Abstract
The Serra Gaúcha region is the most important temperate fruit-producing area in southern Brazil. Despite mealybugs (Hemiptera: Pseudococcidae) infesting several host plants in the region, there is a lack of information about the composition of species damaging different crops. A survey of mealybug species associated with commercial fruit crops (apple, persimmon, strawberry and grapes) was performed in Serra Gaúcha between 2013 and 2015, using both morphology and DNA analyses for species identification. The most abundant species were Pseudococcus viburni (Signoret), found on all four host plant species, and Dysmicoccus brevipes (Cockerell), infesting persimmon, vines and weeds. The highest diversity of mealybug species was found on persimmon trees, hosting 20 different taxa, of which Anisococcus granarae Pacheco da Silva & Kaydan, D. brevipes, Pseudococcus sociabilis Hambleton and Ps. viburni were the most abundant. A total of nine species were recorded in vineyards. Planococcus ficus (Signoret) and Pseudococcus longispinus (Targioni Tozzetti) were observed causing damage to grapes for the first time. A single species, Ps. viburni, was found associated with apples, while both Ps. viburni and Ferrisia meridionalis Williams were found on strawberry. Four of the mealybug species found represent new records for Brazil.
Similar content being viewed by others
Introduction
Brazil is the third largest fruit producer in the world, with a cultivated area of 2.5 million hectares and an estimated production of 40 million tons1,2. Fruit crops have a high social importance in different regions of the country (e.g. nearly six million employees involved2), particularly in the Southern Region, which is responsible for the largest temperate fruit production3. Within this area, the Serra Gaúcha Region of the Rio Grande do Sul State (RS) includes vineyards (Vitis spp.) for wine and juice production, and apple orchards (Malus domestica Borkh) as the most important crops3, but with other crops being of local importance. For example, vineyards in Rio Grande do Sul cover about 50,000 ha, apple trees about 17,493 ha and persimmon (Diospyros kaki L.) around 2,239 ha4.
The presence of agricultural pests is one of the main factors limiting fruit production in Brazil. The value of yield losses caused by insects is around 1.6 billion dollars annually5. Chemical insecticides, applied mainly to control fruit flies and moths, still represent the primary form of control in the Serra Gaúcha Region, although recent progress has been achieved by alternative pest-control technologies, such as the use of pheromones, toxic baits and mass trapping6,7,8. Chemical control through insecticides is often ineffective and it may have an indirect effect on non-target pests, mainly due to its negative impact on natural enemy populations.
Scale insects (Hemiptera: Coccomorpha) are important agricultural pests that can develop on fruits, leaves, branches, trunk and roots, infesting a range of host plants such as vineyards and fruit orchards9,10. The diversity of scale insect species is high, with 148 species recorded on grapevines, 69 on persimmon trees, 28 on strawberry (Fragaria x ananassa Duchesne) and 26 on apple trees11. Most adult females and nymphs extract large amounts of phloem sap while excreting the excess of water and sugar as honeydew12,13. This sugary substance then falls on leaves and fruits, serving as a substrate for the development of sooty mold fungi. Furthermore, cosmetic damage is caused to fruits as a result of scale insect infestation (e.g. by mealybugs (Pseudococcidae)), depreciating its value or even rendering it unmarketable. Mealybugs may hamper international trade to several markets due to quarantine restrictions10. Additionally, mealybugs are responsible for virus transmission of diseases, such as grapevine leafroll-associated viruses (GLRaVs)10,14. Scale insects have been associated with damaged fruit previously in Brazil15,16. In southern Brazil, mealybug population outbreaks are becoming increasingly common17, and preliminary reports have been carried out on vineyards16,18, but the lack of information on mealybugs currently threatens any prospects for integrated pest management in the area.
Correct identification of mealybug species is of utmost importance for the establishment of adequate pest management programs, especially when aiming to implement specific control techniques, such as the use of sex pheromones or biological control. Mealybug identification is usually carried out by microscopic analysis of morphological structures present on the body surface of the adult female. However, the presence of cryptic speciation and significant intraspecific morphological variation makes identification particularly challenging, especially for non-specialists19. In this case, DNA analysis can be an auxiliary tool to facilitate insect identification. The “DNA barcoding” international project proposed by Hebert et al.20 is based on the molecular characterization of one fragment of the Cytochrome oxidase subunit I (COI) mitochondrial gene, and has been used successfully for many animal taxa. However, due to the high intraspecific variation observed in the COI gene for mealybugs, the use of alternative gene regions has been favored21,22. In particular, the 28S ribosomal DNA region has proved useful and complementary to COI due to its ability to discriminate between species while maintaining low intraspecific variation23. In the present study, both morphological and DNA analysis (using COI and 28S gene regions) were used to characterize mealybug species infesting fruit orchards in the Serra Gaúcha region. Different crops, including apples and persimmon orchards, strawberry fields and vineyards were surveyed, including the weeds in the same cropped area. The comprehensive sampling enabled us to identify specific associations between mealybugs and their host plants.
Results
Morphological identification
In total, 412 specimens were studied. Morphological examination of the voucher specimens led to identification of 22 mealybug species (Table 1). Photographs of live adult female specimens were obtained for the main mealybug taxa found in the Serra Gaúcha region (Fig. 1).
Mealybug species found in fruits on Serra Gaúcha region. (a,b) ♀ Anisococcus granarae (2.08–3.28 mm55); (c,d) ♀ Dysmicoccus brevipes (1.3–2.7 mm); (e) ♀ Dysmicoccus sylvarum (up to 4.4 mm25); (f) ♀ Ferrisia meridionalis (3.2–3.5 mm); (g) ♀ Planococcus ficus (1.4–3.2 mm40); (h) ♀ Pseudococcus longispinus (1.0–4.0 mm); (i,j) ♀ Pseudococcus sociabilis (1.9–3.2 mm43); (k,l) ♀ Pseudococcus viburni (1.8–3.5 mm43).
Some specimens identified as Pseudococcus near maritimus were found to be close to Pseudococcus maritimus (Ehrhorn), differing mainly in the absence of one characteristic pair of oral ring tubular ducts next to the anterior ostioles. The concentrations of larger tubular ducts on the body margin and of multilocular disc pores on abdominal segment VII were quite variable in specimens identified as D. sylvarum, differing from the illustration in Williams and Granara de Willink (1992). Some individuals seemed to be close to D. umbambae Granara de Willink24, differing from the original description in (i) the absence of discoidal pores associated with the eyes and (ii) the absence of discoidal pores on the dorsal midline of all abdominal segments. The morphological analyses of Phenacoccus specimens was particularly complex and revealed the need for a much deeper analysis of this genus. Intraspecific differences were observed between our samples and the original descriptions of several taxa. Specimens identified as Ph. gregosus present some morphological divergences from the illustration in Williams & Granara de Willink (1992)25 in (i) the absence of clusters of multilocular disc pores on head, (ii) absence of translucent pores from the hind femur, and (iii) fewer conical setae in the cerarii (2 to 4 instead of 5 to 9). These specimens also seem to be close to Phenacoccus baccharidis Williams, but differ in (i) the type of oral collar tubular ducts present and (ii) the presence of translucent pores on the hind femur (absent in the specimens collected here). For specimens assigned to Ph. near tucumanus, morphological divergence was observed in the (i) absence of quinquelocular pores on venter in this specimens and (ii) the size of the oral collar tubular ducts, there being only one size observed in this specimen.
DNA characterization
In total, 470 DNA sequences, including both COI and 28S gene regions, were obtained from 262 mealybug specimens. The primer pair for 28S provided 260 positive sequences, showing a much higher success rate (99.2%) than the COI gene region (79.4%). The 28S alignment was 804 bp long and resulted in 20 different haplotypes, whereas the COI alignment length was 680 bp long and resulted in 42 haplotypes. The sequence data allowed us to successfully distinguish 41 different multilocus haplotypes, forming 19 different taxonomic groups. The neighbor-joining tree generated using the Tajima-Nei distance based on the 28S sequences only resulted in 3 main clusters, and provides a visual representation of the host plant-mealybug associations present in the dataset (the NJ tree was not generated to provide phylogenetic information) (Fig. 2). Evolutionary analyses were conducted in MEGA726.
Neighbor-joining tree calculated using the Tajima-Nei distance (number of base substitutions per site). The rate of variation among sites was modeled with a gamma distribution (shape parameter = 0.36). All ambiguous positions were removed for each sequence pair and there were a total of 804 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.
DNA sequences could not be obtained from specimens identified based on morphology as C. nakaharai, Ph. gregosus and Ps. rosangelae. Nevertheless, DNA sequences were obtained from the specimens identified as A. granarae, N. jacarandae, Pa. galzerae, Ph. near tucumanus, Ps. nakaharai and Ps. sociabilis for the first time in this study (no previously recorded sequences from these species could be found in public databases). For the remaining species (D. brevipes, D. sylvarum, D. texensis, F. meridionalis, F. terani, Pl. citri, Pl. ficus, Ps. longispinus, Ps. meridionalis and Ps. viburni), the sequences obtained in this work were compared with those present in the NCBI GenBank database, and displayed a percentage similarity of between 99 and 100% with sequences assigned to the same species. The sequences obtained here for Ps. meridionalis are identical to those that were obtained in a previous study and assigned as Pseudococcus near meridionalis 18.
Populations of Dysmicoccus Ferris and Pseudococcus (Westwood) presented intraspecific variation. Dysmicoccus brevipes displaying multilocus haplotypes 28S04-COI05 were found on persimmon fruits and weeds located in two nearby cities, while D. brevipes displaying multilocus 28S04-COI06 were observed only on grape roots in Flores da Cunha. For Dysmicoccus sp., intraspecific variation was observed between mealybugs that developed on persimmon fruits in three different cities (Caxias do Sul, Farroupilha and Pinto Bandeira) (28S20-COI24) and mealybugs that were collected from weeds in Caxias do Sul (28S20-COI25). Four different multilocus haplotypes were obtained from different populations of Ps. meridionalis, two found on weeds and two found on persimmon. For Ps. viburni 11 multilocus haplotypes were observed. While individuals with 28S haplotype 28S06 displayed high variability at COI (10 COI haplotypes), individuals carrying the haplotype 28S08 displayed only one COI haplotype (COI33) even when collected from widely separated sites. Hence, out of the 11 multilocus haplotypes found in Ps. viburni, most genetic diversity was observed in the populations displaying 28S06. Interestingly, intraspecific genetic variation was observed in COI among mealybugs from the same site (on the same host plant and in the same city), e.g. for A. granarae, D. sylvarum and Pa. galzerae. Intraspecific variation was also observed in the 28S gene region for the species N. jacarandae (haplotypes 28S17 and 28S18) and Ps. viburni (haplotypes 28S06 and 28S08).
Incidence of mealybug species among fruit crops
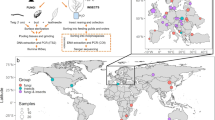
Mealybugs were found in about 50 production areas; four apple, 25 persimmon production areas and 17 vineyards (although in six of these mealybugs were found only on weeds) and only one strawberry field. The geographical distribution of mealybug species is highlighted in Fig. 3. Persimmon orchards revealed the highest incidence of mealybugs, with 56.8% (25 out of 44) of the surveyed orchards being infested. Twenty mealybug species were identified on persimmon trees (Table 1). Mealybugs occurred in 44.7% (17 out of 38) of grape sampling sites. Nine species were found in vineyards; among them, six were found on grapes and six were found on weeds (Table 1). Only two species namely, F. meridionalis and Ps. viburni were observed on strawberries, and only one production area (out of 8) was infested. Mealybugs were observed in only four (out of 34) of the apple orchards surveyed, and Pseudococcus viburni was the only species found.
Summary of mealybug species distribution in the Serra Gaúcha region, RS, Brazil, and their multilocus haplotypes. Base map obtained from www.socioeconomicatlas.rs.gov.br; created in ArcGis 10.2.1 (www.arcgis.com) and modified in Photoshop CC 2014.
Discussion
The obscure mealybug, Ps. viburni, and the pineapple mealybug, D. brevipes, were the most common mealybug species found associated with fruit crops in the Serra Gaúcha region. Pseudococcus viburni was found on all the main fruits species surveyed, commonly in large numbers and widespread in the orchards and vineyards, resulting in aesthetic damage to the fruits. In contrast, D. brevipes was never found in high numbers; this species is an important pest in pineapple fields located in other regions of Brazil, specially due to transmission of pineapple mealybug wilt–associated virus27. In the Serra Gaúcha, D. brevipes was often found associated with grapes and weeds roots. Pseudococcus viburni is an important pest species infesting vineyards in southern part of South America, including Brazil and Chile16,18,22. The species is probably native to Brazil28,29, where it presents substantial morphological variation, in life and in slide-mounted specimens, and intraspecific genetic variation at both the 28S and COI genes18,23. In the Serra Gaúcha region, Ps. viburni was observed at a low infestation level in apple orchards. Nymphs and adults were found often in the calyx and sometimes also near the pedicel of the fruit. When these mealybugs were present, damage was caused by honeydew excretion resulting in the development of sooty molds. In other countries, including Argentina, Italy, New Zealand and South Africa, Ps. viburni is considered the most economically important species found infesting apple and pears30,31,32,33. This species was also the dominant species in strawberry fields (>than 99% of the specimens collected). To date, there have been only three records of mealybugs damaging strawberry plants, Heliococcus bohemicus (Šulc) and Ps. viburni in France34, and Pseudococcus sp. in Canada35. The damage can result in translucent spots on the leaves, which resembles a mosaic; malformation and dwarfing of the leaves; shortening of the petioles; and eventually plant death35.
In this study heavy infestations with the longtailed mealybug, Ps. longispinus, were observed in at least three seedling nurseries but never in commercial vineyards; whereas heavy infestations with the vine mealybug, Planococcus ficus, were observed in two vineyards. The vine and longtailed mealybugs are common species in Californian vineyards10. Pseudococcus longispinus is also found in vineyards in Chile22 and New Zealand36, while Pl. ficus is the main species on vines in other grape production regions10,37,38. However, neither of these species had been recorded causing damages in vineyards in Rio Grande do Sul before16,18. In previous studies, Planococcus citri was considered as the most economically important species in vineyards in southern Brazil39, however, molecular studies have not detected this species in vineyards in Rio Grande do Sul after 201018, as confirmed in this work. Morphological identification of Planococcus species is extremely complex, mainly due to the low number of characters available for reliable identification and the high morphological variation that occurs in some species40. This has favored the use of molecular data to distinguish between similar taxa, such as Pl. citri and Pl. ficus 41,42. Unfortunately, the voucher specimens of previous studies were not saved for later analysis so the disappearance of Pl. citri from southern Brazilian vineyards remains unexplained.
Previous studies recorded mealybug in at least 50% of the persimmon orchards in the Serra Gaúcha region15, with more than one species found even within the same tree or fruit in the majority of the orchards. From the 20 species collected here on persimmon trees, three were found in the majority of orchards and can be considered as pests due to the high infestation rates and the damages caused to fruits: A. granarae, Ps. sociabilis and Ps. viburni. Adult females and nymphs of these species were found on the leaves and fruits (feeding directly on the fruits or in the fruit calyx). Anisococcus granarae was observed in several orchards in Farroupilha and Caxias do Sul, and high infestation rates were observed on the fruits at harvest time in at least two orchards. Pseudococcus sociabilis was the second most important species found in persimmon fruits in terms of abundance and damage. The honeydew excretion of this mealybug results in the development of sooty mold, which was observed coating the fruits at several localities.
The multilocus haplotypes and morphological identifications of the specimens collected in the present study were satisfactorily congruent except for Dysmicoccus. After initial morphological examination, the Dysmicoccus specimens were tentatively identified as D. sylvarum, despite some minor differences from the original description. Unexpectedly, molecular DNA analyses revealed the presence of two separate clades, suggesting the presence of cryptic species. Due to the limited number of adult females collected for the new clade, Dysmicoccus sp., it was not possible to achieve more conclusive results.
Here, several mealybug taxa are reported from Brazil for the first time. This is the first record of C. nakaharai, F. williamsi, Ph. gregosus and Ps. nakaharai in Brazilian territory; also the first record of Ph. gregosus and Ps. nakaharai in South America. Pseudococcus nakaharai has been recorded previously in Mexico, U.S.A., Guatemala and Japan, where it is often associated with at least 50 species of cacti43. Ferrisia williamsi was previously recorded on avocado Persea americana Mill. (Lauraceae) and ornamental trees in Colombia44, and Phenacoccus gregosus was found previously on four different host plant families (Amaranthaceae, Burseraceae, Euphorbiaceae and Fabaceae) in Mexico and Costa Rica25.
Our results revealed a high diversity of mealybug species in southern Brazil, particularly formed by species belonging to the “Pseudococcus maritimus complex” (see Ps. meridionalis, Ps. nakaharai, Ps. rosangelae, Ps. sociabilis, Ps. viburni and Pseudococcus near maritimus). The high level of genetic diversity further corroborates the previous hypothesis of a South American area of origin for Ps. viburni 28,29. New pests were identified for persimmon trees in the Serra Gaúcha region (A. granarae, Ps. sociabilis and Ps. viburni), and for grapevines (Pl. ficus) and strawberry fields (Ps. viburni). Serra Gaúcha fruit production is mainly based on small producers with yield destined for domestic trade; fruits destined for international trade are produced in other regions and states of Brazil. With the exception of exported grapes produced in the state of Pernambuco (where there is a low diversity of mealybugs18,45), little is known about mealybug infestation in the country. Further studies must be conducted in order to identify the mealybugs present in other fruit production areas of Brazil.
Methods
Mealybug sampling
Mealybugs were collected from commercial crops of apple (Rosaceae), grape (Vitaceae), persimmon (Ebenaceae) and strawberries (Rosaceae) chosen randomly in the Serra Gaúcha Region (Antônio Prado, Bento Gonçalves, Caxias do Sul, Farroupilha, Monte Belo and São Valentin do Sul) in RS, Brazil. The number of plants sampled was not standardized, but at least 30 plants per site were examined for mealybug infestations. A total of 78 mealybug populations were sampled; and considered as different populations when collected from different crop areas or different hosts within the same sampling area. Mealybug specimens for each population sample were collected from different parts of the plant (leaves, fruits, trunks and roots). For apples, grapes and persimmons, surveys were carried out between 2013 and 2015, preferably close to the harvesting time during November to April of each year, due to the higher mealybug abundance observed during this period. All developmental stages found (nymphs and/or females with/without ovisacs) were collected. In strawberry fields, sampling was carried out during September 2013 to July 2014 and September 2014 to May 2015. Adult females were stored in 95% ethanol at −20 °C. Nymphs were reared to adulthood for morphological species determination at the Entomology Laboratory of Embrapa Uva e Vinho, Bento Gonçalves, RS, Brazil. Pumpkins (Curcubita maxima Duchesne) and potato sprouts (Solanum tuberosum L.) were used as food substrates. Mealybugs were reared in plastic cages closed with voile tissue to allow ventilation. Paper towels were placed on the bottoms of the cages to prevent the accumulation of honeydew. Cages were kept at 25 ± 1 °C, relative humidity of 70 ± 10% and in a photophase of 14 h light and 10 h darkness.
Morphological Identification
Slide mounting and identification of adult females were carried out at ANSES, Laboratoire de la Santé des Végétaux, Montferrier-sur-Lez, France and Plant Protection Department of Çukurova University, Adana, Turkey. The specimens were slide-mounted using Kosztarab and Kozár’s (1988)46 method with some modification (using distilled water after KOH and cleaning the specimens using a fine brush). Identification was done using an optical microscope (LEICA DM 2500 phase contrast compound microscope) and the keys of von Ellenrieder & Watson (2016)47; Kaydan & Gullan (2012)44; Granara de Willink (2009)24, Granara de Willink & Szumik (2007)48; Williams (2004)49; Gimpel & Miller (1996)43; Williams & Granara de Willink, (1992)25; Cox & Ben-Dov (1986)50 and McKenzie (1967)51. The slide-mounted specimens were stored in the Coccoidea Collection of the Museum Ramiro Gomes Costa Porto Alegre, Brazil (MRGC); Çukurova University Coccomorpha collection, Adana, Turkey (KPTC); Anses, Laboratoire de la Santé des Végétaux, Montferrier-sur-Lez, France (ANSES/LSV) and in the Entomological Collection of Embrapa Uva e Vinho, Bento Gonçalves, Brazil (CEEUV) (listed in the Supplementary Material, Table 1).
Molecular characterization
The molecular characterization of mealybugs was performed at Sophia Agrobiotech Institut – INRA (Institut National de Recherche Agronomique), Sophia Antipolis, France. When possible, we analyzed at least three individuals from each population collected. DNA was extracted using DNeasy Blood and Tissue Kit (QIAGEN, Valencia, CA), based on the non-destructive methodology described by Malausa et al.23. Vouchers were kept in ethyl alcohol 70% for morphological analysis. DNA was amplified from two different loci: the HCO-LCO region of the cytochrome oxidase subunit 1 (mtDNA) and the 28S ribosomal gene (nuclear genome). Polymerase chain reactions were carried out using the Qiagen Multiplex PCR kit (QIAGEN, Valencia, CA), composed by 23 µL of reaction mix (1X Qiagen buffer, primers at 0.2 µM) and 2 µL of diluted DNA (1–20 ng of DNA). The primers (Forward, Reverse) C-28SLong-F 5′GAGAGTTMAASAGTACGTGAAAC3′ and C-28SLong-R 5′TCGGARGGAACCAGCTACTA3′ (28S-D2) were used for amplifying the 28S gene region52 and the primers PCO-F1 5′CCTTCAACTAATCATAAAAATATYAG3′ and Lep-R1 5′TAAACTTCTGGATGTCCAAAAAATCA3′ were used to characterize the COI gene region53. PCR reactions were performed as follows: initial denaturation for 15 minutes at 95 °C, 35 cycles of denaturation – hybridization – elongation (30 seconds at 95 °C for denaturation; 90 seconds at 58 °C for 28S and 54 °C for COI for hybridization; 60S at 72 °C for elongation), and a final extension for 10 minutes at 72 °C. PCR products were sent to Beckman Coulter Genomics (Takeley, United Kingdom) for bidirectional sequencing. Consensus sequences were constructed and checked in Seqscape v.27 (Applied Biosystems, Foster City, CA, USA). Alignments were edited manually in Bioedit v.7.02 (Hall, 1999)54. Sequences were compared between them by direct alignment, and with sequences previously described, using the search option MEGABLAST provided by the GenBank database (http://www.ncbi.nlm.gov/BLAST). New sequences were deposited in GenBank under the accession numbers: KY565026 to KY565046 for 28S and KY687861 to KY687903 for COI. Before carrying out the phylogenetic analyses from the 28S gene region only, all ambiguous positions were removed using GBLOCKS with default parameters, leaving a total of 804 positions in the final dataset. Model selection of nucleotide substitution was performed with MEGA7 Kumar et al.26 according to BIC scores (Bayesian Information Criterion) and AICc value (Akaike Information Criterion, corrected). The Tajima-Nei model, plus rate variation among sites modeled with a gamma distribution (shape parameter = 0.36), was selected for the 28S alignment. A Neighbor-joining (NJ) phylogenetic tree was built using MEGA7. Bootstrap branch support values were calculated with 500 replicates.
References
Retamales, J. B. World temperate fruit production: characteristics and challenges. Rev. Bras. Frutic. 33, 121–130 (2011).
Reetz, E. R., Kist, B. B., Santos, C. E., dos Carvalho, C. de & Drum, M. Anuário Brasileiro da Fruticultura 2014. Santa Cruz do Sul (2015).
Fachinello, J. C., Pasa, M. da. S., Schmitiz, J. D. & Betemps, D. L. Situação e perspectivas da fruticultura de clima temperado no Brasil. Rev. Bras. Frutic. E, 109–120 (2011).
IBGE – Instituto Brasileiro de Geografia e Estatística. Estados. SIDRA: Sistema IBGE de Recuperação Automática. at http://www.ibge.gov.br/estadosat/temas.php (2014).
Oliveira, C. M., Auad, A. M., Mendes, S. M. & Frizzas, M. R. Economic impact of exotic insect pests in Brazilian agriculture. J. Appl. Entomol. 137, 1–15 (2013).
Arioli, C. J. et al. Assessment of SPLAT formulations to control Grapholita molesta (Lepidoptera: Tortricidae) in a Brazilian apple orchard. Chil. J. Agric. Res. 74, 184–190 (2014).
Harter, W. R. et al. Toxicities and Residual Effects of Toxic Baits Containing Spinosad or Malathion to Control the Adult Anastrepha fraterculus (Diptera: Tephritidae). Florida Entomol. 98, 202–208 (2015).
Nondillo, A., Andzeiewski, S., Bello Fialho, F., Bueno, O. C. & Botton, M. Control of Linepithema micans (Hymenoptera: Formicidae) and Eurhizococcus brasiliensis (Hemiptera: Margarodidae) in vineyards using toxic baits. J. Econ. Entomol. 109, 1660–1666 (2016).
Daane, K. M. et al. Vineyard managers and researchers seek sustainable solutions for mealybugs, a changing pest complex (b). Calif. Agric. 62, 167–176 (2008).
Daane, K. M. et al. Biology and management of mealybugs in vineyards. In Arthropod Management in Vineyards (eds Bostanian, N. J., Vincent, C. & Isaacs, R.) 271–306, https://doi.org/10.1007/978-94-007-4032-7 (Springer Netherlands, 2012).
García Morales, M. et al. ScaleNet: A literature-based model of scale insect biology and systematics. at www.scalenet.info (2017).
Franco, J. C., Zada, A. & Mendel, Z. Novel approaches for the management of mealybug pests. In Biorational Control of Arthropod Pests 233–278, https://doi.org/10.1007/978-90-481-2316-2 (2009).
Gullan, P. J. & Martin, J. H. In Encyclopedia of Insects (eds Resh, V. H. & Cardé, R. T.) 957–967 (Elsevier, 2009).
Golino, D. A., Sim, S. T., Gill, R. & Rowhani, A. California mealybugs can spread grapevine leafroll disease. Calif. Agric. 56, 196–201 (2002).
Bavaresco, A., Botton, M., Garcia, M. S. & Nondillo, A. Danos e insetos em frutos de caquizeiro em pomares da Serra Gaúcha. Agropecuária Catarinense 18, 56–59 (2005).
Morandi Filho, W. J., Pacheco da silva, V. C., Granara de Willink, M. C., Prado, E. & Botton, M. A survey of mealybugs infesting South-Brazilian wine vineyards. Rev. Bras. Entomol. 59, 251–254 (2015).
Botton, M., Menezes-Netto, A. C., Arioli, C. J. & Oliveira, J. E. de M. Manejo integrado de insetos e ácaros-praga em uvas de mesa no Brasil. Inf. Agropecuário 36, 57–69 (2015).
Pacheco Da Silva, V. C. et al. Molecular and morphological identification of mealybug species (Hemiptera: Pseudococcidae) in Brazilian vineyards. PLoS One 9, 1–13 (2014).
Beltrà, A. et al. Guiding Classical Biological Control of an Invasive Mealybug Using Integrative Taxonomy. PLoS One 10, e0128685 (2015).
Hebert, P. D. N., Cywinska, A., Ball, S. L. & DeWaard, J. R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 270, 313–321 (2003).
Beltrà, A., Soto, A. & Malausa, T. Molecular and morphological characterisation of Pseudococcidae surveyed on crops and ornamental plants in Spain. Bull. Entomol. Res. 102, 165–172 (2012).
Correa, M. C. G., Germain, J.-F., Malausa, T. & Zaviezo, T. Molecular and morphological characterization of mealybugs (Hemiptera: Pseudococcidae) from Chilean vineyards. Bull. Entomol. Res. 102, 524–530 (2012).
Malausa, T. et al. DNA markers to disentangle complexes of cryptic taxa in mealybugs (Hemiptera: Pseudococcidae). J. Appl. Entomol. 135, 142–155 (2011).
Granara de Willink, M. C. Dysmicoccus de la Región Neotropical (Hemiptera: Pseudococcidae). Rev. la Soc. Entomológica Argentina 68, 11–95 (2009).
Williams, D. J. & Granara de Willink, M. C. Mealybugs of Central and South America. (CAB International, 1992).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, msw054 (2016).
Sether, D. M., Ullman, D. E. & Hu, J. S. Transmission of Pineapple Mealybug Wilt-Associated Virus by Two Species of Mealybug (Dysmicoccus spp.). Phytopathology 88, 1224–30 (1998).
Charles, J. G. Using parasitoids to infer a native range for the obscure mealybug, Pseudococcus viburni, in South America. BioControl 56, 155–161 (2011).
Correa, M. C. G. et al. Mealybug species from Chilean agricultural landscapes and main factors influencing the genetic structure of Pseudococcus viburni. Sci. Rep. 5, 1–10 (2015).
Ciampolini, M., Lupi, D. & Süss, L. Pseudococcus viburni (Signoret) (Hemiptera: Coccoidea) nocivo in frutticoltura nell’Italia centrale. Boll. di Zool. Agrar. e di Bachic. 34, 97–108 (2002).
Wakgari, W. M. & Giliomee, J. H. Mealybugs and their parasitoids in apple and pear orchards in the Western Cape Province, South Africa. African Plant Prot. 10, 7–11 (2004).
Dapoto, G. L., Olave, A., Bondoni, M. & Giganti, H. Obscure mealybug (Pseudococcus viburni) in pear trees in the Alto Valle of Rio Negro and Neuquen, Argentina. Acta Hortic. 909, 497–504 (2011).
Charles, J. G. et al. Evaluation of the synthetic sex pheromone of the obscure mealybug, Pseudococcus viburni, as an attractant to conspecific males, and to females of the parasitoid Acerophagus maculipennis. Entomol. Exp. Appl. 157, 188–197 (2015).
Kreiter, P. et al. Premiers résultats de protection biologique contre deux cochenilles pseudococcines nouvellement présentes sur fraisiers dans le sud de la France. Phytoma 568, 38–40 (2004).
Hildebrand, A. A. Notes on mealy bug injury on strawberry and its resemblance to crinkle. Can. J. Res. 17, 205–211 (1939).
Charles, J. G., Bell, V. A., Lo, P. L., Cole, L. M. & Chhagan, A. Mealybugs (Hemiptera: Pseudococcidae) and their natural enemies in New Zealand vineyards from 1993–2009. New Zeal. Entomol. 33, 84–91 (2010).
Becerra, V., González, M., Herrera, M. E. & Miano, J. L. Dinámica poblacional de Planococcus ficus Sign. (Hemiptera - Pseudococcidae) en viñedos. Mendoza (Argentina). Rev. la Faculdad Ciencias Agrar. 1, 1–6 (2006).
Walton, V. M., Krüger, K., Saccaggi, D. L. & Millar, I. M. A survey of scale insects (Sternorryncha: Coccoidea) occurring on table grapes in South Africa. J. Insect Sci. 9, 1–6 (2009).
Morandi Filho, W. J., Grützmacher, A. D., Botton, M. & Bertin, A. Controle químico da cochonilha-farinhenta Planococcus citri (Risso, 1813) (Hemiptera: Pseudococcidae) em diferentes idades da videira. Arq. Inst. Biol. (Sao. Paulo). 76, 427–435 (2009).
Cox, J. M. The mealybug genus Planococcus (Homoptera: Pseudococcidae). Bull. Br. Museum (Natural Hist. Entomol. 58, 1–78 (1989).
Cavalieri, V., Mazzeo, G., Garzia, G. T., Buenocore, E. & Russo, A. Identification of Planococcus ficus and Planococcus citri (Hemiptera: Pseudococcidae) by PCR-RFLP of COI gene. Zootaxa 1816, 65–68 (2008).
Rung, A., Scheffer, S. J., Evans, G. & Miller, D. Molecular identification of two closely related species of mealybugs of the Genus Planococcus (Homoptera: Pseudococcidae). Ann. Entomol. Soc. Am. 101, 525–532 (2008).
Gimpel, W. F. & Miller, D. R. Systematic analysis of the mealybugs in the Pseudococcus maritimus complex (Homoptera: Pseudococcidae). Int. Contrib. Entomol. 2, 1–163 (1996).
Kaydan, M. B. & Gullan, P. J. A taxonomic revision of the mealybug genus Ferrisia Fullaway (Hemiptera: Pseudococcidae), with descriptions of eight new species and a new genus. Zootaxa 3543, 1–65 (2012).
Lopes, F. S. C. Bioprospecção, identificação e manejo de cochonilhas-farinhentas (Hemiptera: Pseudococcidae) e insetos associados em agroecossistemas de videira no Submédio do vale do São Francisco. Thesis, Universidade Federal Rural de Pernambuco (2017).
Kosztarab, M. & Kozár, F. Scale Insects of Central Europe. (DrW. Junk Publishers, 1988).
Ellenrieder, N. V. O. N. & Watson, G. A new mealybug in the genus Pseudococcus Westwood (Hemiptera: Coccomorpha: Pseudococcidae) from North America, with a key to species of Pseudococcus from the New World. Zootaxa 4105, 65–87 (2016).
Granara de Willink, M. C. & Szumik, C. Phenacoccinae de Centro y Sudamérica (Hemiptera: Coccoidea: Pseudococcidae): Sistemática y filogenia. Rev. la Soc. Entomológica Argentina 66, 29–129 (2007).
Williams, D. J. Mealybugs of southern Asia. (The Natural History Museum/Southdene, 2004).
Cox, J. M. & Ben-Dov, Y. Planococcine mealybugs of economic importance from the Mediterranean Basis and their distinction from a new African genus (Hemiptera: Pseudococcidae). Bull. Entomol. Res. 76, 481–489 (1986).
McKenzie, H. L. Mealybugs of California: with taxonomy, biology and control of North American species. (University of California Press, 1967).
Sequeira, A. S., Normark, B. B. & Farrell, B. D. Evolutionary assembly of the conifer fauna: distinguishing ancient from recent associations in bark beetles. Proc. R. Soc. B-Biological Sci. 267, 2359–2366 (2000).
Park, D.-S. et al. Molecular identification of mealybugs (Hemiptera: Pseudococcidae) found on Korean pears. J. Econ. Entomol. 103, 25–33 (2010).
Hall, T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41, 95–98 (1999).
Pacheco da Silva, V. C., Kaydan, M. B., Germain, J.-F., Malausa, T. & Botton, M. Three new species of mealybug (Hemiptera, Coccomorpha, Pseudococcidae) on persimmon fruit trees (Diospyros kaki) in southern Brazil. Zookeys 584, 61–82 (2016).
Acknowledgements
We are grateful to Elisangela Caroline W. Galzer and the entire team from the Entomology Laboratory of Embrapa Uva e Vinho for their support; to Aurélie Blin and Didier Crochard from INRA - Institut Sophia Agrobiotech for their support with the molecular characterization; to Silvestrin fruit distributor and all the producers for allowing the search for mealybugs on their crops; to SPGG team for the permission to use of the Serra Gaúcha map; to CAPES and CNPq for the scholarship awarded to the first author and for financial support from the FP7 Marie Curie IRSES project “IPRABIO” #269196 and the IAPP project “Colbics” #324475. FP acknowledges a post-doctoral contract funded by the Beatriu de Pinos Programme of the Generalitat de Catalunya.
Author information
Authors and Affiliations
Contributions
V.C.P.S., T.M. and M.B. conceived the project; V.C.P.S., M.B.K. and J.F.G. identified the mealybugs; V.C.P.S. and F.P. analyzed the data; V.C.P.S. collected the data; M.B.K., J.F.G., T.M. and M.B. contributed with reagents/materials/analysis tools; V.C.P.S., M.B.K. and F.P. wrote the paper; and all the authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pacheco da Silva, V.C., Kaydan, M.B., Malausa, T. et al. Integrative taxonomy methods reveal high mealybug (Hemiptera: Pseudococcidae) diversity in southern Brazilian fruit crops. Sci Rep 7, 15741 (2017). https://doi.org/10.1038/s41598-017-15983-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-15983-5
This article is cited by
-
Parasitoids (Hymenoptera) of Mealybug Pests (Hemiptera: Pseudococcidae) from Southern Brazil: Molecular and Morphological Characterization
Neotropical Entomology (2021)
-
Record of new host plants associated to the invasive mealybug species Paracoccus marginatus Williams and Granara de Willink (Hemiptera: Pseudococcidae) in the Center and Littoral regions of Cameroon
International Journal of Tropical Insect Science (2021)
-
Sustainable management of the vine mealybug in organic vineyards
Journal of Pest Science (2021)
-
Temperature Thresholds and Thermal Requirements for Development and Survival of Dysmicoccus brevipes (Hemiptera: Pseudococcidae) on Table Grapes
Neotropical Entomology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.