Abstract
Unlike mammals, in palmipedes de novo lipogenesis from diet takes place mostly in the liver. The French Landes Goose is famous for its high capacity and susceptibility to fatty liver production. While miRNAs play a critical role in the posttranscriptional regulation of gene expression, miRNAs that are involved in the regulation of goose hepatic steatosis have yet to be elucidated. Using high-throughput sequencing, we analyzed miRNAs expression profile of Landes goose liver after overfeeding for 21 days. Aan-miR-122-5p was the most frequently sequenced known miRNA, but it was unchanged after overfeeding. Compared with normal liver, we identified that 16 conserved miRNAs were up-regulated while the other 9 conserved miRNAs were down-regulated in fatty livers. Many of their predicted target genes played key roles in metabolic pathways leading to the development of hepatic steatosis in the goose by KEGG pathways analysis. ACSL1 and ELOVL6 were critical genes in hepatic lipid metabolism and had opposite expression patterns with aan-miR-203a and aan-miR-125b-5p, respectively. And we validated that aan-miR-203a and aan-miR-125b-5p might involve in the regulation of hepatic lipid metabolism by targeting ACSL1 and ELOVL6, respectively. These results add to our current understanding of the regulation network in goose lipid metabolism.
Similar content being viewed by others
Introduction
The livers of animals are important tissues of lipid metabolism. In humans, hepatic steatosis is typically the causal factor for abnormal liver function and is frequently correlated with a variety of metabolic disorders, such as obesity, insulin resistance, and type II diabetes1. In contrast, in wild palmipedes under natural conditions, hepatic steatosis occurs as a consequence of evolutionary adaption to accumulated reserves needed for migration2. In poultry production, this specific capacity is exploited for the production of “foiegras”3. Geese form fatty livers after feeding on a carbohydrate-rich diet for less than two weeks. And the liver weight may increase more than 10-fold and up to 10% of BW4. The French Landes Goose is famous for its high capacity and susceptibility to fatty liver production.
In order to find the mechanism of the development of fatty liver, some researchers have studied the crucial genes involved in hepatic steatosis of waterfowl2,5. MicroRNAs (miRNAs) are 20–24-nt long, endogenous small noncoding RNAs. They can hybridize to sites in the mRNAs of protein-coding genes to direct posttranscriptional regulation of gene expression by inhibiting translation and/or inducing degradation of target mRNAs6,7. To date, evidences indicate that miRNAs are key trans-acting factors and a single miRNA can repress the production of hundreds of protein coding target genes through relatively mild means7. Dysregulated expression of miRNAs has been reported in human fatty liver production8,9. And many miRNAs have been well characterized for their roles in regulating lipid metabolism, such as miR-12210. Nevertheless, so far there is a scarcity of information available regarding the abnormal expression miRNAs and their target genes in fatty liver of goose. Thus, the studies on lipid metabolism-related miRNAs and their mechanisms in goose liver could serve as an important reference for poultry breeding and human liver disease research.
The goal of this study was to identify differentially expressed miRNAs in the fatty liver of the Landes goose by using high-throughput technologies, deep-sequencing analysis, and qRT-PCR. Furthermore, we have predicted the target genes of the miRNAs and preliminarily validated some miRNA-mRNA pairs involved in the development of goose fatty liver. These findings provided insights into the role of miRNAs in the hepatic steatosis of goose.
Results
Effects of overfeeding
As shown in Fig. 1, the body weight and liver weight of overfed geese increased 1.5-fold and 4.0-fold (P < 0.01), respectively. The liver accounted for 3.92% of total body weight in the control geese and 10.17% in overfed geese. The increased liver weight accounted for 21.80% of total increase in body weight. By histological examination, the livers from control geese and overfed geese displayed marked differences. In Fig. 1d, the cell boundaries of hepatocytes were visible, while the nuclei of hepatocytes were centrally located. However, the cell boundaries were blurry, as shown in Fig. 1e. In contrast, we clearly observed many lipid droplets, and the nuclei were compressed to the edges of the cells.
miRNA expression profiles in A. anser liver
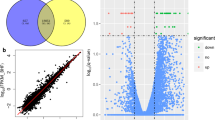
By sequencing analysis, the distributions of all small RNA classes in the two groups were displayed in Table S1. The information of a total 1410 miRNAs (995 from control group library, 1021 from overfed group library and 606 in both two groups) were obtained and their scatter plot were analyzed (Fig. 2). We listed some miRNAs, which showed expression with high predominance in Table 1. Of these miRNAs, the most frequently sequenced known miRNA was aan-miR-122-5p, which represented 70.34% and 72.01% of total miRNA reads in the control and overfed groups, respectively. However, the expression of aan-miR-122-5p was unchanged after overfeeding. In order to elucidate the role of miRNAs in the hepatic steatosis of geese, we compared miRNAs expression between normal livers and fatty livers. As shown in Table 2, 25 known conserved miRNAs were found differentially expressed, among which 16 conserved miRNAs were up-regulated while the other 9 conserved miRNAs were down-regulated in fatty liver.
Validation of selected miRNAs by qRT-PCR
We confirmed 6 miRNAs using qRT-PCR (Fig. 3). Aan-miR-122-5p, the most abundant miRNA in the liver, had similar level in the two groups. Aan-miR-222 was the most abundant upregulated miRNA in overfed geese liver. The relative expression of aan-miR-222 was increased 2.20-fold after overfeeding, while the relative expression of aan-miR-203a was increased 3.84-fold (P < 0.05). Conversely, the levels of aan-miR-30d, aan-miR-125b-5p, and aan-miR-146a-5p in overfed groups were only 42.60%, 62.09%, and 54.08% of those in the control groups (P < 0.05), respectively. The fold-change was differential between high-throughput sequencing and qRT-PCR. However, the variation trend was consistent between the two detection methods.
miRNA function analysis
To understand the biological functions of miRNAs in A. anser, we analyzed the putative target genes of differentially expressed miRNAs and classified the putative targets according to their pathways by KEGG pathway analyses. Figure 4 shows some pathways related to lipid metabolism. The network of some miRNAs and their predicted genes involved in glucose and lipid metabolism were shown in Fig. 5. ACSL1 and ELOVL6 transcripts may be the target genes of aan-miR-203a (Fig. 6a) and aan-miR-125b-5p (Fig. 6b), respectively. ACSL1 and ELOVL6 had opposite expression patterns with aan-miR-203a and aan-miR-125b-5p (Fig. 6c) after overfeeding, respectively. The recombined reporter vectors with the 3′-UTRs of ACSL1/ELOVL6 were co-transfected into goose primary hepatocytes with aan-miR-203a/aan-miR-125b-5p mimic, respectively. The luciferase activities of the two recombined reporters were both significantly suppressed by aan-miR-203a/aan-miR-125b-5p (P < 0.05) (Fig. 6d). Compared with blank control (BC) and negative control (NC), the mRNA expression level of ELOVL6 decreased in goose primary hepatocytes at 48 h after transfection with the aan-miR-125b-5p mimic. And ACSL1 mRNA level had no significant change in hepatocytes transfected with miR-203a mimic (Fig. 6e). The intracellular lipid contents of hepatocytes transfected with the aan-miR-203a mimic or aan-miR-125b-5p mimic had no significant difference compared with BC group and NC group (Fig. 6f).
Validation of selected target genes. (a) Binding sites for aan-miR-203a in the ACSL1 3′UTR of different species; (b) Binding sites for aan-miR-125b-5p in the ELOVL6 3′UTR of different species; (c) Relative expression of ACSL1 and ELOVL6 in control and overfed liver samples; (d) Luciferase activity assay of the recombined Dual-Luciferase reporter vectors with 3′UTR of ACSL1 or ELOVL6 co-transfected with aan-miR-203a mimic or aan-miR-125b-5p mimic; (e) Relative expression of ACSL1 and ELOVL6 in hepatocytes transfected with aan-miR-203a mimic or aan-miR-125b-5p mimic; (f) The relative content of intracellular lipid in hepatocytes transfected with aan-miR-203a mimic or aan-miR-125b-5p mimic. **Means P < 0.01.
Discussion
To date, many researchers have focused on liver diseases in mammals11, such as nonalcoholic fatty liver disease (NAFLD)8,12, alcoholic fatty liver (AFL)13, etc. miRNAs have been reported to play important roles in the regulation of glucose and lipid metabolism in the liver14,15. Domestic Palmipedes are particularly susceptible to hepatic steatosis, and unlike human NAFLD, the fatty liver is nonpathological and reversible16. Goose liver has been widely studied by researchers as a good model to investigate the mechanism of lipid metabolism17. Since the miRNAs involved in fatty liver production in geese remain to be elucidated, an analysis of the functional miRNAs involved in goose hepatic steatosis was performed on.
In our study, the size, color, weight and histology of the fatty liver showed that the livers of geese produced obvious hepatic steatosis after 21 days overfeeding. It provides a good animal model for fatty liver research. In addition, our lab possesses the A. anser genome database for matching analysis18 and the miRNAs profile of goose hypothalamus for reference19. It provides a more accurate platform for studying the roles of miRNAs in fatty liver production in geese. In this study, we found that 25 conserved miRNAs were differentially expressed in goose liver after overfeeding. In addition, we verified several miRNAs by qRT-PCR. Although some of the fold changes obtained by qRT-PCR were not identical to those obtained by deep sequencing, the trend in the changes was consistent across the two techniques.
Some dysregulated miRNAs in this study were consistent with known changes observed in other studies focused on lipid metabolism. MIR222 was upregulated in NASH and its predicted target genes are known to be involved in carbohydrate metabolism20. MIR221 was upregulated in adipose tissue of obesity individuals21. Similarly, the expression of aan-miR-222 and aan-miR-221 was increased as indicated by our study. MIR30d was a key regulator of human adipocyte development22. MIR148a controlled the expression of key proteins involved in cholesterol-lipoprotein trafficking23. Mir29 was downregulated in fatty livers of ob/ob mice24. Consistent with these studies, the expressions of aan-miR-30d, aan-miR-148a and aan-miR-29a significantly reduced in fatty liver of goose.
Aan-miR-122-5p is a miRNA with tissue-specific expression and showed predominance in the liver. Consistent with mammals, it accounted for at least 70% in the two groups. Many studies have suggested that MIR122 is important for the regulation of hepatic lipid metabolism25. Mir122 knockdown in mice reduced the cholesterol levels in the circulation and liver, decreased fatty acid synthesis, and increased fatty acid oxidation in the liver26. Mir122 was downregulated in rats treated with high-fat diet and enriched with or without fructose27. However, in our study, the expression of aan-miR-122-5p showed no significant change. And this result is consistent with the change in duck with high fat diet28. MIR375 is involved in the regulation of insulin release and glucose homeostasis29. Inconsistent with the underexpression of MIR375 in NASH30, aan-miR-375 was significantly increased in the fatty liver of goose. It is worth noting that some miRNAs have different change in same processes in different species. For example, MIR125b was abnormal expression in GBM5 cell line treated with ARA and DHA but not in the other, although all the studied cell lines were glioblastoma cells31. These differences can be attributed to the presence of different signaling pathways and can be species-specific. Elucidation of the specific functions of these dysregulated miRNAs in the development of goose fatty liver requires further experimental data.
In order to understand the biological function of differential expression miRNAs, we predicted their target genes and classified these genes according to their pathways by KEGG pathway analyses. Many of these target genes play key roles in metabolic pathways leading to the development of hepatic steatosis in the goose fatty liver. Mir203a can be used as the potential biomarker for type 2 diabetes32 and alcoholic steatohepatitis33 in rat models. Mir125b has been shown related to adipocyte differentiation and adiposity34,35. However, few studies described their function in hepatic lipid metabolism. ACSL1 and ELOVL6 are predicted target genes of aan-miR-203a and aan-miR-125b-5p, respectively. ACSLs play a critical role in the first committed step in fatty acid metabolism by the thioesterification of long-chain fatty acids into their acyl-CoA derivatives36. ELOVL6 is responsible for the elongating C12-C16 to longer fatty acids37. In addition, ACSL1 and ELOVL6 had opposite expression patterns with aan-miR-203a and aan-miR-125b-5p, respectively. Relative luciferase activity values of recombined Dual-Luciferase reporter vectors with 3′-UTR of ACSL1 and ELOVL6 were significantly decreased by aan-miR-203a and aan-miR-125b-5p mimics, respectively. These results suggest that aan-miR-203a and aan-miR-125b-5p may implicate in the regulation of hepatic lipid metabolism by targeting ACSL1 and ELOVL6, respectively. But because anti-ELOVL6 and anti-ACSL1 antibodies specific for goose were lacking, the effect of miRNAs on the protein expression level of their target genes were not measured. However, the overexpression of aan-miR-203a and aan-miR-125b-5p in primary hepatocyte had no significant impact on lipid accumulation. Therefore, the specific function of aan-miR-203a and aan-miR-125b-5p on lipid metabolism of goose need further research.
Goose liver can be a good model to investigate the mechanism of lipid metabolism. In summary, we built fatty liver model of the A. anser and identified some conserved miRNAs with significant differences in expression in the fatty liver that may play an important role in hepatic steatosis. These results add to our current understanding of the regulation network in goose lipid metabolism.
Materials and Methods
Ethics Statement
All experimental protocols were approved by Animal Ethics Committees of Nanjing Agricultural University and Zhejiang Academy of Agricultural Sciences, China. The slaughter and sampling procedures complied with the “Guidelines on Ethical Treatment of Experimental Animals” (2006) No. 398 set by the Ministry of Science and Technology, China. All efforts were made to minimize animal suffering.
Animals and experiment design
The experiment was carried out using 90 days-old male Landes Geese hatched on the same day. 60 animals were divided into two groups and bred under natural conditions of light and temperature. All geese were housed in individual cages during the trial period. The carbohydrate-rich diet contains 3500 kcal, 4.8 g fat, and 100 g protein/kg. For 1–2 days of overfeeding, the animals were fed 150 g each time and 2 times per day. For 3–7 days of overfeeding, the animals were fed 150 g at a time and 3 times per day. At the end of overfeeding, they were fed 300 g at a time and 4 times per day. As control, geese were given a growing diet (containing 2800 kcal and 150 g protein/kg) and ad libitum during the trial period. Animals had free access to water at all times. Ten liver samples, including five control and five experimental geese (overfed for 21 d), were prepared for histological studies and analyzed for the expression of miRNAs. Prior to sampling, the geese underwent fasting for 12 h and only water was provided. The workflow can be found as Supplementary Figure S1.
Histological studies
The geese were overfed for 21 days and the control geese were sacrificed at the same time. The livers of 10 geese were collected and fixed in formalin solution for 24 h, and then embedded in paraffin and sectioned. The slices were stained with hematoxylin-eosin (HE) and observed by microscopy.
High-throughput sequencing and computational analysis
Total RNA of each sample was extracted with Trizol (Invitrogen, USA). Using a Novex 15% TBE-Urea gel, 20–30-nt long RNA fragments were isolated and purified from 10 μg of total RNA. The small RNA molecules were prepared for high-throughput sequencing according the method described previously19. High-throughput sequencing was performed with Illumina HiSeq. 2000 system. After removing the adaptors, low quality tags and contaminants reads, the clean reads were aligned with the goose genome (https://www.ncbi.nlm.nih.gov/nuccore/AOGC00000000). Then we filtered the known non-miRNA reads, such as rRNA, tRNA, snRNA, and snoRNA by screening against ncRNA deposited in the GenBank and Rfam databases. The remaining reads were searched against the miRbase 21 to identify the conserved miRNAs (http://www.miRbase.org/). The conserved miRNAs were named according to the guidelines for the miRNA terminology38. The expressions of miRNAs were normalized by Fragments per Kilobase of Transcript per Million Fragments Mapped (FPKM)39. Differentially expressed miRNAs were identified based on FPKM ≥ 100.00 in either of the two groups, fold change ≥ 1.50 or ≤ 0.67, and P < 0.001. Potential target genes of differential expressed miRNAs were predicted by matching miRNA-3′-UTR sequence and assessing their energy stability using Miranda algorithm40. And then GO Term (http://geneontology.org/) and KEGG pathway analyses (http://www.genome.jp/kegg/) of target genes were performed. The miRNA-mRNA interaction network was constructed using Cytoscape 3.5.1 software.
Validation of miRNAs by qRT-PCR
Total miRNAs from the livers were extracted using the miRcute miRNA isolation kit (TIANGEN, China). miRNAs were reverse transcribed to cDNA with the miRcute miRNA First-Strand cDNA Synthesis Kit (TIANGEN, China) and specific stem-loop primers. mRNAs were reverse transcribed to cDNA with PrimeScript RT reagent Kit with gDNA Eraser (Takara, Japan). Primers used in the qRT-PCR were listed in Supplementary Table S1. qRT-PCR was performed on ABI 7300 system (Applied Biosystems, CA) with THUNDERBIRD SYBR qPCR Mix (TOYOBO, Japan). U6 and β-actin were used as the reference gene, respectively. The relative expression levels of miRNAs and mRNAs were indicated by 2−△Ct.
Dual-luciferase reporter assay
The 3′-UTRs of goose ACSL1 and ELOVL6 were amplified and recombined to pmirGLO Dual-Luciferase miRNA Target expression Vector (Promega, USA) according to the manufacturer’s introductions. The verified vector plasmid was co-transfected into goose primary hepatocytes with miRNA mimics or negative control (GenePhama, China) by lipofectamine 2000 (Life Technologies, USA). After 48 h of transfecting, the luciferase activities were measured with Dual-Luciferase Reporter Assay System (Promega, USA).
Cell culture and transfection
Goose hepatocytes were isolated from 1–2 week-old Landes goose following the perfusion method41. Cells were cultured in RPMI1640 medium containing 10% FBS (Gibco, USA) and incubated at 5% CO2, and 37 °C on 24-well cell culture dishes (Sangon, China) at a density of 3 × 105/cm2. Medium was changed every 24 h. Cells were distributed into 4 groups after 24 h growth: blank control group (BC), negative control group (NC), aan-miR-203a mimic group and aan-miR-125b-5p mimic group. Transfections were performed using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s introductions. Cells were collected after 48 h of tranfection to detect the expression of ACSL1 and ELOVL6. The intracellular lipid contents of hepatocytes were monitored using multimode reader (Perkinelmer, Singapore) after Nile Red staining.
Statistical analysis
Differences between two groups were analyzed using the SPSS software based on two-tailed Student’s t-test. Differences were considered significant when P < 0.05. All data are expressed as the mean ± standard deviation (S.D.).
Data availability
Our miRNAs dates of high-throughput sequencing are available in NCBI (GEO99200, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE99200). All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
References
Gluchowski, N. L., Becuwe, M., Walther, T. C. & Farese, R. V., Jr. Lipid droplets and liver disease: from basic biology to clinical implications. Nature reviews. Gastroenterology & hepatology (2017).
Zhu, L. H. et al. Gene expression profile in the liver tissue of geese after overfeeding. Poultry science. 90, 107–117 (2011).
Hermier, D., Salichon, M. R., Guy, G. & Peresson, R. Differential channelling of liver lipids in relation to susceptibility to hepatic steatosis in the goose. Poult Sci. 78, 1398–1406 (1999).
Hermier, D., Rousselot-Pailley, D., Peresson, R. & Sellier, N. Influence of orotic acid and estrogen on hepatic lipid storage and secretion in the goose susceptible to liver steatosis. Biochim Biophys Acta. 1211, 97–106 (1994).
He, H. et al. Molecular cloning of the goose ACSL3 and ACSL5 coding domain sequences and their expression characteristics during goose fatty liver development. Molecular biology reports. 41, 2045–2053 (2014).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell. 136, 215–233 (2009).
Selbach, M. et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 455, 58–63 (2008).
Jin, X. et al. MicroRNA expression pattern in different stages of nonalcoholic fatty liver disease. Dig Liver Dis. 41, 289–297 (2009).
Cermelli, S., Ruggieri, A., Marrero, J. A., Ioannou, G. N. & Beretta, L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PloS one. 6, e23937 (2011).
Szabo, G. & Bala, S. MicroRNAs in liver disease. Nature reviews. Gastroenterology & hepatology. 10, 542–552 (2013).
Kerr, T. A., Korenblat, K. M. & Davidson, N. O. MicroRNAs and liver disease. Translational research: the journal of laboratory and clinical medicine. 157, 241–252 (2011).
Ceccarelli, S., Panera, N., Gnani, D. & Nobili, V. Dual Role of MicroRNAs in NAFLD. Int J Mol Sci. 14, 8437–8455 (2013).
Liu, Y., Chen, S. H., Jin, X. & Li, Y. M. Analysis of differentially expressed genes and microRNAs in alcoholic liver disease. Int J Mol Med. 31, 547–554 (2013).
Aryal, B., Singh, A. K., Rotllan, N., Price, N. & Fernandez-Hernando, C. MicroRNAs and lipid metabolism. Current opinion in lipidology. 28, 273–280 (2017).
Latorre, J. et al. Decreased lipid metabolism but increased FA biosynthesis are coupled with changes in liver microRNAs in obese subjects with NAFLD. International journal of obesity. 41, 620–630 (2017).
Han, C. et al. The role of insulin and glucose in goose primary hepatocyte triglyceride accumulation. The Journal of experimental biology. 212, 1553–1558 (2009).
Osman, R. H. et al. Fads1 and 2 are promoted to meet instant need for long-chain polyunsaturated fatty acids in goose fatty liver. Molecular and cellular biochemistry. 418, 103–117 (2016).
Lu, L. et al. The goose genome sequence leads to insights into the evolution of waterfowl and susceptibility to fatty liver. Genome biology. 16, 89 (2015).
Chen, F. et al. Identification of differentially expressed known and novel miRNAs in broodiness of goose. Mol Biol Rep. 41, 2767–2777 (2014).
Cheung, O. et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 48, 1810–1820 (2008).
Meerson, A. et al. Human adipose microRNA-221 is upregulated in obesity and affects fat metabolism downstream of leptin and TNF-alpha. Diabetologia. 56, 1971–1979 (2013).
Zaragosi, L. E. et al. Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol. 12, R64 (2011).
Wagschal, A. et al. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nature medicine. 21, 1290–1297 (2015).
Li, S. et al. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. Journal of lipid research. 50, 1756–1765 (2009).
Naderi, M. et al. Two Triacylglycerol Pathway Genes, CTDNEP1 and LPIN1, are Down-Regulated by hsa-miR-122-5p in Hepatocytes. Archives of Iranian medicine. 20, 165–171 (2017).
Esau, C. et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98 (2006).
Alisi, A. et al. Mirnome analysis reveals novel molecular determinants in the pathogenesis of diet-induced nonalcoholic fatty liver disease. Lab Invest. 91, 283–293 (2011).
He, J. et al. Analysis of miRNAs and their target genes associated with lipid metabolism in duck liver. Scientific reports. 6, 27418 (2016).
Pandey, A. K., Agarwal, P., Kaur, K. & Datta, M. MicroRNAs in diabetes: tiny players in big disease. Cell Physiol Biochem. 23, 221–232 (2009).
Guo, Y. et al. A micro-RNA expression signature for human NAFLD progression. Journal of gastroenterology. 51, 1022–1030 (2016).
Farago, N., Feher, L. Z., Kitajka, K., Das, U. N. & Puskas, L. G. MicroRNA profile of polyunsaturated fatty acid treated glioma cells reveal apoptosis-specific expression changes. Lipids Health Dis. 10, 173 (2011).
Delic, D. et al. Characterization of Micro-RNA Changes during the Progression of Type 2 Diabetes in Zucker Diabetic Fatty Rats. International journal of molecular sciences. 17 (2016).
Chen, Y. P., Jin, X., Xiang, Z., Chen, S. H. & Li, Y. M. Circulating MicroRNAs as potential biomarkers for alcoholic steatohepatitis. Liver international: official journal of the International Association for the Study of the Liver. 33, 1257–1265 (2013).
Xie, H., Lim, B. & Lodish, H. F. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 58, 1050–1057 (2009).
Ortega, F. J. et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PloS one. 5, e9022 (2010).
Kanter, J. E., Tang, C., Oram, J. F. & Bornfeldt, K. E. Acyl-CoA synthetase 1 is required for oleate and linoleate mediated inhibition of cholesterol efflux through ATP-binding cassette transporter A1 in macrophages. Biochimica et biophysica acta. 1821, 358–364 (2012).
He, J. et al. MiR-144 affects fatty acid composition by regulating ELOVL6 expression in duck hepatocytes. Cell biology international (2017).
Desvignes, T. et al. miRNA Nomenclature: A View Incorporating Genetic Origins, Biosynthetic Pathways, and Sequence Variants. Trends in genetics: TIG. 31, 613–626 (2015).
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 5, 621–628 (2008).
Enright, A. J. et al. MicroRNA targets in Drosophila. Genome biology. 5, R1 (2003).
He, J. et al. Expression pattern of L-FABP gene in different tissues and its regulation of fat metabolism-related genes in duck. Molecular biology reports. 40, 189–195 (2013).
Acknowledgements
This study was supported by the International Science & Technology Cooperation Program of China (2013DFR30980), National Waterfowl-industry Technology Research System (CARS-43-2), The Project of Hubei Innovation Center of Agricultural Science and Technology (2016-620-000-001-028) and Science Foundation for Yong Scientists of Hubei Province (2016CFB214).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: F.C., H.Z., J.J. Y.T. and L.L. Preformed the experiments: F.C., H.Z. and J.X. Analysed the data: F.C., J.W., N.Z., X.H. and W.Z. Contributed reagents/materials/analysis tools: F.C. and L.L. Wrote the paper: F.C. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, F., Zhang, H., Li, J. et al. Identification of differentially expressed miRNAs in the fatty liver of Landes goose (Anser anser). Sci Rep 7, 16296 (2017). https://doi.org/10.1038/s41598-017-16632-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-16632-7
This article is cited by
-
Effects of the breeder age on the egg yield and egg quality traits of Landes geese (Anser anser)
Tropical Animal Health and Production (2022)
-
Exploring the genetic basis of fatty liver development in geese
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.