Abstract
A series of 5-aryl-4-(4-arylpiperazine-1-carbonyl)-2H-1,2,3-triazol derivatives were designed as potential microtubule targeting agents. The regioselective alkylation of 5-aryl-4-(4-arylpiperazine-1-carbonyl)-2H-1,2,3-triazole was predicted by computations and confirmed by an unambiguous synthetic route. The antiproliferative activity of the synthesized compounds was tested in vitro using three human cancer cell lines and some compounds exhibited significant antiproliferative activity, which suggested the reasonability of introduction of the 1,2,3-triazole fragment. Among them, compound 7p showed highest activity with the IC50 values at nanomolar level towards all three cell lines, which were comparable to the positive control, CA-4. Tubulin polymerization assay, immunofluorescence studies, cell cycle analysis and competitive tubulin-binding assay strongly proved that 7p is a colchicine binding site inhibitor of tubulin. Thus, 7p was identified as a promising drug candidate for further development of colchicine binding site inhibitors.
Similar content being viewed by others
Introduction
Microtubules play crucial roles in cellular structure and various aspects of cell functions. The polymerization dynamics of microtubules are vital during mitosis, which is regarded as a promising target for cancer therapy since it can be profoundly affected by even small molecules1. Microtubule targeted agents can be classified into two major classes: microtubule-stabilizing agents which stimulate microtubule polymerization, such as paclitaxel (taxol); microtubule-destabilizing agents which inhibit polymerization of tubulin, include vinblastine and colchicine (1, Fig. 1).
Although colchicine can significantly inhibit the polymerization of tubulin, its therapeutic application is limited by the low therapeutic index. To overcome the disadvantages of colchicine, it is desired to find novel compounds that target at the colchicine binding site and exert their biological effects by inhibiting tubulin assembly and suppressing microtubule formation. Those compounds are classified as colchicine binding site inhibitors (CBSIs)2. Over decades, a large number of CBSIs have been reported, such as combretastatin A-4 (CA-4, 2)3, ABT-751 (3)4 and TN16 (4)5. XRP44X (5)6 and other piperazine based CBSIs reported by Wasylyk and Chopra et al.7, showed potential effects on the inhibition of microtubules polymerization and the morphology of the actin cytoskeleton.
1,2,3-Triazoles attracted our attention since tremendous compounds containing this fragment have been reported to show potent antitumor activity8,9. For example, 4,5-disubstituted 1,2,3-triazole (6) was designed as an analogue of CA-410, which showed low IC50 value at nanomolar level. Therefore, we designed a series of 5-aryl-4-(4-arylpiperazine-1-carbonyl)-2H-1,2,3-triazole derivatives (7) and 2-alkyl-5-aryl-4-(4-arylpiperazine-1-carbonyl)-2H-1,2,3-triazole derivatives (8) by replacing the pyrazole of XRP44X with 1,2,3-triazole. Meanwhile, those compounds showed well performance on molecular modeling studies. To access 2-alkylated derivatives, the regioselective alkylation of 5-aryl-4-(4-arylpiperazine-1-carbonyl)-2H-1,2,3-triazole was investigated by computational before actual synthesis. Antiproliferative activity of all compounds was evaluated, and several compounds with considerable activity had been found. The compound with the highest antiproliferative activity was thought as a colchicine binding site inhibitor by further investigation.
Result and Discussion
Molecular design
By replacing the pyrazole of XRP44X with 1,2,3-triazole, 7b was designed. XRP44X possess a methyl group on the five-member ring, and previously study suggested that the methyl group was critical for bioactivity11. Thus, 8b that possess a methyl group on N-2 of 1,2,3-triazole was designed.
We further carried out the molecular docking for 7b and 8b (Fig. 2). Both of them were superimposed well with 5. The carbonyl group contributes a hydrogen bond to β-Ala317 for these three ligands. Although 7b lacked a methyl group, its 2-H on 1,2,3-triazoe ring could contribute a hydrogen bond to β-Thr353. It suggested the desired compounds are promising for inhibition of tubulin polymerization and a series of derivates (7, 8) were further designed and synthesized for biological investigation.
Chemistry
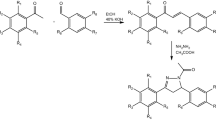
All desired compounds (7, 8) were obtained by following the synthetic route outlined in Fig. 3. Commercially available arylacetylene derivatives (9) were used as the starting materials, which were transferred into arylpropiolaldehyde derivatives (10) by the formulation of acetylides with DMF12. Then, azide–alkyne 1,3-dipolar cycloadditions was carried out for obtaining 5-aryl-2H-1,2,3-triazole-4-carbaldehyde (11), followed by oxidation into 5-aryl-2H-1,2,3-triazole-4-carboxylic acid (12) with hydrogen peroxide13. Finally, amidations were carried by EDCI and HOBt to generate the desired compounds 7a-7t.
Synthetic route of all compounds. Reagents and conditions: (a) 1) n-BuLi (2.5 M in hexane), THF, −40 °C, 30 min; 2) DMF, −40 °C, 3) r.t., 1 h; (b) NaN3, DMSO, r.t., 30 min; (c) H2O2, KOH, MeOH, 30 min; (d) arylpiperazine, EDCI, HOBt, ethyl acetate, r.t., overnight; (e) (R3)2SO4, K2CO3, acetone, r.t., 1~3 h. or R3Br, K2CO3, KI, acetone, r.t., 2~4 h.
Direct alkylation of 7 may produce three different products since the tautomerism of 1,2,3-triazole. Previously studies suggested that similar substrates14,15,16 prone to generate N-2 alkylated products. In our case, density functional theory (DFT) computations were carried out for three tautomers of 7 to further predict the alkylated products. It showed that 2H-tautomers always have lowest energy in acetone. The results of computations were obtained for the all 7 substrates, as shown in Fig. 4. The energy of 2H-tautomers, lower than 1H- and 3H-tautomers, suggested N-2 alkylation products would be the principal products.
To further verify the structures of the alkylated products, we synthesized several representative compounds by an unambiguous synthetic route (Fig. 5). 4,5-Dibrome-2-methyl-2H-1,2,3-triazole (13) was synthesized by the reported methods17. Then, Grignard reaction was carried out to yield 14 18. 5-Aryl was installed by Suzuki coupling to obtain 15 19, followed by oxidation and amidation to generate 8g, 8h, and 8i. These three compounds were proven as identical as the principal products obtained from the synthetic route in Fig. 3.
In vitro antiproliferative activity and structure-activity relationship
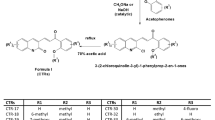
The synthesized compounds (7, 8) were investigated for their antiproliferative activity against cancer cells by MTT assay. Three human carcinoma cell lines including gastric adenocarcinoma SGC-7901 cells, lung adenocarcinoma A549 cells and fibrosarcoma HT-1080 cells were used. Two of the most well-known CBSIs, colchicine (1) and CA-4 (2) were evaluated as positive controls (Table 1).
At first, the effect of alkyl on N-2 of 1,2,3-triazole (R3) was evaluated. All methylated compounds showed no significant change on the activity compared with corresponding compounds without methyl (7a vs. 8a, 7b vs. 8b, 7h vs. 8g, 7j vs. 8h, 7l vs. 8k, ect.). These results indicated that methyl on N-2 is unnecessary, however, activity severely declined as bulky alkyl group was introduced (8c-8f vs. 8b, 8j vs. 8i). Then, the effect of substituents on the aryl ring linked to the piperazine fragment (R2) was investigated. Compared with 7b (R2 = 3-chloro), both 7a (R2 = H) and 7c (R2 = 4-methyl) showed dramatically decreased activity, while 7d (R2 = 3-methoxy) also show considerable activity, which indicated steric factor is crucial on R2 rather than electronegativity factor. Furthermore, di-meta-substituted compounds showed increased activity, for instance, 7j (R2 = 3,5-dimethoxy) showed evident higher activity than 7d (R2 = 3-methoxy). Finally, we investigated the effect of substituents on the aryl ring directly linked to 1,2,3-triazole core (R1) and found that the position and electronegativity had minimum effects on the activity. Among the designed compounds, 7p (R1 = 2-chloro) showed an IC50 value of 5~52 nM towards all three cell lines, which was comparable to CA-4 (7~48 nM).
Tubulin polymerization
7p, the compound with the highest antiproliferative activity, was assessed its effect on the tubulin polymerization in vitro. CA-4 (2) was used as a positive control and Taxol as a negative control. As shown in Fig. 6, 7p exhibited a dose-dependent inhibition of tubulin polymerization. The inhibitory activity of 7p towards tubulin polymerization (IC50 value, 2.06 µM) was more potent than the positive control CA-4.
Immunofluorescence studies
To visualize the effects of 7p toward microtubule network, immunofluorescence studies were carried out. SGC-7901 cell lines were treated for 24 h with CA-4 and 7p at their IC50. As shown in Fig. 7, both CA-4 and 7p lead to radical changes in cell morphology of structured microtubules. These results indicated that 7p can destabilize the microtubules of SGC-7901 cells.
Immunofluorescence studies. CA-4 (48.1 nM) and 7p (52.0 nM) induced depolymerization of the microtubule networks of SGC-7901 cancer cells, while untreated cells also used as control. After 24 h, cells were fixed and stained with anti-α-tubulin-FITC specific antibodies followed by DAPI. Microtubules and unassembled tubulin are shown in green, and nuclei, which were stained with DAPI, are shown in blue.
Cell cycle analysis
To evaluate the effects of 7p on the cell mitosis, we have analyzed the effect of 7p on the cell cycle of SGC-7901 cells by flow cytometry (Fig. 8). CA-4 was also examined as a positive control. Similar as CA-4, 7p arrested the cell cycle in G2/M phase significantly, increasing the percentage of cells in G2/M phase at 12 h and 24 h. Then, population of G2/M declined while the population of Sub-G1 raised, that indicated part of cells underwent apoptosis.
Competitive tubulin-binding assay
To confirm that 7p binds to colchicine site of tubulin, we further assessed the competition between 7p and colchicine for binding to tubulin via competitive binding assays (Fig. 9). CA-4 (2) and Taxol were used as the positive control and negative control, respectively20. The fluorescence of a colchicine-tubulin complex was reduced in the presence of 7p in a dose-dependent manner, which indicated that 7p binds at the colchicine binding site.
In this study, we designed and synthesized a series of 5-aryl-4-(4-arylpiperazine-1-carbonyl)-2H-1,2,3-triazole derivatives. The N-2 regioselective alkylation of 1,2,3-triazole was predicted by computations and confirmed by an unambiguous synthetic route. Most compounds showed potent antiproliferative activity at sub-micromolar or nanomolar, which suggested the reasonability of introduction of 1,2,3-triazole fragment. 7p displayed highest activity against three human carcinoma cell lines, which was comparable to the positive control, CA-4. 7p also highly inhibited tubulin polymerization in vitro with IC50 value was 2.06 µM. The immunofluorescence studies observed that 7p induced depolymerization of the microtubule networks. Cell cycle analysis displayed evident G2/M arrest and induction of apoptosis. Furthermore, 7p could compete with colchicine in tubulin binding site. All those experiments suggested that 7p should be a colchicine binding site inhibitor of tubulin.
Methods
Reagents and equipment
All reagents were commercially available and were used without further purification. The silica gel plate (HSGF-254) and silica gel (H, 200–300 mesh) from Yantai Jiangyou silicone Development Co., Ltd. was used for preparative TLC and column chromatography, respectively. Visualization was made with UV light (254 nm and 365 nm). Mass spectra (MS) were obtained from Agilent Co. Ltd. on an Agilent 1100-sl mass spectrometer with an electrospray ionization source. 1H-NMR and 13C-NMR spectra were measured in CDCl3 or d6-DMSO with TMS as the internal reference on a Bruker AVANCE spectrometer operating at 400 MHz or 600 MHz (1H at 400 or 600 MHz, 13C at 100 MHz).
Chemistry
The detailed information is in Supplementary Information.
Cell line and culture conditions
The human gastric adenocarcinoma SGC-7901 cells, lung adenocarcinoma A549 cells and fibrosarcoma HT-1080 cells were purchased from Shanghai Institute of Cell Resources Center of Life Science (Shanghai, China). All cells were cultured in RPMI-1640 medium (Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone, USA), streptomycin and penicillin at 37 °C in humidified atmosphere with 5% CO2.
MTT assay
MTT assays were used to measure the cell viability after treatment. Briefly, 4~10 × 103 cells/well were seeded in 96-well plates (Corning, NY, USA), cultured for 24 h, and treated with various concentrations of compounds for 72 h or incubated with SNP (10 mM) or Haemoglobin (10 mM) for 2 h, then treated with 4d (25 nM) for the indicated times. The DMSO concentration was kept below 0.05% in cell culture so it did not effect on cell growth. Then, MTT solution (5 mg/mL in PBS) was added (20 mL/well) to each well and incubated for another 4 h at 37 °C. The purple formazan crystals were dissolved in 100 mL dimethyl sulfoxide, and the plates were read on a plate reader (MK3, Thermo, German) at 492 nm. Experiments were repeated three times.
Tubulin polymerization assay
In vitro tubulin polymerization assays were conducted as described in the manufacturer’s protocol (Cytoskeleton, Cat.#BK011P) using 96-well plates. Briefly, 7p, CA-4 or Taxol were incubated with purified porcine tubulin (2 mg/mL) and buffer containing 10% glycerol and 1 mM GTP at 37 °C, and the effects of these compounds on tubulin polymerization were monitored kinetically for 82 min using a plate reader (Biotek Synergy HT, Winoo-skin, VT, USA). The increase in the relative fluorescence unit (RFU) was measured at an excitation of 340 ± 20 nm and emission of 415 ± 20 nm every minute. Experiments were repeated three times. The detail information of IC50 calculation is in Supplementary Information.
Immunofluorenscence assay
One thousand SGC-7901 cells were cultured for 24 h in 96-well plates, followed by treatment with 7p, CA-4 or 0.05% DMSO for the indicated times. After treatment, the cells were fixed with 4% formaldehyde in PBS, washed twice with PBS and permeabilised with 0.1% (v/v) Triton X-100 in PBS for 5 min. Then, cells were blocked with 5% bovine serum albumin (BSA) in PBS for 10 min. Microtubules were detected using a monoclonal anti-a-tubulin antibody diluted 1:50 in PBS overnight at 4 °C and a FITC conjugated secondary antibody diluted 1:200 in PBS for 1 h at 37 °C. After nuclear counter staining with DAPI (1 mg/mL), the cells were imaged using a fluorescence microscope. Experiments were repeated three times.
Cell cycle analysis
One million SGC-7901 cells were incubated with 7p (52 nM), CA-4 (48 nM) or 0.05% DMSO (control) for the indicated times. The cells were collected by centrifugation, washed with PBS and fixed in ice-cold 70% ethanol. The fixed cells were harvested by centrifugation and resuspended in 500 mL of PBS containing 50 mg/mL RNase. After 30 min incubation at 37 °C, cells were stained with 50 mg/mL PI at 4 °C in the dark for 30 min. Then, the samples were analysed by FACS can flow cytometry (BectoneDickinson, Franklin Lakes, NJ, USA). Experiments were repeated at least three times.
Competitive tubulin-binding assay
7p, CA-4 and Taxol were carried out at various concentrations containing 5 μM of tubulin and colchicine for 60 min at 37 °C. Fluorescence values are normalized to DMSO (control), F/F0 represents inhibition rate (IR = F/F0) whereas F0 refers to fluorescence of the 5 μM colchicine–tubulin complex, and F describes the fluorescence of a given concentrations (1.5 μM, 5 μM and 15 μM) of 7p, CA-4 and Taxol competition with the 5 μM colchicine–tubulin complex.
DFT computations
The molecules was prepared by Avogadro (Version 1.20)21, and optimized by molecular dynamic forcefield MMFF9422,23,24,25,26. Then, all molecules were fully optimized by DFT basic set B3LYP/6-31 G(d) on Gaussian 0927. Solvent effects were computed using the CPCM28,29 model. After fully optimization, the energy of three tautomer was obtained for compare. The raw data with unit a.u. are transformed to kJ/mol (1 a.u. = 2625.5 kJ/mol). All date are provided on Supplementary Information.
Molecular modeling studies
Molecular docking was carried out by Discovery Studio 3.5 with LibDock program. The 3D structure of 3HKC in docking study was downloaded from Protein Data Bank. The docking poses were selected according to previously studies11.
References
Jordan, M. A. & Wilson, L. Microtubules as a target for anticancer drugs. Nat Rev Cancer 4, 253–265, https://doi.org/10.1038/nrc1317 (2004).
Lu, Y., Chen, J., Xiao, M., Li, W. & Miller, D. D. An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm Res 29, 2943–2971, https://doi.org/10.1007/s11095-012-0828-z (2012).
Pettit, G. R. et al. Antineoplastic Agents. 291. Isolation and Synthesis of Combretastatins A-4, A-5, and A-6. Journal of Medicinal Chemistry 38, 1666–1672, https://doi.org/10.1021/jm00010a011 (1995).
Risinger, A. L. et al. ELR510444, a novel microtubule disruptor with multiple mechanisms of action. J Pharmacol Exp Ther 336, 652–660, https://doi.org/10.1124/jpet.110.175331 (2011).
Roach, M. C. & Luduena, R. F. The effect of TN-16 on the alkylation of tubulin. Biochemical and Biophysical Research Communications 129, 200–205, https://doi.org/10.1016/0006-291x(85)91422-6 (1985).
Wasylyk, C. et al. Inhibition of the Ras-Net (Elk-3) pathway by a novel pyrazole that affects microtubules. Cancer Res 68, 1275–1283, https://doi.org/10.1158/0008-5472.CAN-07-2674 (2008).
Chopra, A., Anderson, A. & Giardina, C. Novel Piperazine-based Compounds Inhibit Microtubule Dynamics and Sensitize Colon Cancer Cells to Tumor Necrosis Factor-induced Apoptosis. Journal of Biological Chemistry 289, 2978–2991, https://doi.org/10.1074/jbc.M113.499319 (2014).
Dheer, D., Singh, V. & Shankar, R. Medicinal attributes of 1,2,3-triazoles: Current developments. Bioorg Chem 71, 30–54, https://doi.org/10.1016/j.bioorg.2017.01.010 (2017).
Feng, D. et al. Synthesis and antiproliferative activity of 2-aryl-4-(3,4,5-trimethoxybenzoyl)-1,2,3-triazol derivatives as microtubule-destabilizing agents. RSC Adv. 7, 29103–29111, https://doi.org/10.1039/c7ra02720f (2017).
Madadi, N. R. et al. Synthesis and biological evaluation of novel 4,5-disubstituted 2H-1,2,3-triazoles as cis-constrained analogues of combretastatin A-4. Eur J Med Chem 103, 123–132, https://doi.org/10.1016/j.ejmech.2015.08.041 (2015).
Choi, M. J. et al. Synthesis and biological evaluation of aryloxazole derivatives as antimitotic and vascular-disrupting agents for cancer therapy. J Med Chem 56, 9008–9018, https://doi.org/10.1021/jm400840p (2013).
Journet, M., Cai, D., Kowal, J. J. & Larsen, R. D. Highly efficient and mild synthesis of variously 5-substituted-4-carbaldehyde-1,2,3-triazole derivatives. Tetrahedron Letters 42, 9117–9118, https://doi.org/10.1016/s0040-4039(01)01923-2 (2001).
Cong, Z. Q., Wang, C. I., Chen, T. & Yin, B. Z. Efficient and Rapid Method for the Oxidation of Electron‐Rich Aromatic Aldehydes to Carboxylic Acids Using Improved Basic Hydrogen Peroxide. Synthetic Communications 36, 679–683, https://doi.org/10.1080/00397910500408456 (2006).
Trofimov, B. et al. Click Chemistry with 2-Ethynyl-4,5,6,7-tetrahydroindoles: Towards Functionalized Tetrahydroindole-Triazole Ensembles. Synthesis 45, 678–682, https://doi.org/10.1055/s-0032-1318149 (2013).
Chen, Y., Liu, Y., Petersen, J. L. & Shi, X. Conformational control in the regioselective synthesis of N-2-substituted-1,2,3-triazoles. Chem Commun (Camb), 3254–3256, https://doi.org/10.1039/b805328f (2008).
Chen, B. et al. Metal- and Base-Free Three-Component Reaction of Ynones, Sodium Azide, and Alkyl Halides: Highly Regioselective Synthesis of 2,4,5-Trisubstituted 1,2,3-NH-Triazoles. Synlett 2010, 1617–1622, https://doi.org/10.1055/s-0030-1258086 (2010).
Wang, X. J., Zhang, L., Krishnamurthy, D., Senanayake, C. H. & Wipf, P. General solution to the synthesis of N-2-substituted 1,2,3-triazoles. Org Lett 12, 4632–4635, https://doi.org/10.1021/ol101965a (2010).
Krasovskiy, A., Straub, B. F. & Knochel, P. Highly efficient reagents for Br/Mg exchange. Angew Chem Int Ed Engl 45, 159–162, https://doi.org/10.1002/anie.200502220 (2005).
Miyaura, N., Yamada, K. & Suzuki, A. A new stereospecific cross-coupling by the palladium-catalyzed reaction of 1-alkenylboranes with 1-alkenyl or 1-alkynyl halides. Tetrahedron Letters 20, 3437–3440, https://doi.org/10.1016/s0040-4039(01)95429-2 (1979).
Liu, Z. L. et al. A novel sulfonamide agent, MPSP-001, exhibits potent activity against human cancer cells in vitro through disruption of microtubule. Acta Pharmacol Sin 33, 261–270, https://doi.org/10.1038/aps.2011.156 (2012).
Hanwell, M. D. et al. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminform 4, 17, https://doi.org/10.1186/1758-2946-4-17 (2012).
Halgren, T. A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. Journal of Computational Chemistry 17, 490–519, https://doi.org/10.1002/(sici)1096-987x(199604)17:5/6<490::aid-jcc1>3.0.co;2-p (1996).
Halgren, T. A. Merck molecular force field. II. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions. Journal of Computational Chemistry 17, 520–552, https://doi.org/10.1002/(sici)1096-987x(199604)17:5/6<520::aid-jcc2>3.0.co;2-w (1996).
Halgren, T. A. Merck molecular force field. III. Molecular geometries and vibrational frequencies for MMFF94. Journal of Computational Chemistry 17, 553–586, https://doi.org/10.1002/(sici)1096-987x(199604)17:5/6<553::aid-jcc3>3.0.co;2-t (1996).
Halgren, T. A. & Nachbar, R. B. Merck molecular force field. IV. conformational energies and geometries for MMFF94. Journal of Computational Chemistry 17, 587–615, https://doi.org/10.1002/(sici)1096-987x(199604)17:5/6<587::aid-jcc4>3.0.co;2-q (1996).
Halgren, T. A. Merck molecular force field. V. Extension of MMFF94 using experimental data, additional computational data, and empirical rules. Journal of Computational Chemistry 17, 616–641, https://doi.org/10.1002/(sici)1096-987x(199604)17:5/6<616::aid-jcc5>3.0.co;2-x (1996).
Gaussian 09 (Gaussian, Inc., Wallingford CT, 2009).
Barone, V. & Cossi, M. Quantum Calculation of Molecular Energies and Energy Gradients in Solution by a Conductor Solvent Model. The Journal of Physical Chemistry A 102, 1995–2001, https://doi.org/10.1021/jp9716997 (1998).
Cossi, M., Rega, N., Scalmani, G. & Barone, V. Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J Comput Chem 24, 669–681, https://doi.org/10.1002/jcc.10189 (2003).
Acknowledgements
We gratefully acknowledge the National Natural Science Foundation of China (81673293, 81502932, 81602969, 81641128), Program for Liaoning Excellent Talents in University (LR2013046) and State Key Laboratory of Natural Medicines and Active Substance (No. GTZK201603) for their generous financial support.
Author information
Authors and Affiliations
Contributions
Yue Wu, D. Feng, M. Gao, P. Yan, Z. Gu performed the experiments. Yue Wu, D. Feng, Z. Wang, Q. Guan, D. Zuo, K. Bao, J. Sun, Yingliang Wu, W. Zhang analyzed, interpreted the data and wrote the paper.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, Y., Feng, D., Gao, M. et al. Design and synthesis of 5-aryl-4-(4-arylpiperazine-1-carbonyl)-2H-1,2,3-triazole derivatives as colchicine binding site inhibitors. Sci Rep 7, 17120 (2017). https://doi.org/10.1038/s41598-017-17449-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-17449-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.