionary stability and conservation of the genetic factors that control the CENH3 structure in the genus Secale.
Methods
Plant material and plant growth
We selected 13 accessions of weedy/wild rye and cultivated rye representing the most commonly recognized taxa ranked as species or subspecies in the genus Secale according to Sencer and Hawkes17 (Table 1). Seeds were kindly provided by the Leibniz Institute of Plant Genetics and Crop Plant Research (Germany), the United States Department of Agriculture (USA) and N.I.Vavilov Research Institute of Plant Industry (Russia) from their respective germplasm collections. Triticum and Aegilops seeds were kindly provided by Dr. B. Kilian. Accessions are listed and characterized in Table 1. All plants were grown in a greenhouse at 22 °C: 18 °C, day: night with a 16-h day length.
Screening of databases
Pyrosequenced rye cDNAs (GABI-RYE EXPRESS project, accession: PRJEB2219 ID:203975) from the NCBI SRA database (http://www.ncbi.nlm.nih.gov/sra/) were analyzed using the WUBLAST software (http://blast.wustl.edu) and reads with high similarity to αCENH3 of H. vulgare (JF419328.1) and βCENH3 of T. urartu (KM507184) were revealed. These reads were used for generation of rye αCENH3 and βCENH3 contigs by the CodonCodeAligner software program (http://www.codoncode.com/aligner). The search for rye genomic CENH3 sequences was performed in the DNA database (European Bioinformatics Institute sequence read archive, accession ID ERP001745) obtained from sorted rye chromosomes 1R-7R24.
RNA isolation and PCR amplification
Total RNA was isolated from leaves of 12-day-old seedlings using the TRI Reagent (MRC, Inc., USA) and treated by RQ-RNase-Free DNase (Promega, Madison, WI) according to the manufacturer’s instructions. RNA was reverse-transcribed to cDNA using a RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Specific primers used to amplify CENH3 and its domains, NTT and HFD, from rye cDNA are presented in Supplementary Table S3. For amplification of HFD CENH3 from Triticum and Aegilops species, we used a set of degenerated primers designed for monocotyledons38.
Sequencing and sequence alignment
RT-PCR products were purified using a Qiagen Purification Kit (Qiagen) and cloned using an InsTAclone PCR Cloning Kit (Thermo Fisher Scientific). Both strands of 12–20 clones of each accession were sequenced using an ABI 3130 × 1 Genetic Analyzer (Applied Biosystems Inc., CA) and an ABI BigDye Kit according to a standard protocol. Similarity searches between the obtained rye CENH3 sequences and their orthologous from other species were carried out using the TBLASTN software39 in the NCBI database (http://blast.ncbi.nlm.nih.gov/Database/). Multiple alignments of amino acids and coding sequences were performed online using Clustal Omega40 (http://www.ebi.ac.uk/Tools/msa/clustalo). Alignments were further refined manually and used for downstream analysis with the aid of statistical, phylogenetic programs and for visualization41 (http://www.jalview.org). The deduced protein sequences were examined for potential posttranslational modifications using NetPhos 2.0 (www.cbs.dtu.dk/services/NetPhos).
Phylogenetic analysis, tests for positive selection
Phylogenetic trees were drawn using MEGA642. Mean pairwise amino acid and nucleotide distances were also calculated using MEGA6 according to the Poisson and T92 + G models. Bootstrap values were calculated from at least 1,000 replications.
Sequences were analyzed for deviations from neutrality with the McDonald–Kreitman22 test using DnaSP43. Analysis of the ratios of nonsynonymous (Ka) to synonymous (Ks) substitutions (ω) was performed using DnaSP. The statistical significance of positive selection was calculated by Fisher’s exact test as implemented in MEGA6. MEME and SLAC (with a significance level cutoff of 0.05 and 0.1, correspondingly) analyses were performed through the Datamonkey server (http://datamonkey.org/).
Data availability
The sequence data described are available in GenBank under accession numbers MG384763–MG384788.
References
De Rop, V., Padeganeh, A. & Maddox, P. S. CENP-A: the key player behind centromere identity, propagation, and kinetochore assembly. Chromosoma 121, 527–538 (2012).
Comai, L., Maheshwari, S. & Marimuthu, P. A. Plant centromeres. Curr. Opin. Plant. Biol. 36, 158–167 (2017).
Allshire, R. C. & Karpen, G. H. Epigenetic regulation of centromeric chromatin: old dogs, new tracks? Nat. Rev. Genet. 9, 923–937 (2008).
Maheshwari, S. et al. Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids. PLoS Genet. 11(1), e1004970, https://doi.org/10.1371/journal.pgen.1004970 (2015).
Neumann, P. et al. Centromeres off the hook: massive changes in centromere size and structure following duplication of CenH3 gene in Fabeae species. Mol. Biol. Evol. 32, 1862–1879 (2015).
Vermaak, D., Hayden, H. S. & Henikoff, S. Centromere targeting element within the histone fold domain of Cid. Mol. Cell Biol. 22, 7553–7561 (2002).
Malik, H. S. & Henikoff, S. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157, 1293–1298 (2001).
Talbert, P. B., Masuelli, R., Tyagi, A. P., Comai, L. & Henikoff, S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14, 1053–1066 (2002).
Hirsch, C. D., Wu, Y., Yan, H. & Jiang, J. Lineage-specific adaptive evolution of the centromeric protein CENH3 in diploid and allotetraploid Oryza species. Mol. Biol. Evol. 26, 2877–2885 (2009).
Finseth, F. R., Dong, Y., Saunders, A. & Fishman, L. Duplication and adaptive evolution of a key centromeric protein in Mimulus, a genus with female meiotic drive. Mol. Biol. Evol. 32, 2694–2706 (2015).
Zhong, C. X. et al. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14, 2825–2836 (2002).
Nagaki, K. et al. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36, 138–145 (2004).
Yuan, J., Guo, X., Hu, J., Lv, Z. & Han, F. Characterization of two CENH3 genes and their roles in wheat evolution. New Phytol. 206, 839–851 (2015).
Sanei, M., Pickering, R., Kumke, K., Nasuda, S. & Houben, A. Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc. Nat. Acad. Sci. USA 108, 498–505 (2011).
Ishii, T. et al. The differential loading of two barley CENH3 variants into distinct centromeric substructures is cell type and development-specific. Chromosome Res. 23, 277–284 (2015).
Tang, Z. X. et al. Secale in Wild Crop Relatives: Genomic and Breeding Resources: Cereals (ed Kole, C.) 367–396 (Springer-Verlag Berlin Heidelberg, https://doi.org/10.1007/978-3-642-14228-4_8, (2011).
Sencer, H. A. & Hawkes, J. G. On the origin of cultivated rye. Biol. J. Linn. Soc. 13, 299–313 (1980).
Frederiksen, S. & Petersen, G. A taxonomic revision of Secale (Triticeae, Poaceae). Nord. J. Bot. 18, 399–420 (1998).
Chikmawati, T., Schovmand, B. & Gustafson, J. P. Phylogenetic relatioships in Secale revealed by amplified fragment length polymorphism. Genome 48, 792–801 (2005).
Chebotar, S. et al. Molecular studies on genetic integrity of open-pollinating species rye (Secale cereale L.) after long-term genebank maintenance. Theor. Appl. Genet. 107, 1469–1476 (2003).
Bell, G. D. H. The comparative phylogeny of the temperate cereals in Essays on Crop PlantEvolution (ed Hutchinson, J.) 70–102 (Cambridge Univ. Press, London 1965).
McDonald, J. H. & Kreitman, M. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351, 652–654 (1991).
Black, B. E. et al. Structural determinants for generating centromeric chromatin. Nature 430, 578–582 (2004).
Martis, M. et al. Reticulate Evolution of the Rye Genome. Plant Cell 25, 3685–3698 (2013).
Keren, H., Lev-Maor, G. & Ast, G. Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 11, 345–355 (2010).
Murat, F., Pont, C. & Salse, J. Paleogenomics in Triticeae for translational research. Curr. Plant Biol. 1, 34–39 (2014).
Karimi-Ashtiyani, R. et al. Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc. Nat. Acad. Sci. USA 112, 11211–11216 (2015).
Blattner, F. R. Phylogenetic analysis of Hordeum (Poaceae) as inferred by nuclear rDNA ITS sequences. Mol. Phylogenet. Evol. 33, 289–299 (2004).
Middleton, C. P. et al. Sequencing of chloroplast genomes from wheat, barley, rye and their relatives provides a detailed insight into the evolution of the Triticeae tribe. PLoS ONE 9(3), e85761 (2014).
Chalupska, D. et al. Acc homoeoloci and the evolution of wheat genomes. Proc Natl Acad Sci USA 105, 9691–9696 (2008).
Martis, M. et al. Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc Natl Acad Sci USA 109, 13343–13346 (2012).
Khush, G. S. Cytogenetic and evolutionary studies in Secale. III. Cytogenetics of weedy ryes and origin of cultivated rye. Econ. Bot. 17, 60–71 (1963).
Stutz, H. C. On the origin of cultivated rye. Am. J. Bot. 59, 59–70 (1972).
Escobar, J. S. et al. Multigenic phylogeny and analysis of tree incongruences in Triticeae (Poaceae). BMC Evol. Biol. 11, 181, https://doi.org/10.1186/1471-2148-11-181 (2011).
Tachiwana, H., Kagawa, W. & Kurumizaka, H. Comparison between the CENP-A and histone H3 structures in nucleosomes. Nucleus 3, 6–11 (2012).
Kriventseva, E. V. et al. Increase of functional diversity by alternative splicing. Trends Genet. 19, 124–128 (2003).
Pingault, L. et al. Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome. Genome Biol. 16, 29, https://doi.org/10.1186/s13059-015-0601-9 (2015).
Nagaki, K., Kashihara, K. & Murata, M. Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula nivea. Plant Cell 17, 1886–1893 (2005).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Sievers, F. et al. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539, https://doi.org/10.1038/msb.2011.75 (2011).
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Librado, P. & Rozas, J. DnaSPv5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009).
Acknowledgements
We thank B. Kilian for the provided Aegilops and Triticum accessions used in this work. This research was financially supported by Russian Fundamental Scientific Research Program on the project 0310-2016-0005, the Russian Foundation for Basic Research (grant 17-04-00748a) and the Deutsche Forschungsgemeinschaft (DFG, HO 1779/15-1).
Author information
Authors and Affiliations
Contributions
E.V.E., S.S.G., Y.A.L. performed all experimental work, E.A.E. together with E.V.E. performed the bioinformatics analysis. A.V.V., A.H. and E.V.E. wrote the manuscript. A.H. and A.V.V. provided guidance. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Evtushenko, E.V., Elisafenko, E.A., Gatzkaya, S.S. et al. Conserved molecular structure of the centromeric histone CENH3 in Secale and its phylogenetic relationships. Sci Rep 7, 17628 (2017). https://doi.org/10.1038/s41598-017-17932-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-17932-8
This article is cited by
-
Uncovering natural allelic and structural variants of OsCENH3 gene by targeted resequencing and in silico mining in genus Oryza
Scientific Reports (2023)
-
CENH3 mediated haploid induction: application and future perspectives in crop plants
Horticulture, Environment, and Biotechnology (2023)
-
Decoding allelic diversity, transcript variants and transcriptional complexity of CENH3 gene in Brassica oleracea var. botrytis
Protoplasma (2023)
-
The origin and evolution of a two-component system of paralogous genes encoding the centromeric histone CENH3 in cereals
BMC Plant Biology (2021)
-
Unequal contribution of two paralogous CENH3 variants in cowpea centromere function
Communications Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.