Abstract
Detecting how historical and contemporary factors contribute to genetic divergence and genetic structure is a central question in ecology and evolution. We examine this question by intergrating population genetics with ecological niche modelling of Litsea auriculata (Lauraceae), which is endangered and native to east China. Geographical and environmental factors including climatic fluctuations since the last glacial maximum (LGM) have also contribute to population demography and patterns of genetic structure. L. auriculata populations underwent expansion after divergence and dramatically decreased to the current small size with relative population bottlenecks due to climate changes. Populations separated by physical geographical barrier including geographic distance and Yangtze River, as a result contemporary gene flow among L. auriculata populations showed drastic declines in comparison with historical gene flow, resulting in a high level of population divergence. Thus, patterns of genetic structure of L. auriculata can result from both geographic and environmental factors including climate changes. This information is helpful in forming conservation strategies for L. auriculata in China.
Similar content being viewed by others
Introduction
Litsea auriculata (Lauraceae) is restricted to eastern China, occurring within a limited range from 30°21′–33°33′N and 112°1′–119°26′E. The current populations of L. auriculata are only scattered and distributed in a few mountain locations, mainly due to habitat loss1. This species is listed as endangered in the Chinese Plant Red Book1. Most extant populations are small and consist of less than 40 individuals. Previous studies of L. auriculata focused on its habitat characteristics2 and seedling biology3,4, but nothing is known about its population genetics. Determining effective conservation actions for this endemic species often requires knowledge of contemporary population structure, along with an understanding of the historical processes by which populations were shaped. Although its historical geographic distribution is unknown, extant Eastern populations are thought to represent a remnant of a formerly more widespread distribution. Therefore, these geographic discrete populations of L. auriculata represent an ideal system to elucidate the roles of historical vs. contemporary processes in determining the current patterns of disconnected population remnants and genetic variation5.
Pleistocene climatic cycling is one of the most important drivers of contemporary diversity and genetic structure in many temperate species and communities6. The distributions of various organisms experienced concomitant expansions and contractions caused by the global cyclical cooling-warming events in the Quaternary glacial cycles6. Therefore, these cyclical events have played a dominant role in shaping the current distribution and genetic structure of many temperate species in the Northern Hemisphere7. East Asia has harboured the most diverse temperate flora in the world and was the most important glacial refuge for “Tertiary relics” throughout the Quaternary ice-age cycles. This region was never covered by massive ice-sheets during these glacial periods8; however, it experienced severe climatic oscillations throughout the Quaternary, with a dramatic effect on the evolution and distribution of both plants and animals9. Apart from historical factors such as Pleistocene range fragmentation and past demographic changes, more recent human-caused range fragmentation and isolation have also contributed to contemporary patterns of genetic variation within and between populations of species10,11. Evaluation of the relative influences of historical and contemporary elements on genetic diversity of forest trees has been a major topic in forest conservation for many years5,12.
Various approaches have been used to evaluate these influences, including using different types of analytical methods to test historical and contemporary gene flow or effective population sizes (N e) based on extant samples13,14. Recently, a very powerful and flexible approach, Approximate Bayesian computation (ABC), has been used to estimate demographic and historical parameters (e.g., effective population size, divergence time) and quantitatively compare alternative scenarios15. This approach can be used to resolve many important questions in ecology and evolutionary biology15. In addition, it should be emphasized that molecular markers with different rates of substitution can capture the signatures of historical and contemporary processes and could therefore help to determine historical demographic patterns16,17,18. Nuclear DNA (nDNA) is recombined, bi-parentally inherited and affected by both pollen and seeds, which can complicate attempts to reveal species phylogeographic, evolutionary history and gene flow patterns19. By contrast, chloroplast DNA (cpDNA) in angiosperms is usually maternally inherited, and thus, phylogeographical patterns revealed by cpDNA-based markers are solely due to seed dispersal19,20. In addition, the mutation rates in cpDNA are low and cpDNA markers are not affected by recombination21,22. Therefore, comparisons of genetic structure detected by nDNA and cpDNA marker analyses can also assess differences between pollen and seed flow dynamics. Furthermore, phylogeographic studies has been integrated ecological niche modelling (ENM), which provided relevant information about the geographic distributions of species or genetic lineages and their relationship with the spatial distribution of genetic diversity23,24.
Microsatellites (SSRs) are known to be highly polymorphic, codominant markers that have been widely used in studies of population genetics25,26. In this study, we first to analyse the population genetic structure and phylogeographic history of L. auriculata using nuclear (nSSR) and chloroplast (cpSSR) microsatellite markers to assess historical and contemporary evolutionary processes, investigate the causes of the endangerment of this species and provide basic guidelines for its conservation. Second, we also use approaches for genetic variation to detect changes in population size over different time scales in the present study. Third, we used ENM to infer the climatic potential niche of L. auriculata under current and Last Glacial Maximum (LGM) conditions. The aims of this study were as follows: (i) to evaluate the genetic structure of L. auriculata; (ii) to assess historical and contemporary evolutionary processes in shaping the genetic structure; (iii) to infer the species population demography using an ABC approach; (iv) to investigate how climatic and geographical variation over space and time to shape the patterns of genetic structure; and (v) to propose conservation and management of L. auriculata populations based on genetic structure and evolutionary history.
Results
Genetic diversity and network
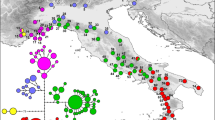
The results of sequencing indicated that variations in allele size were due to the increase or decrease in the number of repeats. The seven cpSSR loci yielded a total of 20 alleles and 30 different haplotypes (H1 to H30), among 231 individuals of L. auriculata. The number of haplotypes per population yielded 3 to 17 haplotypes in the eight populations, and the mean number of haplotypes for the eight populations was 6.5. The most polymorphic populations were seen in AY in Anhui province, with 17 haplotypes. The HY population had only 3 haplotypes. The most common haplotypes were H1 (24.7%) and H2 (26.5%), found in all populations, followed by H6 (11.4%) and H7 (11.9%). The haplotype H2 (26.5%) showed the highest frequency in all populations. A high level of genetic diversity at the population level was detected. The number of different alleles (N a) ranged from 1.571 to 2.429, and the number of effective alleles (N E) ranged from 1.112 to 1.590 (Table 1). The average Shannon’s information index (I), diversity (h) and unbiased diversity (uh) values were 0.268 (0.160–0.511), 0.159 (0.088–0.315) and 0.167 (0.090–0.323), respectively (Table 1). Overall, the cpSSR data revealed high within-population diversity (h S = 0.667, v S = 0.698) and total diversity (h T = 0.855, v T = 0.851) in L. auriculata populations.
Median-joining haplotype network reflecting topology and haplotype frequency was constructed for cpSSRs (Fig. 1). The distribution of haplotypes of L. auriculata showed no obvious geographic structure (Fig. 1). These results were further confirmed as the R ST (0.180) was significantly less than G ST (0.221) (P < 0.05), indicating a lack of significant phylogeographic structure in L. auriculata populations.
Map of China showing the sampling locations and geographic distribution of the chloroplast haplotypes found in Litsea auriculata populations [The original map was created by using Google Earth Pro 7.1.2.2041, which was free downloaded from https://google-earth-pro.en.softonic.com/ and the edge of China was drew by using AutoCAD 2010 (https://www.autodesk.com/education/free-software/autocad)]. (a) Pie charts on the map represent the haplotype composition of samples from the corresponding populations. The colour in each chart represents the haplotype, as indicated in the cluster tree; (b) Cluster analysis of 30 haplotypes detected in 231 samples based on the median-joining haplotype network developed using NETWORK 5.0. Haplotype circle sizes are proportional to the frequency of each Haplotype.
Across all nuclear microsatellite loci, a total of 76 alleles were detected, and high levels of gene diversity (H T = 0.657, H S = 0.530) were observed. Genetic diversity at the population level, the mean number of alleles per locus (A) and the allelic richness (A R) for each population ranged from 3.38 to 6.38 and from 3.03 to 4.16, respectively (Table 1). The observed (H O) and expected (H E) heterozygosity for each population ranged from 0.329 to 0.506 and from 0.404 to 0.642, respectively (Table 1). The mean number of alleles and expected heterozygosity for each population were highest in ZD (A = 6.38, H E = 0.642) and lowest in HY (A = 3.38, H E = 0.404). In total, 34 private alleles (P A) were detected within eight populations, and P A ranged from 0.00 to 0.38 in eight populations. The highest P A values were found in AY and HX populations. Inbreeding coefficients (F IS) ranged from 0.098 to 0.271 (Table 1), six of which significantly deviated from zero indicating that most of the populations experienced inbreeding. Effective population size (N e) values for all L. auriculata populations were less than 50 (Table 1). The values of allelic richness (A r) and expected heterozygosity (H E) were significantly negative correlated to the latitude (R 2 = 0.709, P = 0.009 and R 2 = 0.674, P = 0.012; Fig. 2), while there was significantly positive correlation to the longitude (R 2 = 0.743, P = 0.006 and R 2 = 0.712, P = 0.008; Fig. 2).
Genetic structure
The AMOVA tests indicated significant differentiation among all populations. For the cpSSR data, 28.54% of the total variation was attributed to differences among populations, and 71.46% within populations (Table 2). For nSSR, 16.50% of the total variation was attributed to differences among populations, and 83.5% within populations (Table 2). Relatively high genetic differentiation among the eight populations (F ST(c) = 0.285, wright F ST(n) = 0.165) was also observed based on cpSSR and nSSR data, respectively. Pairwise G ST values based on cpSSR data among populations ranged from 0.004 to 0.220. The highest G ST value was found between populations ZQ and AS (G ST = 0.220), and the lowest between populations HJ and HY (G ST = 0.004) (Table 3). Meanwhile, pairwise F ST values based on nSSR data among populations ranged from 0.061 to 0.340. The highest F ST value was observed between populations HX and HY (F ST = 0.340), and the lowest between populations HJ and HY (F ST = 0.061, Table 3). All G ST and F ST estimates were significant (P < 0.05, Table 3) except for that between the two populations HJ and HY, which confirmed that genetic differentiation among the populations was quite high.
The mantel test indicated that no significant correlations between geographic and genetic distance were observed from the cpSSR (y = −3E-05x + 0.1022, R2 = 0.0199, P = 0.241) and nSSR (y = 0.0001x + 0.2229, R2 = 0.0771, P = 0.093) analysis (Fig. 3).
Geographic distribution of the genetic groups detected from STRUCTURE analysis of Litsea auriculata (ΔK = 4) [The original map was created by using Google Earth Pro 7.1.2.2041, which was free downloaded from https://google-earth-pro.en.softonic.com/ and the edge of China was drew by using AutoCAD 2010 (https://www.autodesk.com/education/free-software/autocad)] .
In the STRUCTURE analysis of L. auriculata using eight nSSR loci, the ad hoc statistic ∆K recovered the highest likelihood at K = 4 (Supplementary Fig. S1). All L. auriculata individuals were assigned to four clusters (Fig. 4). Cluster I (pink) contained ZD, ZT and ZQ populations from Tianmu Mountain. Cluster II (red) included AS and HX populations and AS showed evidence of extensive admixture. Cluster III (blue) only contained the AY population and Cluster IV (green) contained the remaining two populations (HJ and HY) from Qinglin Mountain.
ABC-based inferences of population divergence
Among the ten predesigned scenarios in DIYABC analysis (Fig. 5), scenario 1 had the highest posterior probability (0.9767, 95% CI: 0.9745, 0.9788); it was much higher than other scenarios and did not overlap with those obtained for the other scenarios (Table 4, Supplementary Fig. S2). Based on model performance, we discriminated scenario 1 from the other scenarios. This is corroborated by model checking: PCA showed that the observed data point was centred around the cluster of points for the simulated data based on posterior distributions (Supplementary Fig. S3), and no summary statistics showed a significant difference between the observed and simulated data based on posterior distributions (Supplementary Table S1). This suggested that scenario 1 was still fitted to the observed data. Analyses to estimate confidence in scenario choice, based on 500 PODs, indicated that type I and Type II errors for the best-supported scenario 1 were 0.038 and 0.036, respectively; these are low errors. Under scenario 1, posterior mean parameter estimates indicated that the simultaneous divergence occurred 3,010 (95% CI: 519–10 900) generations ago, corresponding to 60,200 years (95% CI: 10,380–218,000 years with a generation time of 20 years). For scenario 1, the median values of the effective population size were 29,600 (95% CI: 6,610–86,900) for N1 (ZT, ZQ and ZD population), 19,600 (95% CI: 3,930–78,100) for N2 (AS and HX population), 14,000 (95% CI: 2,720–70,200) for N3 (AY population), 9,210 (95% CI: 1,720–61,200) for N4 (HJ and HY population), and 3,020 (95% CI: 127–37,900) for NA before the divergence at t3 (Supplementary Fig. S4, Supplementary Table S2). The posterior parameter estimation indicated that the effective population size of ancestral populations were hundreds of times higher than those of current populations (N e = 13.5), which suggest that species have undergone major contraction of their ranges. Moreover, the N1 group has the largest effective population size, and the N4 group has the smallest effective population size (N1 > N2 > N3 > N4). The estimated mean mutation rate of nSSRs was 1.74 × 10−4 (95% CI: 1.05 × 10−4–5.65 × 10−4).
Contemporary and historical gene flow
The results of the migration rates estimated by BAYESASS suggested a significantly lower level of contemporary gene flow (m c) throughout all populations. The average migration rates in all pairwise comparisons ranged from 0.007 to 0.176, with standard deviations less than 0.05 (Table 5). Each value represents the proportion of individuals derived from a corresponding source population for each generation. However, historical migration rates (m h) estimated by Migrate-N were significantly high and ranged from 0.037 (ZD to HJ) to 0.367 (from HJ to HY) (Table 6). Estimates of contemporary gene flow demonstrated a lack of significant gene flow among populations, which suggesting that gene flows were affected by the recent habitat fragmentation of L. auriculata.
Population bottleneck test
The inbreeding coefficients ranged from 0.056 to 0. 265, and all eight populations showed significant deviation from mutation-drift equilibrium under IAM assumptions and a mode-shift test (Table 7). Furthermore, six (ZT, ZD, AY, HX, HJ and HY) and three populations (AY, HX and HY) also showed significant deviation from mutation-drift equilibrium under SMM and TPM assumptions, respectively (Table 7). These results indicate that all eight populations appear to have undergone a recent bottleneck, resulting in a significant excess in heterozygosity compared with the heterozygosity expected.
Ecological niche modelling
ENM were reconstructed for distribution of the present and the LGM (Fig. 6a–c). Models performance showed that the average values of all models are 0.998, indicating good performance of the models. The potential distribution for the present range of L. auriculata, predicted by Maxent, is very similar to the actually known geographic distribution of the species (Fig. 6a) in the central-east China. Palaeodistribution modelling suggested a relatively larger area during the LGM than that predicted under current conditions (319 grid numbers, P ≥ 0.75), based on both CCSM (425 grid numbers, P ≥ 0.75) and MIROC (608 grid numbers, P ≥ 0.75, Fig. 6b,c). The CCSM indicated that optimum suitable habitat was in the central-east China, which was decreased after LGM about 24.9%. Based on MIROC, the range decreased 47.5%. Both models suggested that central-east China experienced habitat contraction and declines after LGM, although at different levels, which suggest that species experienced considerable population decreases, and suffered many population extinctions. The suitable distribution of L. auriculata flowed from the central-east area to the southeast of China (Fig. 6), which may contribute to the climate change. The potential distribution of current was more scattered, while LGM was more concentrated based on two models (Fig. 6). In addition, the suitable distribution area of L. auriculata is separated by the Yangtze River from LGM to current (Fig. 6).
Predicted distribution of Litsea auriculata based on Ecological niche distribution model. (a) Predicted distribution based on current data; (b) distribution during the Last Glacial Maximum (LGM) based on community climate system model (CCSM); (c) distribution during LGM model based on model for interdisciplinary research on climate (MIROC). Ecological niche models were established with current bioclimatic variables on the basis of extant occurrence points of the species using Maxent version 3.4.1. Predicted distribution probabilities are shown in each 2.5 arc-min pixel. The map of China is free downloaded from Global Administrative Areas (http://www.gadm.org/country). The map is made by ArcGIS 10.3 software (http://www.arcgis.com/features/index.html), and then cut by Visio Pro for Office 365 (Trial) (https://products.office.com/zh-cn/visio/visio-professional-free-trial-flowchart-software).
Discussion
Population demography relative to climatic changes
Based on the coalescent theory, the most frequent and widespread haplotypes are likely to be the ancestral haplotypes27. Median-joining haplotype network analyses revealed that haplotype H1 and H2 may be ancestral haplotypes, which are distributed in all populations. The widespread occurrence of ancestral haplotypes (H1 and H2) appeared among the widely disjunct populations (Table 1, Fig. 1), which may result from historical migration events and/or shared ancestral polymorphism due to incomplete lineage sorting rather than long-distance gene flow28. Similar conclusions have also been drawn for other species, such as N. sericea 28.
Mainland China, as part of the Sino-Japanese Floristic Region, harbours the most diverse of the world’s temperate flora and was the most important glacial refuge for its Tertiary representatives (“relics”) throughout Quaternary ice-age cycles9,29,30. Although this region has never been directly impacted by extensive ice sheets29,31, it experienced severe climatic oscillations throughout the Quaternary, resulting in repeated drastic environmental changes that profoundly shaped the current distributions and genetic structures of many plant and animal species32,33. The results of the phylogeographic analysis are consistent with the basic Quaternary expansion-contraction (EC) model, which also proved to be consistent with fossil pollen evidence in indicating extensive latitudinal range shifts, typically in the form of southward retreats during glacial events, followed by rapid expansions northward during inter-/postglacial events9,34,35. The tropical southern populations from potential refugia perhaps maintain higher levels of genetic diversity, consistent with the EC model9. The climatic fluctuations throughout inter-/postglacial events would have altered the distribution of habitats for L. auriculata, leading to variation in population continuity and isolation. The values of H E and A r were higher in the southern area than the northern area, and higher in the eastern area than western area. These values significantly decreased as latitude increased, but increased as longitude increased in the present study (Fig. 2). This result is consistent with the reconstruction of species distribution at LGM.
The STRUCTURE analysis revealed that eight L. auriculata populations were assigned to four groups (Fig. 4). These four groups located at three mountains distant from each other or between the Yangtze River and diverged at the same time (60,200 years), corresponding to the beginning of the last ice age; this was strongly supported by the ABC analyses. This scenario is also consistent with the reconstruction of species distribution at LGM, which is supported by both CCSM and MIROC (Fig. 6b,c). The models predicted the occurrence of discontinuous suitable areas between south (Tianmu Mountain) and north regions (Dabie Mountain, Qinling Mountain and Funiu Mountian) along both banks of Yangtze River (Fig. 6). Moreover, it is notable that paleoenvieronmental reconstructions also showed that the suitable distribution of L. auriculata was scattered especially based on MIROC model (Fig. 6c). Although mainland China was not covered by ice sheets during the ice age, the four groups of L. auriculata occupy geographic mountains and Yangtze River separated by habitat type, driving isolation and genetic divergence between groups of populations enhanced by environmental differences.
In the genetic pattern observed, we also found the signature for past demographic changes in L. auriculata populations. The demographic history inferred from the ABC analyses is that the four groups expansion starting about 60,200 years BP, that is, during the last glacial phase including the LGM. Although the lack of a calibrated molecular clock demands caution, paleoenvironmental reconstructions of ENM at the LGM support the hypothesis that populations of this species expanded during this period.
ENM of habitat changes following the LGM show clearly that the current distribution of L. auriculata experienced severe habitat shifting and fragmentation, which is supported by both CCSM and MIROC (Fig. 6). The suitable distribution of L. auriculata experienced a northwest-to-southeast migration in China and scattered. Compare the ENM of LGM and current, AY population (Dabie Mountian) exists in optimum suitable habitats wherever in LGM or current, which contains the highest haplotype diversity, with the most (unique) haplotypes and private alleles (Table 1). Moreover, Dabie Mountains is the north passageway of plant spread, where floristic elements can spread from southwest China and central China to east China or from east China to west China and central China36.
Our study supports hypothesis that range and distribution of suitable habitat are the key factors determining demographic histories. The suitable habitats of a species are influenced by both climatic and geographic features (such as Yangtze River, Mountains). Climate fluctuations have enormous impacts on changes in a species’ suitable habitat, however these impacts are also influenced by geographic features. This result supports the idea that climatic and geographic features can impact the evolutionary history of a species.
Impact of habitat fragmentation on L. auriculata populations
Our ABC and ENM analysis through time suggested that populations likely experienced expansion and retraction into and out of local refugia, and Quaternary climatic fluctuation may have played role in genetic structure as well9,37. As a result, the distribution area of L. auriculata is drastic decreased and scattered in several mountains along the Yangtze River. Therefore, L. auriculata populations were expected to experience erosion of genetic variation, significant population bottlenecks, reduced remnant population size and increased population genetic divergence due to increased random genetic drift, high levels of inbreeding and reduced gene flow38,39. In the present study, L. auriculata did exhibit genetic signs of a high level of population divergence (F ST(c) = 0.285, wright F ST(n) = 0.165), a significant population bottleneck (Table 7), a relatively small effective population size (N e = 13.5), significant increases in inbreeding coefficient (F IS = 0.184) and reduced gene flow—i.e., the contemporary gene flow (m c: 0.007–0.176) is lower than historical gene flow (m h: 0.037–0.367) in L. auriculata populations. Nevertheless, high levels of genetic diversity were observed in L. auriculata at both the species (h T = 0.855, 30 haplotypes based on cpSSR loci; H T = 0.657 based on nSSR loci) and population level (H E = 0.515, h = 0.159).
The significant population bottlenecks may be explained by a recent decline in population size due to habitat destruction. Eight populations appear to have undergone recent bottlenecks (Table 7), resulting in a significant excess of heterozygosity. The number of alleles decreases more rapidly than heterozygosity during a population decline, this will result in an excess heterozygosity for this particular population40. This is consistent with increasing human activity during the past century, including agriculture, urbanization and deforestation2, which resulted in a dramatic reduction in the number of sites and population sizes of L. auriculata. In addition, in the present study estimates for N e values were all less than 30, with a mean of 13.5 (Table 1). Although different populations may lose different alleles, a large number of alleles may still be maintained across the species as a whole41. However, these L. auriculata populations remain relatively rich in genetic diversity (at species level: h T = 0.855, 30 haplotypes based on cpSSR loci; H T = 0.657 based on nSSR loci; at population level: H E = 0.515, h = 0.159) even when experiencing drastic habitat fragmentation. Genetic variation may decrease with reduced remnant population size, but not all fragmentation events lead to genetic losses and different types of genetic variation38. Thus, genetic drift does not appear to lead to decreased genetic diversity, but still increased population differentiation42. Furthermore, L. auriculata is a perennial species, and this habit may delay time between generations and buffer the effect of habitat fragmentation on genetic erosion43. Similar results were found in other endangered species in the same family, such as L. szemaois (AFLP: H T = 0.1947; ISSR: H T = 0.2498) in SW China42, Machilus thunbergii (H T = 0.165, 9 haplotypes) in Southeast Asia44, M. kusanoi (H T = 0.159, 9 haplotypes) in southeast China44, and Neolitsea sericea (H T = 0.761, 9 haplotypes) in Southeast Asia28.
Contemporary gene flow (m c = 0.007–0.176) among populations was lower than historical gene flow (m h = 0.037–0.367, Table 5). This finding demonstrates that the recent habitat fragmentation in these L. auriculata populations is accompanied by decreased gene flow. This reduced gene migration between L. auriculata populations (Tables 5 and 6) might be attributed to poor dispersal ability, combined with isolation; and this is consistent with expectations for insect-pollinated and fleshy-fruited (bird-mediated seed dispersal) species, for which pollen and seed dispersal is relatively limited45. This conclusion was confirmed by several studies in Lauraceae, such as L. szemao 42 and Neolitsea sericea 46,47. Reduced contemporary gene flow can further decrease genetic diversity and contribute to high levels of among-population genetic differentiation.
Another expectation for fragmentation population is increased inbreeding (F IS = 0.184), which is influenced by the mating system and the limited gene flow48. We found that inbreeding coefficients ranged from 0.056 to 0.265, and all populations significantly deviated from mutation-drift equilibrium under IAM assumptions and a mode-shift test (Table 7), indicating that all L. auriculata populations experienced inbreeding. Since L. auriculata is a dioecy species with insect-pollinated outcrossing trees, inbreeding could be due to mating between relatives, such as half-sibs, rather than selfing49.
Fragmentation usually decreases the habitat sizes and increases spatial distances between populations, leading to high levels of genetic differentiation (F ST(c) = 0.285, wright F ST(n) = 0.165). Similar results have been reported in other endangered or endemic species of the same family, such as L. szemaois (Φ st = 0.2420)42 and N. sericea (F ST(c) = 0.355; F ST(n) = 0.141)28. In addition, hierarchical AMOVA (28.54% for cpSSR and 16.50% for nSSR, Table 2) also showed the similar level of genetic differentiation among populations. Furthermore, all pairwise G ST and F ST values among populations were also significant (P < 0.05, Table 3) except between HY and HJ, which further confirmed that genetic differentiation among populations was quite high. Meanwhile, a lack of significant correlations between geographic and genetic distance based on cpSSR and nSSR analysis were detected (Fig. 3), which may be attributed to a higher impact of genetic drift. At very small spatial scales gene flow may not be sufficiently unidirectional to cause a significant correlation50.
Conservation implications
L. auriculata is listed in the “Lower Risk/near-threatened” category of the IUCN red list of threatened species, which means that this species is close to being qualified as vulnerable. Information on the genetic diversity, genetic structure, and demographic history of endangered species intergrate with ecological niche modeling can establish effective and efficient strategies for their conservation. One of our aims for the conservation of threatened plants is to preserve their genetic diversity51. Although L. auriculata has a relatively higher level of genetic diversity, its distribution, population size and effective population size are fairly small. Therefore, this species faces an extinction risk in the short term. An effective population size (N e) greater than 50 is proposed to prevent inbreeding depression in the short-term (5 generations) and a N e greater than 500 was proposed to maintain the evolutionary potential in perpetuity52. However, all of the estimated N e in populations of L. auriculata were less than 30 (Table 1). Therefore, we suggest that protection zones in the distribution areas of L. auriculata should be established to protect the habitat for this species (in situ).
Our study showed significant genetic differentiation among populations based on cpSSR and nSSR data, which resulted in geographically defined genetic clusters of populations. Four groups were detected in L. auriculata, which could be managed as four evolutionary units and assigned the highest priority protection. Considering the highest number of private alleles and haplotypes detected in the AY population, special attention should be paid to genetic resources from this region. To preserve all extant populations and their habitats, comprehensive ex situ conservation is viable by collecting seeds or seedling individuals from different populations from the three mountains, especially from the AY population.
Furthermore, artificial pollination is necessary to augment in situ populations because wild populations lack effective pollen53. Prohibition on deforestation in L. auriculata distribution areas should also be implemented. Taken together, these measures could protect L. auriculata from extinction.
Conclusions
In summary, by using population genetics and ecological niche modeling analysis together, we were able to identify the many factors, historical and contemporary, have shaped population genetic structure in this endangered species. Moreover, geographical, environmental and climatic factors also contribute to patterns of genetic structure. Contemporary gene flow among L. auriculata populations showed drastic declines in comparison with historical gene flow, resulting in a high level of population divergence. L. auriculata populations underwent expansion after divergence and dramatically decreased to the current small size with relative population bottlenecks due to climate changes. Thus, both geographic environmental factors including climate changes have shaped patterns of genetic structure in L. auriculata, along with small population sizes, significant recent population bottlenecks, significant increases in the inbreeding coefficient and contemporary limited gene flow. Information on the genetic diversity, genetic structure, demographic history and integrated with ENM of L. auriculata can establish effective and efficient strategies for their conservation.
Material and Methods
Sample collection
Litsea auriculata occurs in deciduous broad-leaved forests or mixed coniferous broad-leaved forests at 500–1,250 m elevation, preferentially on valleys covered by deep and nutrient-rich soil, and their most important associated species are Cyclocarya paliurus (Juglandaceae), Nyssa sinensis (Nyssaceae), Tilia tuan Szyszyl. (Tiliaceae), and Emmenopterys henryi (Rubiaceae)1. Litsea auriculata is a diploid, dioecious and insect-pollinated tree (up to 25 m tall), with dark purple, one-seeded fruits (drupes) that are primary dispersed by birds1,28,42,54.
For this study samples were collected from their natural communities throughout east China including three provinces: Zhejiang, Anhui and Henan (Table 7). A total of 231 individuals of L. auriculata were collected from eight locations. Sample size ranged from 7 to 40 individuals per location (Table 7). Three populations (ZT, ZQ and ZD) are distributed in the Tianmu Mountains of Zhejiang Province; three populations (AS, AY and HX) were found in the Dabie Mountains of Anhui and Henan Province, and the last two populations (HY and HJ) occur in the Qinling Mountains of Henan Province (Fig. 1).
At least forty individuals within each location at an interval of at least 10 m were sampled. If the number of individuals in a particular site was less than 40, all available trees were then samples. Young leaves were randomly sampled from trees and immediately placed into plastic sealed bags with silica gel for fast drying. Subsequently these samples were stored at room temperature until use.
DNA extractions and microsatellite analysis
Total genomic DNA from each of the samples was extracted from approximately 20 mg of silica-gel-dried leaves using a modified cetyltrimethyl ammonium bromide (CTAB) method55. After extraction, DNA were dissolved in 200 μL of sterilized water and kept at −30 °C until use.
For cpDNA analysis, we employed 33 universal chloroplast SSR (cpSSR) markers56,57; however, only seven of them (ccmp3, ccmp4, ccSSR-3, ccSSR-5, ccSSR-8, ccSSR-15 and ccSSR-22) showed polymorphism across all samples.
For nuclear DNA analysis, eight nSSR markers were polymorphic in L. auriculata. Three of these nuclear markers (Nese8, Nese9 and Nese10) were developed by58 and five of them (Litsea-10, Litsea-11, Litsea-36, Litsea-80 and Litsea-86) were obtained by Chiang, et al.59.
To amplify the SSR loci of L. auriculata, we used a tailed primer method to perform PCR: a U19 (5′-GGTTTTCCCAGTCACGACG-3′) was tailed to the 5′ end of the forward primer, and the U19 primer labelled with 6-FAM, VIC, NED or PET, was added to the PCR reaction mix. PCR amplification was performed in a reaction mixtures (10 μL) containing approximately 10 ng of template DNA, 0.2 mM of each dNTP, 1 × PCR buffer (16 mM (NH4)2SO4, 67 mM Tris–HCl (pH 8.8), 0.01% Tween-20), 1.35 mM of Mg2+, 0.5 U of BIOTaq DNA polymerase (Bioline, USA), and 0.5 mM of each of the three primers: a forward primer with a tail of the U19 sequence, a non-tailed reverse primer, and a 5′-labeled U19 primer.
The PCR reactions were conducted using a Veriti Thermal Cycler (Applied Biosystems). The PCR reaction was performed using the following cycling conditions: denaturation at 94 °C for 1 min; followed by 30 cycles of 30 s at 94 °C, 30 s at annealing temperature of primer pair and 1 min at 72 °C; and final extension at 72 °C for 5 min. The PCR products were genotyped on an ABI 3730 Genetic Analyzer (Applied Biosystems). Allele sizes were determined using the GeneMapperTM analysis software version 4.0 (Applied Biosystems) according to a GeneScanTM 500 LIZTM Size Standard (Applied Biosystems).
To investigate allele-size variation at the seven cpSSR loci, PCR products of each allele at each locus was subjected to DNA sequencing on an ABI 3730XL DNA Analyzer (Beijing Genomics Institute, Shenzhen, China).
Genetic diversity and haplotype network construction
For cpSSRs, haplotype composition, number of haplotypes and number of unique haplotypes were calculated for each population. A unique haplotype was defined as one found in only that population. We constructed a haplotype frequency map to examine whether haplotype distribution was geographically structured. The haplotype diversity as defined by cpSSRs in each population was quantified in terms of the number of observed alleles per locus (N a), the effective number of alleles (N E), the Shannon index (I), diversity index, and unbiased diversity (uh) using GenAlEx 6.560. Mean within-population genetic diversity, species total genetic diversity, and population genetic differentiation were calculated treating alleles as unordered (h S, h T and G ST) and ordered (v S, v T and R ST), following the methods described by Pons & Petit (1996) using PermutCpSSR 2.061.
Phylogeographical structure was analysed by comparing two coefficients of population divergence (G ST and R ST). G ST is only based on haplotype frequencies, while R ST also takes into account haplotype similarities. These two parameters were compared via a permutation test, using 10,000 permutations by PermutCpSSR 2.061. A higher R ST than the estimated G ST indicates the presence of a phylogeographical structure. To examine the phylogenetic relationships of haplotypes, a haplotype tree was constructed in Network 5.0 using a median-joining haplotype network algorithm62. The distance between each pair of haplotypes was the sum of nucleotide differences between them over seven chloroplast microsatellite loci.
For nSSRs, genetic diversity was assessed at the species level, as gene diversity in the total population (H T) and average gene diversity within populations (H S)63, and at the population level, as the mean number of alleles per locus (A), allelic richness (A R), private alleles (P A), observed (H O) and expected (H E) heterozygosity. These calculations were performed using FSTAT version 2.9.3.264 and GenAlEx 6.560. Inbreeding coefficients (F IS) were calculated using FSTAT version 2.9.3.264. The deviation of F IS from zero was tested in each population using 1,000 permutation tests with a sequential Bonferroni correction65. The effective population sizes of each population (N e) were estimated at three levels of the lowest allele frequency (0.01, 0.02 and 0.05) with a 95% confidence interval using the program LDNe66. Significant tests of a linear parametric regression were performed for the two parameters (A r and H E) against the latitude and longitude of the eight populations.
Population genetic structure
Analysis of molecular variance [AMOVA, Excoffier, et al.67] of cpSSRs and nSSRs data was performed to estimate the total percentage variance attributable to difference within and among populations of L. auriculata. Analyses were performed using Arlequin version 3.568, and significance tests were conducted using 10,000 permutations. Genetic differentiation among populations (F ST(c) for cpSSRs, wright F ST(n) for nSSRs) was also obtained using Arlequin version 3.568.
For cpSSRs, Nei’s unbiased genetic distance for all population pairs [G ST, Nei69] was calculated using PopGene 1.3270. For nSSRs, pairwise F ST between populations was calculated and tested for significant differences using FSTAT version 2.9.3.2 [1000 bootstrap permutation, Goudet64].
Correlations between the pairwise G ST (Nei’s unbiased genetic differentiation, cpSSRs) or F ST/(1 − F ST) (nSSRs) values and the geographic distance matrix were evaluated by the Mantel test71 in GenAlEx 6.5 software with 9,999 random permutations60,72.
To determine the optimal number of clusters that compose the studied populations, we implemented the Bayesian algorithm analysis in STRUCTURE 2.3 software73. Fifteen independent runs were performed for each K between 1 to 8 without prior information using the admixture model and assuming correlated allele frequencies. Each run consisted of 1,000,000 MCMC (Markov Chain Monte Carlo) repetitions after burn-in with 100,000 iterations based on the LOCPRIOR model74, an admixture model and the correlated allele frequencies model (hereafter, the F-model) described by Falush, et al.75. The optimal K was determined using the ΔK method implemented in STRUCTURE HARVESTER76. CLUMPP version 1.177 were used to summarize parameters across 15 runs at the optimal K value. DISTRUCT 1.1 software78 was used to visualize the STRUCTURE results after processing with CLUMPP.
Approximate Bayesian computation (ABC)
To further understand the population demographic history of L. auriculata, we employed an approximate Bayesian computation (ABC) analysis in DIYABC v 2.179 to test evolutionary scenarios based on nuclear loci. Populations covering the entire species distribution were analysed to infer the history of the genetic structure indicated by nSSR analysis. To simplify the scenarios in the ABC analysis, we pooled all populations into four large populations based on the results of STRUCTURE analysis of this core data set: Pop1 (ZT, ZQ and ZD population), Pop2 (AS and HX population), Pop3 (AY population) and Pop4 (HJ and HY population). Considering the result of haplotype diversity of L. auriculata, Pop3 (AY population) was inferred as the ancestry population. A set of ten evolutionary scenarios were built and tested (see Fig. 5): either a Simple split model (scenario 1), Hierarchical split model (scenarios 2–6), or Isolation with admixture model (admixture, scenarios 7–10). In these scenarios, t# represents time scale measured in number of generations and N# represents effective population size of the corresponding populations (Pop1, Pop2, Pop3 and Pop4). The range and distribution of priors for parameters used to describe these scenarios (effective population size, time of splitting or merging events, and rates of admixture in the case of merging events) are shown in Supplementary Table S3. For each scenario, 9 × 106 simulated data sets were run and the most likely scenario was determined by comparing the posterior probabilities (with 95% confidence intervals) using the logistic regression method80. For each set of scenarios, the most likely scenario was the one with the highest posterior probability value. To check the goodness of fit of the scenario, principal component analysis (PCA) was performed on the first 100,000 simulated datasets of the reference table in the summary statistics using the “model checking” option in DIYABC. Finally, to assess confidence in the model choice, type I and type II error rates was estimated with 500 pseudo-observed datasets (PODs) using the option “evaluate confidence in scenario choice”.
Detection of recently and historical migrants
To evaluate the direction and rates of recent migration (i.e., within the last several generations) among populations, the BAYESASS 3.0.4 program was used81. Preliminary runs were performed to adjust the MCMC mixing parameters of migration rates (m), allele frequencies (a) and inbreeding coefficients (f), to ensure proposed acceptance rates approximately 20–60%81. For the final runs, the mixing parameters m = 0.15, a = 0.4 and f = 0.5 were used. Ten independent replicates with different random starting seeds were performed with a burn-in of 5,000,000 iterations followed by 50,000,000 MCMC iterations and a sampling frequency of 2,000.
To investigate historical migration rates (m h) among populations, we used Migrate-N version 3.6 (http://popgen.sc.fsu.edu/Migrate/Migrate-n.html)82. MIGRATE is based on coalescent theory to estimate effective population sizes (Theta = θ, θ = 4 Neµ, where Ne is historical effective population size) and symmetrical migration rates (M) [(M = m h/µ, µ is mutation rate per generation (10−2)] between population pairs. Migrate-N assumes that populations are in migration-drift equilibrium, that population sizes and migration rates are constant through time, and that populations are randomly sampled. We ran three replicates of Migrate-N using Microsatellite mode using a simulation of the single stepwise mutation model with constant mutation rates and starting parameters based on F ST calculations. The program estimates the parameters θ and M using a Bayesian method83,84, both of which could be used to estimate the number of migrants per generation (N m) into each population using the equation 4 N m = θ* M. We estimated θ and M with slice sampling and uniform prior distribution (for θ, range = 0–1000.0, delta = 100; for M, range = 0–2000.0, delta = 200). The three long chains were performed with 50,000-recorded genealogies at a sampling interval of 20 increments after discarding the first 10,000 genealogies (burn-in) for each chain. We used a static heating scheme at four temperatures (1.0, 1.5, 3.0, 1,000,000) to efficiently search the genealogy space. The confidence interval for θ and the migration parameter M was calculated using a percentile approach84.
Bottleneck test
In a recently bottlenecked population, the observed heterozygosity is higher than the expected equilibrium heterozygosity estimated from the observed number of alleles85. To evaluate whether the population had undergone recent bottlenecks, we adopted Wilcoxon’s signed-rank test with 10,000 iterations using BOTTLENECK software version 1.2.0286. We ran BOTTLENECK using three possible mutation models: the Infinite Allele Model (IAM), the Stepwise Mutation Model (SMM) and the Two Phase Model (TPM; 30% IAM and 70% SMM).
Ecological niche modelling
ENM were constructed using current climatic conditions and those of the LGM through the principle of maximum entropy model in Maxent version 3.4.187. Maxent uses occurrence data along with environmental data to estimate the probability of species occurrence on the basis of a uniform probability distribution (maximum entropy) and on the presence data provided by the user88. L. auriculata occurrence points in the study were based on GPS data taken during our field work and from Nature Reserve records. We used all georeferenced localities. For occurrence points without geographic coordinates, which were then georeferenced by using the software Google Earth (http://www.ditu7.com/). A total of 26 occurrence points of L. auriculata were obtained to be used for ENM construction (Supplementary Table S4).
We used 19 bioclimatic variables with a resolution of 2.5 min (~5 km) from WorldClim database (http://www.worldclim.org/)89. These variables represent summaries of means and variation in temperature and precipitation, and characterize dimensions of climate considered particularly relevant in determining species distributions89.
We used random seed to test the ENM of L. auriculata. To obtain accurate prediction, we run the model making 100 replicates under the crossvalidation form of replication, 0.01 regularization multiplier, 10,000 background points, jackknife tests of variable importance and a maximum of 500 iterations. Finally, 75% of the localities were used to build the model and remaining 25% were randomly selected to test it.
In order to construct niche model for LGM (21,000 years ago- 21 kyr BP) conditions, we projected the current species’ bioclimatic niche onto past climate layers, which were downloaded from the WorldClim database at a resolution of 2.5 min as in the analysis above. Both the community climate system model (CCSM4)90 and model for interdisciplinary research on climate (MIROC-ESM)91 were used. Both models were run in Maxent using the settings chosen for current conditions. The area under receiver operating characteristic (ROC) curve (AUC) was then used to evaluate the model performance. An AUC score above 0.75 represent good model prediction92. All ENM predictions were then visualized in ArcGis 10.3 (http://www.arcgis.com/features/index.html), and the map of China (shapefile) is free downloaded from Global Administrative Areas version 2.8 (http://www.gadm.org/country).
References
Fu, L. G. & Jin, J. M. China Plant Red Data Book: Rare and Endangered Plants Volume 1. 350–351 (Science press, 1992).
Deng, Y. & Pu, F. G. Conservation actualities of rare and endangered plants in Anhui Tianma National Nature Reserve and coping strategies. Anhui For. Sci. Tech. 41, 35–38 (2015).
Wang, F. Z., Xie, F., Zhan, M. D. & Yuan, S. H. Preliminary study on Litsea auriculata Chien et Cheng tree twig cottage under full sunshine and automatic spray. Journal of Henan For. Sci. Tech. 31, 7–17 (2011).
Cheng, H. Y., Hu, X. L., Zhou, M. Y. & Li, M. Q. Seedling techniques by sowing of Litsea auriculata. Anhui For. Sci. Tech. 33–34 (2004).
Yuan, N., Sun, Y., Comes, H. P., Fu, C. X. & Qiu, Y. X. Understanding population structure and historical demography in a conservation context: population genetics of the endangered Kirengeshoma palmata (Hydrangeaceae). Am. J. Bot. 101, 521–529 (2014).
Zhou, W. et al. Mountains too high and valleys too deep drive population structuring and demographics in a Qinghai-Tibetan Plateau frog Nanorana pleskei (Dicroglossidae). Ecol. Evol. 7, 240–252 (2017).
Hewitt, G. M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58, 247–276 (1996).
Sun, X. J. & Chen, Y. S. Palynological records of the last 11,000 years in China. Quaternary Sci. Rev. 10, 537–544 (1991).
Qiu, Y. X., Fu, C. X. & Comes, H. P. Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evo. 59, 225–244 (2011).
Dixo, M., Metzger, J. P., Morgante, J. S. & Zamudio, K. R. Habitat fragmentation reduces genetic diversity and connectivity among toad populations in the Brazilian Atlantic coastal forest. Biol. Conserv. 142, 1560–1569 (2009).
Wang, Y. et al. Effects of habitat fragmentation on genetic diversity and population differentiation of Liposcelis bostrychophila badonnel (Psocoptera: Liposcelididae) as revealed by ISSR markers. J.Stored Prod. Res. 68, 80–84 (2016).
Harris, L. N. & Taylor, E. B. Pleistocene glaciations and contemporary genetic diversity in a Beringian fish, the broad whitefish, Coregonus nasus (Pallas): inferences from microsatellite DNA variation. J Evolution Biol. 23, 72–86 (2010).
Johnson, J. A. et al. Long-term survival despite low genetic diversity in the critically endangered Madagascar fish-eagle. Mol. Ecol. 18, 54–63 (2009).
Chiucchi, J. E. & Gibbs, H. L. Similarity of contemporary and historical gene flow among highly fragmented populations of an endangered rattlesnake. Mol. Ecol. 19, 5345–5358 (2010).
Bertorelle, G., Benazzo, A. & Mona, S. ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Mol. Ecol. 19, 2609–2625 (2010).
Wang, I. J. Recognizing the temporal distinctions between landscape genetics and phylogeography. Mol. Ecol. 19, 2605–2608 (2010).
Wang, I. J. Choosing appropriate genetic markers and analytical methods for testing landscape genetic hypotheses. Mol. Ecol. 20 (2011).
Bohonak, A. J. & Vandergast, A. G. The value of DNA sequence data for studying landscape genetics. Mol. Ecol. 20, 2477–2479 (2011).
Tsuda, Y. & Ide, Y. Chloroplast DNA phylogeography of Betula maximowicziana, a long-lived pioneer tree species and noble hardwood in Japan. J. Plant Res. 123, 343–353 (2010).
Amane, M. et al. Chloroplast-DNA variation in cultivated and wild olive (Olea europaea L.). Theor. Appl. Genet. 99, 133–139 (1999).
Palme, A. E., Su, Q., Rautenberg, A., Manni, F. & Lascoux, M. Postglacial recolonization and cpDNA variation of silver birch. Betula pendula. Mol. Ecol. 12, 201–212 (2003).
Heuertz, M. et al. Nuclear microsatellites reveal contrasting patterns of genetic structure between western and southeastern European populations of the common ash (Fraxinus excelsior L.). Evolution 58, 976–988 (2004).
Lira-Noriega, A. & Manthey, J. D. Relationship of genetic diversity and niche centrality: a survey and analysis. Evolution 68, 1082–1093 (2014).
Alvarado-Serrano, D. F. & Knowles, L. L. Ecological niche models in phylogeographic studies: applications, advances and precautions. Mol. Ecol. Resour. 14, 233–248 (2014).
Zane, L., Bargelloni, L. & Patarnello, T. Strategies for microsatellite isolation: a review. Mol. Ecol. 11, 1–16 (2002).
Abdul-Muneer, P. M. Application of microsatellite markers in conservation genetics and fisheries management: recent advances in population structure analysis and conservation strategies. Genet. res. int. 2014, 691759 (2014).
Posada, D. & Crandall, K. A. Intraspecific gene genealogies: trees grafting into networks. Trends Ecol. Evol. 16, 37–45 (2001).
Zhai, S.-N., Comes, H. P., Nakamura, K., Yan, H.-F. & Qiu, Y.-X. Late Pleistocene lineage divergence among populations of Neolitsea sericea (Lauraceae) across a deep sea-barrier in the Ryukyu Islands. J. Biogeogr. 39, 1347–1360 (2012).
Liu, K. B. Quaternary history of the temperate forests of China. Quaternary Sci. Rev. 7, 1–20 (1988).
Ying, J. Species diversity and distribution pattern of seed plants in China. Chin. Biodiv. 9, 393–398 (2001).
Shi, Y., Ren, B., Wang, J. & Derbyshire, E. Quaternary glaciation in China. Quaternary Sci. Rev. 5, 503–507 (1986).
Harrison, S. P., Yu, G., Takahara, H. & Prentice, I. C. Palaeovegetation: diversity of temperate plants in east Asia. Nature 413, 129–130 (2001).
Axelrod, A. I., Shehbaz, I. A. & Raven, P. H. History of the modern flora of China. 43–45 (Springer, 1996).
Tsuda, Y. & Ide, Y. Wide-range analysis of genetic structure of Betula maximowicziana, a long-lived pioneer tree species and noble hardwood in the cool temperate zone of Japan. Mol. Ecol. 14, 3929–3941 (2005).
Fujii, N. et al. Chloroplast DNA phylogeography of Fagus crenata (Fagaceae) in Japan. Plant Syst. Evol. 232, 21–33 (2002).
Yang, H. & Yang, G. F. Research and analyse on rare and endangered plants in Liankang Mountain in Natural Reserve. J.Xinyang Norm. Univ. (Natural Science Edition) 20, 328–330 (2007).
Qiu, Y. X., Guan, B. C., Fu, C. X. & Comes, H. P. Did glacials and/or interglacials promote allopatric incipient speciation in East Asian temperate plants? Phylogeographic and coalescent analyses on refugial isolation and divergence in Dysosma versipellis. Mol. Phylogenet Evol. 51, 281–293 (2009).
Young, A., Boyle, T. & Brown, T. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol. 11, 413–418 (1996).
Aguilar, R., Quesada, M., Ashworth, L., Herrerias-Diego, Y. & Lobo, J. Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Mol. Ecol. 17, 5177–5188 (2008).
Kang, M., Wang, J. & Huang, H. W. Demographic bottlenecks and low gene flow in remnant populations of the critically endangered Berchemiella wilsonii var. pubipetiolata (Rhamnaceae) inferred from microsatellite markers. Conserv. Genet. 9, 191–199 (2008).
Neigel, J. E. Estimation of effective population size and migration parameters from genetic data. 329–346 (Oxford University Press, 1996).
Ci, X. Q., Chen, J. Q., Li, Q. M. & Li, J. AFLP and ISSR analysis reveals high genetic variation and inter-population differentiation in fragmented populations of the endangered Litsea szemaois (Lauraceae) from Southwest China. Plant Syst. Evol. 273, 237–246 (2008).
Honnay, O. & Bossuyt, B. Prolonged clonal growth: escape route or route to extinction? Oikos 108, 427–432 (2005).
Wu, S. H. et al. Contrasting phylogeographical patterns of two closely related species, Machilus thunbergii and Machilus kusanoi (Lauraceae), in Taiwan. J.Biogeogr. 33, 936–947 (2006).
Oddou-Muratorio, S., Petit, R. J., Le Guerroue, B., Guesnet, D. & Demesure, B. Pollen- versus seed-mediated gene flow in a scattered forest tree species. Evolution 55, 1123–1135 (2001).
Chung, M. G., Chung, M. Y., Oh, G. S. & Epperson, B. K. Spatial genetic structure in a Neolitsea sericea population (Lauraceae). Heredity 85, 490–497 (2000).
Wang, Z. S. et al. Genetic structure of the endangered plant Neolitsea sericea (Lauraceae) from the Zhoushan Archipelago using RAPD markers. Annals of Botany 95, 305–313 (2005).
Keller, L. F. & Waller, D. M. Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241 (2002).
Hamrick, J. L. Isozymes and the analysis of genetic structure in plant populations. 87–105 (Chapman and Hall, 1990).
Hellberg, M. E. Footprints on water: the genetic wake of dispersal among reefs. Coral Reefs 26, 463–473 (2007).
Montalvo, A. M. et al. Restoration biology: A population biology perspective. Restor. Ecol. 5, 277–290 (1997).
Frankham, R., Bradshaw, C. J. A. & Brook, B. W. Genetics in conservation management: Revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol. Conserv. 170, 56–63 (2014).
Yang, H., Li, P. X., Dai, H. T. & Liu, D. The endangered mechanism and protective measures to the rare and endangered plants in Jigongshan. J. Henan For. Sci. Tech. 30, 37–39 (2010).
Fan, Y. B., Wang, R. M., Pan, F. J. & Yang, P. S. Study of the floral and pollination biology of Cinamomum camphora and Litsea cubeba (Lauraceae). J. Taiwan Mus. 59, 75–90 (2006).
Zhou, Z. H., Miwa, M. & Hogetsu, T. Analysis of genetic structure of a Suillus grevillei population in a Larix kaempferi stand by polymorphism of inter-simple sequence repeat (ISSR). New Phytol. 144, 55–63 (1999).
Chung, S. M. & Staub, J. E. The development and evaluation of consensus chloroplast primer pairs that possess highly variable sequence regions in a diverse array of plant taxa. Theor. Appl. Genet. 107, 757–767 (2003).
Weising, K. & Gardner, R. C. A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42, 9–19 (1999).
Zhai, S.-N., Yan, X.-L., Nakamura, K., Mishima, M. & Qiu, Y.-X. Isolation of compound microsatellite markers for the endangered plant Neolitsea sericea (Lauraceae). Am. J. Bot. 97, E139–E141 (2010).
Chiang, Y.-C., Shih, H.-C., Huang, M.-C., Ju, L.-P. & Hung, K.-H. Characterization of microsatellite loci from Litsea hypophaea (Lauraceae), a tree endemic to Taiwan. Am. J. Bot. 99, E251–E254 (2012).
Peakall, R. & Smouse, P. E. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295 (2006).
Pons, O. & Petit, R. J. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144, 1237–1245 (1996).
Bandelt, H. J., Forster, P. & Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Nei, M. Molecular Evolutionary Genetics. 159–164 (Columbia University Press, 1987).
Goudet, J. FSTAT (Version 1.2): A computer program to calculate F-statistics. J. Hered. 86, 485–486 (1995).
Rice, W. R. Analyzing tables of statistical tests. Evolution 43, 223–225 (1989).
Waples, R. S. & DO, C. IDNe: a program for estimating effective population size from data on linkage disequilibrium. Mol. Ecol. Resour. 8, 753–756 (2008).
Excoffier, L., Smouse, P. E. & Quattro, J. M. Analysis of molecular variance inferred from metric distances among DNA haplotypes - application to human mitochondrial-DNA restriction data. Genetics 131, 479–491 (1992).
Excoffier, L. & Lischer, H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol.Resour. 10, 564–567 (2010).
Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89, 583–590 (1978).
POPGENE Version 1.32: Software Microsoft Window-Based Freeware for Population Genetic Analysis (University of Alberta, Edmonton, Canada, 1997).
Mantel, N. Detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220 (1967).
Rousset, F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145, 1219–1228 (1997).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Hubisz, M. J., Falush, D., Stephens, M. & Pritchard, J. K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 9, 1322–1332 (2009).
Falush, D., Stephens, M. & Pritchard, J. K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620 (2005).
Jakobsson, M. & Rosenberg, N. A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806 (2007).
Rosenberg, N. A. DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Notes 4, 137–138 (2004).
Cornuet, J. M. et al. DIYABCv2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30, 1187–1189 (2014).
Cornuet, J. M. et al. Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24, 2713–2719 (2008).
Wilson, G. A. & Rannala, B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163, 1177–1191 (2003).
Beerli, P. & Palczewski, M. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics 185, 313–326 (2010).
Beerli, P. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22, 341–345 (2006).
Beerli, P. & Felsenstein, J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. P. Natl. Acad. Sci. USA 98, 4563–4568 (2001).
Cornuet, J. M. & Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144, 2001–2014 (1996).
Piry, S., Luikart, G. & Cornuet, J. M. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 90, 502–503 (1999).
Phillips, S. J., Anderson, R. P. & Schapire, R. E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259 (2006).
Porretta, D. et al. The integration of multiple independent data reveals an unusual response to Pleistocene climatic changes in the hard tick Ixodes ricinus. Mol. Ecol. 22, 1666–1682 (2013).
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G. & Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005).
Gent, P. R. et al. The Community Climate System Model Version 4. J. Climate 24, 4973–4991 (2011).
Watanabe, S. et al. MIROC-ESM 2010: model description and basic results of CMIP5-20c3m experiments. Geosci. Model Deve. Discuss. 4, 845–872 (2011).
Elith, J. Quantitative methods for modeling species habitat: comparative performance and an application to Australian plants. In: Quantitative Methods for Conservation Biology (eds Ferson, S. & Burgman, M.) 39–58 (Springer, 2000).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31100270). We thank Prof. Javier Francisco Ortega of Florida International University for his kind comments and suggestions on this manuscript. We are also grateful to Dr. Yunpeng Zhao of Zhejiang University, Dr. Na Yuan of Jiangsu Province Academy of Agricultural Sciences and Zhongyuan Yang of Nanjing University for assistance with data analysis.
Author information
Authors and Affiliations
Contributions
Q.F.G. and Z.S.W. conceived the study; Q.F.G. analysed the data and wrote the manuscript. L.S. collected samples, conducted laboratory work and processed the data. P.H.Z. helped to collect samples; Y.X.Q., H.L. and C.L.L. provided advice and assisted with writing, which was approved by all authors.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Geng, Q., Sun, L., Zhang, P. et al. Understanding population structure and historical demography of Litsea auriculata (Lauraceae), an endangered species in east China. Sci Rep 7, 17343 (2017). https://doi.org/10.1038/s41598-017-16917-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-16917-x
This article is cited by
-
Morpho-physiological and demographic responses of three threatened Ilex species to changing climate aligned with species distribution models in future climate scenarios
Environmental Monitoring and Assessment (2023)
-
Localized environmental heterogeneity drives the population differentiation of two endangered and endemic Opisthopappus Shih species
BMC Ecology and Evolution (2021)
-
Population genetic variation characterization of the boreal tree Acer ginnala in Northern China
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.