Abstract
The aim of this study was to identify metabolite biomarkers associated with acute rejection after heart transplantation in rats using a LC-MS-based metabolomics approach. A model of heterotopic cardiac xenotransplantation was established in rats, with Wistar rats as donors and SD rats as recipients. Blood and cardiac samples were collected from blank control rats (Group A), rats 5 (Group B) and 7 days (Group C) after heart transplantation, and pretreated rats 5 (Group D) and 7 days (Group E) post-transplantation for pathological and metabolomics analyses. We assessed International Society for Heart and Lung Transplantation (ISHLT) grades 0, 3B, 4, 1 and 1 rejection in groups A to E. There were 15 differential metabolites between groups A and B, 14 differential metabolites between groups A and C, and 10 differential metabolites between groups B and C. In addition, four common differential metabolites, including D-tagatose, choline, C16 sphinganine and D-glutamine, were identified between on days 5 and 7 post-transplantation. Our findings demonstrate that the panel of D-tagatose, choline, C16 sphinganine and D-glutamine exhibits a high sensitivity and specificity for the early diagnosis of acute rejection after heart transplantation, and LC-MS-based metabolomics approach has a potential value for screening post-transplantation biomarkers.
Similar content being viewed by others
Introduction
Heart transplantation has increasingly become the primary treatment for end-stage heart diseases1,2,3. Rejection is recognized as a major risk factor affecting the survival in patients undergoing heart transplantation4,5,6. Untimely diagnosis and treatment of rejection may cause irreversible damages to transplanted organs and even be life-threatening5. Immunosuppressant treatment has been proved to greatly reduce the incidence of acute rejection; however, acute rejection remains a major cause that affects the early survival of the transplanted organs post-transplantation7,8,9. Early diagnosis of acute rejection and the subsequent adjustment of immunosuppressive protocols is therefore of great importance to improve the survival in patients undergoing transplantation.
Currently, endomyocardial biopsy remains the “golden” standard for the diagnosis of post-transplantation rejection10,11,12; however, endomyocardial biopsy is an invasive procedure, which may cause a high incidence of complications and have a low compliance13,14. In addition, the transplanted organs often undergo irreversible damages upon on diagnosis of acute rejection by pathological examinations15. A search for non-invasive approaches with high sensitivity and specificity is urgently needed for the diagnosis of post-transplantation rejection.
Metabolomics, a quantitative measurement of the dynamic multiparametric metabolic response of living systems to pathophysiological stimuli or genetic modification16, has been widely used for the screening of biomarkers for disease diagnosis17,18,19,20. Metabolomics is able to reveal metabolites, which have the most direct associations with biological phenotypes21. The aim of this study was to identify metabolite biomarkers associated with acute rejection after heart transplantation in rats using non-targeted metabolomics, so as to provide potential candidates for the early diagnosis of post-transplantation acute rejection.
Results
Pathology of the myocardial tissues of the donor heart
International Society for Heart and Lung Transplantation (ISHLT) grade 0 rejection was assessed in the normal myocardium (Fig. 1A)22. In Group B, mild edema and necrosis of myocardial cells, and massive interstitial lymphocyte infiltration were seen, and ISHLT grade 3B rejection was assessed (Fig. 1B). In Group C, mild edema and necrosis of myocardial cells, massive interstitial lymphocyte and granulocyte infiltration, and a smaller amount of thrombus were observed, and ISHLT grade 4 rejection was assessed (Fig. 1C). In addition, no myocardial cell necrosis and a little massive interstitial lymphocyte infiltration were seen in groups D and E, with ISHLT grade 1 rejection assessed (Fig. 1D and E).
HE staining of rat myocardium (×100). (A) Normal myocardium; (B) myocardial tissues sampled 5 days after organ transplantation; (C) myocardial tissues sampled 7 days after organ transplantation; (D) rats are intraperitoneally injected with cyclosporine A at a dose of 20 mg/kg one day pre-transplantation and at a dose of 10 mg/kg post-transplantation for 5 days, and then myocardial tissues are sampled; (E) rats are intraperitoneally injected with cyclosporine A at a dose of 20 mg/kg one day pre-transplantation and at a dose of 10 mg/kg post-transplantation for 7 days, and then myocardial tissues are sampled.
Data quality assessment
Quality control (QC) and quality assurance (QA) are required in order to obtain reliable and high-quality data from liquid-chromatography-tandem-mass-spectrometry (LC-MS)-based metabolomics analysis, and the QA samples cluster is considered reliable if the error in QC is within two standard deviations (SDs). All QC samples were found to cluster in the principal component analysis (PCA) plot and within the 95% confidential intervals (CIs), and the error in QC was within two SDs (Fig. 2). Total ion chromatograms (TICs) and base peak chromatograms (BPCs) showed no drifting retention time or chromatographic shapes during the whole run-sequence (Fig. 3), indicating the LC-MS results are qualified for statistical analyses.
Multivariate statistical analysis
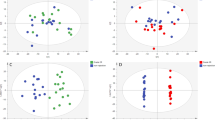
Following pretreatment, the LC-MS data were subject to PCA and partial least square-discriminant analysis (PLS-DA), and permutation tests were performed to prevent the PLS-DA model overfitting23,24. In order to investigate the changes of the global metabolite profiles in the serum of rats given immunosuppressive therapy at acute rejection, the samples in the 5 groups were combined for PCA and PLS-DA. The results showed that the samples from the 5 groups were all distributed in the 95% CI PCA scores and appeared remarkable clusters and grouping (Fig. 4), indicating that immunosuppressive therapy exhibits a remarkable metabolic disturbance during acute rejection. The LC-MS data sets captured from the positive- and negative-ion modes were subject to the same grouping and statistical analyses, and significant clusters were found in the clustering of LC-MS spectral features among the groups A, B and C, and the groups A, B and D in the positive- and negative-ion modes, and among the groups A, C and E in the positive-ion mode. Taking the pathological examinations of the rat myocardial tissues, it is indicated that the metabolite profiles change during acute rejection.
Principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) score plots. (A) PCA score plot. The horizontal ordinate indicates the score of samples in the first principle component, and the vertical ordinate indicates the score of samples in the second principle component. R2X1, the interpretable degree of the first principle component; R2X2, the interpretable degree of the second principle component. (B) PLS-DA score plot; (C), permutation tests for the PLS-DA score plot. The horizontal ordinate indicates the score of samples in the first principle component, and the vertical ordinate indicates the score of samples in the second principle component. R2X1, the interpretable degree of the first principle component; R2X2, the interpretable degree of the second principle component.
Identification and characterization of potential biomarkers
According to the PCA and PLS-DA results, we identified potential biomarkers based on a combination of an independent t test (P < 0.05), variable importance in project (VIP, ≥1), S-plot. pcorr and receiver operating characteristic (ROC) curve analysis of the differential metabolites. There were 15 differential metabolites identified between groups A and B, and ROC curve analysis revealed area under curve (AUC) values of >0.90 in 6 metabolites. We identified 14 differential metabolites between groups A and C, and 5 metabolites had AUC values of >0.90. In addition, there were 10 differential metabolites identified between groups B and C, and 2 metabolites had AUC values of >0.90 (Table 1).
On day 5 after heart transplantation, there were 14 differential metabolites identified between groups B and D, and ROC curve analysis revealed AUC values of >0.90 in 4 metabolites, including D-glutamine, choline, C16 sphinganine and D-tagatose (Table 2). We identified 13 differential metabolites between groups C and E 7 days after heart transplantation, and ROC curve analysis revealed AUC values of >0.90 in 5 metabolites, including D-tagatose, choline, lysopc (15:0), C16 sphinganine and D-glutamine. There were 15 differential metabolites between groups A and D, with AUC values of >0.90 in 5 metabolites, and there were 13 differential metabolites between groups A and E, with AUC values of >0.90 in 5 metabolites. In addition, there were 10 differential metabolites between groups D and E, without AUC values of >0.90 in any metabolites.
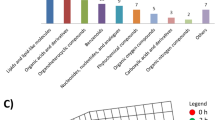
Hierarchical clustering and KEGG metabolic pathways of the differential metabolites in the five groups are shown in Fig. 5. Low expression of threonate, 2-deoxyuridine and 3-pyrimindin-2-yl-propionic acid was seen in groups B and D 5 days post-transplantation, indicating no obvious alterations seen in these metabolites expression 5 days post-transplantation; however, the levels of these metabolites significantly increased 7 days post-transplantation. Moderate and low expression of C16 sphinganine, L-phenylalanine and triiodothyronine, and moderate and high expression of D-glutamine, 15-keto latanoprost and cholic acid was seen in groups D and E relative to in groups B and C. The data indicate that these metabolites are greatly affected by cyclosporine A, and may be indicative of the development of acute rejection. In addition, these metabolites are mainly involved in amino acid, fatty acid and bile acid metabolism, demonstrating that acute rejection may mainly affect substance metabolism pathways, but have few effects on energy metabolism pathways.
Hierarchical clustering and KEGG metabolic pathways of differential metabolites. (A) Heat maps of differential metabolites; (B) heat maps of KEGG metabolic pathways. Pathway activity profiling (PAPi) is used to calculate the activity score of each metabolic pathway. The heat maps are based on metabolic process and classified, and the scattergrams of the metabolic pathways are statistical analyses of the KEGG metabolic pathways (P < 0.05 as revealed by ANOVA).
By comparing the differential metabolic biomarkers identified between on days 5 and 7 post-transplantation, there were four common differential metabolites, including D-tagatose, choline, C16 sphinganine and D-glutamine. During acute rejection, the serum D-tagatose level was significantly elevated, while the serum levels of choline, C16 sphinganine and D-glutamine were markedly decreased. These four potential metabolites were analyzed by logistic regression analysis and ROC curve analysis to build a panel of biomarkers. The results showed that the panel of the 4 potential metabolic biomarkers (D-tagatose, choline, C16 sphinganine and D-glutamine) had an AUC value of 0.983, and the sensitivity and specificity of the biomarker panel were 90.2% and 92.3%, respectively, at the best cut-off value. Our findings demonstrate that the panel of D-tagatose, choline, C16 sphinganine and D-glutamine may provide a higher value for the diagnosis of acute rejection after heart transplantation.
Discussion
Acute rejection remains a major problem in heart transplantation until now, which may be life-threatening if timely therapy is not given25. However, there are no effective approaches for the early identification of acute rejection to date. A search for non-invasive, rapid and accurate early diagnosis of acute rejection after heart transplantation is therefore urgently needed. Proteomics has been widely used for screening transplantation-related biomarkers26,27,28,29,30; however, proteomics suffers from problems of acquisition of huge amounts of information and high detection costs31.
The term metabolomics was firstly introduced in 199932. Until now, approximately 2500 metabolites have been identified in the human body33, which greatly reduces the screening difficulty and workload by using metabolomics. Based on the platforms such as LC-MS, nuclear magnetic resonance (NMR) spectroscopy ion-mobility spectrometry and electrochemical detection34, metabolomics is able to non-invasively, more accurately and more rapidly investigate drug metabolism and in-vivo metabolic changes in disease models with lower costs35,36,37,38.
Metabolomics approaches have been employed in organ transplantations39,40,41. NMR spectroscopy-based metabolomics analysis of the whole blood samples from a patient who underwent two consecutive liver transplantations showed distinctive metabolic changes 2 h after the first transplant surgery when no other variable or conventional laboratory tests indicated poor graft function, indicating the great value of metabolomics in the identification of transplantation-related biomarkers42,43,44. Currently, a huge lack of donors is a major crisis in organ transplantation; however, there are two key issues that remain unaddressed in organ transplantation, which are how to assess whether a donors after cardiac death (DCD) liver is damaged beyond repair, and whether machine perfusion has rendered an injured organ sufficiently viable for transplantation45. A metabolic analysis of the transient responses of cadaveric rat livers during normothermic machine perfusion revealed that four metabolites ornithine, arginine, albumin and tyrosine that varied significantly for ischemic livers enabled the evaluation of the organ ischemic injury level, and may be used to identify whether an organ is feasible for transplantation46. The metabolomic approach based on ultra performance liquid chromatography (UPLC) and quadrupole time-of-flight MS was considered as a feasible tool to investigate the metabolic abnormality in the acute graft rejection in renal transplantation40. In addition, metabolomics has shown promising values in the storage of donor kidneys47, cancers48 and heart diseases49.
In the current study, a rat model of heterotopic heart transplantation was established, and the post-transplantation heart can be considered as a tissue with ejection function alone. Therefore, the impact of liver metabolism on endogenous metabolites may be excluded relative to liver transplant models. Pathological examinations confirmed the successful modeling of heart transplantation on day 5 post-transplantation in Group B and on day 7 post-transplantation in Group C. Previous studies have shown that mild lymphocyte infiltration occurs in animal models of heart transplantation on day 2 post-transplantation, and peaks on day 8 post-transplantation. During this period, myocardial edema, degeneration and necrosis, and inflammatory cell infiltration are seen50. Ono and Lindsey also observed acute rejection 5 to 7 days post-transplantation in untreated rats51, which is consistent with the findings from the present study.
Currently, the mechanisms of acute rejection have not been completely demonstrated52. The primary mechanism for acute rejection is a T lymphocytes-mediated cellular immune response. Namely, antigen-activated T cells are aggregated in transplants and release active cytokines or cause a direct killing effect, resulting in immune injuries; in addition, ischemia-reperfusion injury is also a mechanism of acute rejection52. In this study, we investigated the metabolic profiles in the serum of rats undergoing heart transplantation using non-targeted metabolomics, and PCA, PLS-DA and ROC curve analysis was employed to compare the metabolic profiles between groups. The difference in metabolites levels between groups may indicate the apparent alterations of metabolites during acute rejection, and serum metabolomics may identify biomarkers for predicting and diagnosing acute rejection. In this study, there were 14 differential metabolites identified between groups B and D, including 5 types of amino acids (D-glutamine, muricholic acid, 3-oxo-chol-11-enic acid, cholic acid and creatinine), 3 types of lipids (15-keto latanoprost, mycinamicin VII and 16,17-epoxy-DHA), 2 types of alcohols (C16 sphinganine and sideridiol), 2 types of organic bases (choline and 2-methylbutyroylcarnitine), one saccharide (D-tagatose), and one indole derivative (serotonin). There were 11 metabolites down-regulated and 3 metabolites up-regulated in Group B, and these metabolites were found to be involved in amino acid (D-glutamine, 3-oxo-chol-11-enic acid, serotonin, choline, sideridiol and creatinine), fatty acid (16,17-epoxy-DHA, 2-methylbutyroylcarnitine and C16 sphinganine), bile acid (muricholic acid and cholic acid), drug (15-keto latanoprost and mycinamicin VII) and saccharide (D-tagatose) metabolism. The results demonstrate that cyclosporine A exhibits multiple effects on acute rejection in rats, and has the greatest impact on amino acid metabolism. As a polypeptide, cyclosporine A exhibits immunosuppressive functions through impacting T cell activity via the amino acid-protein interactions. Our data demonstrate that four metabolites, including choline, C16 sphinganine, D-tagatose and D-glutamine, may be used as biomarkers for the prediction and diagnosis of acute rejection after heart transplantation. Choline, which is involved in lipid metabolism, up-regulates the expression of HSP70 and Cox-2, two effectors of ischemic preconditioning, through activating M3 receptor, thereby alleviating myocardial ischemia-reperfusion injury53. In this study, choline was found to be significantly reduced in Group B than in Group D on day 5 post-transplantation, and in Group C than in Group E on day 7 post-transplantation. C16 sphinganine has been found active to protect ischemia/reperfusion-induced arrhythmias in the isolated heart, prevent allograft rejection during heart transplantation, promote cardiovascular regeneration and maintain myocardial cell survival during ischemia54. In addition, low C16 sphinganine was reported to promote the development of rejection54. We detected a significant reduction in serum C16 sphinganine in Group B than in Group D on day 5 post-transplantation, and in Group C than in Group E on day 7 post-transplantation. D-tagatose, which is involved in energy metabolism55, was found to significantly reduce in Group B than in Group D on day 5 post-transplantation, and in Group C than in Group E on day 7 post-transplantation. Metabolomics analysis revealed that glutamine, glycine and methionine were of great importance to induce immune tolerance after liver transplantation in rats, while the metabolites urocanic acid and etiocholanolone may affect the induction of immune tolerance56.
In this study, we identified two metabolic biomarkers, C16 sphinganine and oxoglutaric acid, by comparing the metabolic profiles between groups B and C. C16 sphinganine was found to significantly reduce on day 7 (Group C) in relative to day 5 (Group B) after heart transplantation, while oxoglutaric acid significantly increased. Oxoglutaric acid is involved in energy metabolism and tricarboxylic acid cycle to produce amino acid. As a precursor of glutamine, oxoglutaric acid may replace the function of glutamine in enteral nutrition and immune response, and it may enhance immune functions during liver transplantation57. It is hypothesized that elevation of oxoglutaric acid may aggravate acute rejection after heart transplantation through increasing immune functions. However, further studies are required to test the hypothesis. Pathological examinations showed aggravation of rejection in Group C than in Group B, suggesting that these two metabolites may be useful for identifying the degree of acute rejection.
KEGG pathway provides integration and interpretation of metabolic pathways, including metabolism of carbohydrates, nucleosides and amino acids and biodegradation of organic compounds, which may reveal the possible metabolic pathways58. In this study, KEGG pathway analysis revealed abnormal metabolism of amino acid, purine, pyrimidine, oxidative phosphorylation, and D-glutamine in rats during acute rejection.
The results of the present study demonstrate that the panel of D-tagatose, choline, C16 sphinganine and D-glutamine exhibits a high sensitivity and specificity for the early diagnosis of acute rejection after heart transplantation. The current study is the first to employ LC-MS to investigate the metabolomic alterations in the serum of rats undergoing heart transplantation, which provides basis for examining the role of related molecules in rejection following heart transplantation. The LC-MS-based metabolomics technique, a novel approach that may identify biomarkers associated with acute rejection after heart transplantation, has a potential value for screening post-transplantation biomarkers.
Materials and Methods
Ethical statement
This study was approved by the Ethical Review Committee of the Union Hospital of Fujian Medical University (approval number: FJXHYY2013-00127). All animal experimentations were performed according to the international and national guidelines for the management and care of laboratory animals.
Animals
Forty donor male Wistar rats, each weighing 224.61 ± 12.10 g, and forty recipient male SD rats, each weighing 316.26 ± 11.61 g, were purchased from Wushi laboratory animal Co., Ltd. (Minhou, China). All animals were housed in a clean laboratory animal facility and given free access to food and water.
Animal modeling and grouping
All Wistar and SD rats were randomly assigned into 5 groups, of 8 animals in each group. A rat model of heterotopic cardiac xenotransplantation was established using a modified Heron’s technique59, with Wistar rats as donors and SD rats as recipients. Blood and myocardial samples were collected from blank control rats (Group A). In two acute rejection groups, the recipient rats were not given any pretreatment, and blood and myocardial samples were collected 5 (Group B) and 7 days (Group C) after heart transplantation. In two pretreatment groups, the recipient rats were intraperitoneally injected with cyclosporine A (Novartis Pharmaceuticals Corporation; East Hanover, NJ, China) at a dose of 20 mg/kg one day prior to transplantation and at a daily dose of 10 mg/kg post-transplantation. Briefly, cyclosporine A (250 mg/5 ml) was added into 95 ml normal saline to produce a solution at 2.5 mg/ml. For a rat weighing 300 g, 2.4 ml of cyclosporine A solution was given by intraperitoneal injection prior to transplantation and 1.2 ml was given post-transplantation. Blood and cardiac samples were collected 5 (Group D) and 7 days (Group E) post-transplantation.
Blood collection
Rats were anesthetized with intraperitoneal injection of 3% pentobarbital sodium (Maixin Biological Technology Development Co., Ltd.; Fuzhou, China) at a single dose of 45 mg/kg. Then, rats were sterilized, and the abdominal wall was incised along the median line of the abdomen. The inferior cava vena was exposed, and 3 ml of blood samples were collected by puncture of the inferior vena cava. The collected blood samples were slowly transferred to additive-free blood collection tubes, stored at 4 °C for 2 h, and centrifuged at 4 °C, 3000 r/min for 10 min. The supernatant was carefully collected, and stored at −80 °C for the subsequent experiments.
Pathological examinations
Following blood collection, the skin over the neck was incised along the original incision in rats receiving heart transplantation, and the transplanted heart was rapidly dissociated and extracted. In control rats, an incision was made on the median chest, and the heart was rapidly dissociated and extracted. Cardiac specimens were fixed in 10% neutral buffered formalin for more than 24 h, embedded in paraffin, cut into 5 µm sections, and subject to hematoxylin & eosin (HE) staining. The rejection was graded according to the criteria proposed by the International Society for Heart and Lung Transplantation22.
Metabolomics analysis
All samples were thawed at 4 °C, and 100 µL of each sample was transferred to 1.5 ml centrifuge tubes. Then, each tube was added with 200 µL of methanol (pre-cooled at −20 °C) and vortexed for 60 s. The samples were centrifuged at 4 °C, 12000 r/min for 10 min, and the supernatant was collected into another 1.5 mL centrifuge tubes. The samples were then filtered through a 0.22 µm membrane, and the extracts were collected for LC-MS analysis, while 20 µL from each extract and mixture served as QC samples. These QC samples were used to monitor deviations of the analytical results from these pool mixtures and compare them to the errors caused by the analytical instrument itself.
Chromatographic separation was accomplished in an Acquity UPLC system (Waters Corporation; Milford, MA, USA) equipped with an ACQUITY UPLC® HSS T3 (150 × 2.1 mm, 1.8 µm) column maintained at 35 °C. The temperature of the autosampler was 8 °C. Gradient elution of analytes was carried out with 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) at a flow rate of 0.3 ml/min. Injection of 3 μL of each sample was done after equilibration. An increasing linear gradient of solvent B (v/v) was used as follows: 0 to 1 min, 2% B; 1 to 11 min, 2% to 50% B; 11 to 17 min, 50% to 98% B; 17 to 18 min, 98% B; 18 to 19 min, 98% to 2% B; 19 to 20 min, 2%.
The ESI-MSn experiments were executed on the Thermo LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Inc.; Waltham, MA, USA) with the spray voltage of 4.8 kV and −4.5 kV in positive and negative modes, respectively. Sheath gas and auxiliary gas were set at 45 and 10 arbitrary units, respectively. The capillary temperature was 325 °C. The voltages of capillary and tube were 35 V and 50 V, −15 V and −50 V in positive and negative modes, respectively. The Orbitrap analyzer scanned over a mass range of m/z 50 to 1000 for full scan at a mass resolution of 60000. Data dependent acquisition (DDA) MS/MS experiments were performed with CID scan. The normalized collision energy was 30 eV. Dynamic exclusion was implemented with a repeat count of 2, and exclusion duration of 15 s.
Data management and multivariate statistical analysis
The original LC-MS data were transformed into mzXML format using the software ProteoWizard version 3.0.8789 60. Peaks identification, peaks filtration and peaks alignment were done using the XCMS program in R package version 3.1.3, to obtain data matrix containing mass-to-charge ratio (m/z), retention time and peak intensity. Then, the data matrix was subject to the following multivariate statistical analyses, including PCA and PLS-DA, and permutation tests were conducted to avoid the PLS-DA model overfitting.
The criteria for a differential metabolite included a P value of < 0.05 as revealed by an independent t test, VIP ≥ 1, and S-plot. pcorr ≥ 0.8 61, a variable with VIP ≥ 1 indicated that the importance of this variable in the project was higher than the mean importance62.
Metabolite identification and characterization
All metabolites were validated and annotated in the Human Metabolome Database (HMDS, http://www.hmdb.ca/), METLIN metabolite database (http://metlin.scripps.edu/landing_page.php?pgcontent=mainPage) and the standard sample database built by Yancheng Zhijian Biotechnology Cooperation (Yancheng, China), and the identified differential metabolites were evaluated for potential biomarkers using ROC curve analysis. An AUC value of >0.9 indicated a high accuracy for the prediction.
References
Andrew, J. & Macdonald, P. Latest developments in heart transplantation: a review. Clin. Ther. 37, 2234–2241 (2015).
Argenziano, M., Michler, R. E. & Rose, E. A. Cardiac transplantation for endstage heart disease. Heart. Vessels. 7, 23–27 (1997).
Heroux, A. L. et al. Heart transplantation as a treatment option for end-stage heart disease in patients older than 65 years of age. J. Heart. Lung. Transplant. 12, 573–578 (1993).
Toyoda, Y., Guy, T. S. & Kashem, A. Present status and future perspectives of heart transplantation. Circ. J. 77, 1097–1110 (2013).
Almenar, L. et al. Risk factors affecting survival in heart transplant patients. Transplant. Proc. 37, 4011–4013 (2005).
Bove, A. A. et al. Factors affecting survival after heart transplantation: comparison of pre- and post-1999 listing protocols. J. Heart. Lung. Transplant. 25, 42–47 (2006).
Cole, E. et al. A pilot study of steroid-free immunosuppression in the prevention of acute rejection in renal allograft recipients. Transplantation. 72, 845–850 (2001).
Bhorade, S. M. et al. New combination of immunosuppressive therapy decreases acute rejection in lung transplantation. J. Heart. Lung. Transplant. 22, S120 (2003).
Garcia, M. et al. Campath-1H immunosuppressive therapy reduces incidence and intensity of acute rejection in intestinal and multivisceral transplantation. Transplant. Proc. 36, 323–324 (2004).
Billingham, M. E. Endomyocardial biopsy diagnosis of acute rejection in cardiac allografts. Prog. Cardiovasc. Dis. 33, 11–18 (1990).
Cooper, L. T. et al. The role of endomyocardial biopsy in the management of cardiovascular disease: a scientific statement from the American Heart Association, the American College of Cardiology, and the European Society of Cardiology. Circulation. 116, 2216–2233 (2007).
From, A. M., Maleszewski, J. J. & Rihal, C. S. Current status of endomyocardial biopsy. Mayo. Clin. Proc. 86, 1095–1102 (2011).
Winters, G. L. The challenge of endomyocardial biopsy interpretation in assessing cardiac allograft rejection. Curr. Opin. Cardiol. 12, 146–152 (1997).
Patel, J. K. & Kobashigawa, J. A. Should we be doing routine biopsy after heart transplantation in a new era of anti-rejection? Curr. Opin. Cardiol. 21, 127–131 (2006).
Zwang, N. A. & Turka, L. A. Transplantation immunology in 2013: New approaches to diagnosis of rejection. Nat. Rev. Nephrol. 10, 72–74 (2014).
Nicholson, J. K., Connelly, J., Lindon, J. C. & Holmes, E. Metabonomics: a platform for studying drug toxicity and gene function. Nat. Rev. Drug. Discov. 1, 153–161 (2002).
Lindon, J. C., Holmes, E. & Nicholson, J. K. Metabonomics and its role in drug development and disease diagnosis. Expert. Rev. Mol. Diagn. 4, 189–199 (2004).
Lindon, J. C., Holmes, E., Bollard, M. E., Stanley, E. G. & Nicholson, J. K. Metabonomics technologies and their applications in physiological monitoring, drug safety assessment and disease diagnosis. Biomarkers. 9, 1–31 (2004).
Ng, J. Y., Pasikanti, K. K. & Chan, E. C. Y. Trend analysis of metabonomics and systematic review of metabonomics-derived cancer marker metabolites. Metabolomics. 7, 155–178 (2011).
Trezzi, J. P., Vlassis, N. & Hiller, K. The role of metabolomics in the study of cancer biomarkers and in the development of diagnostic tools. Adv. Exp. Med. Biol. 867, 41–57 (2015).
Nicholson, J. K. & Lindon, J. C. Systems biology: Metabonomics. 455, 1054–1056 (2008).
Stewart, S. et al. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J. Heart. Lung. Transplant. 24, 1710–1720 (2005).
Zhou, B., Xiao, J. F., Tuli, L. & Habtom, W. LC-MS-based metabolomics. Mol. Biosyst. 8, 470–481 (2012).
Foster, J. C., Nan, B., Shen, L., Kaciroti, N. & Taylor, J. M. Permutation testing for treatment-covariate interactions and subgroup identification. Stat. Biosci. 8, 77–98 (2016).
Delgado, J. F., Sánchez, V. & de la Calzada, C. S. Acute rejection after heart transplantation. Expert. Opin. Pharmacother. 7, 1139–1149 (2006).
Cohen-Freue, G. V. et al. Computational biomarker pipeline from discovery to clinical implementation: plasma proteomic biomarkers for cardiac transplantation. PLoS. Comput. Biol. 9, e1002963 (2013).
Schaub, S., Wilkins, J. A. & Nickerson, P. Proteomics and renal transplantation: searching for novel biomarkers and therapeutic targets. Contrib. Nephrol. 160, 65–75 (2008).
Liang, S. L. & Clarke, W. Urine proteomic profiling for biomarkers of acute renal transplant rejection. Methods. Mol. Biol. 641, 185–191 (2010).
Mas, V. R., Mueller, T. F., Archer, K. J. & Maluf, D. G. Identifying biomarkers as diagnostic tools in kidney transplantation. Expert. Rev. Mol. Diagn. 11, 183–196 (2011).
Anglicheau, D., Naesens, M., Essig, M., Gwinner, W. & Marquet, P. Establishing biomarkers in transplant medicine: a critical review of current approaches. Transplantation. 100, 2024–2038 (2016).
Bohra, R. et al. Proteomics and metabolomics in renal transplantation-quo vadis? Transpl Int. 26, 225–241 (2013).
Nicholson, J. K., Lindon, J. C. & Holmes, E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 29, 1181–1189 (1999).
Wishart, D. S. et al. HMDB 3.0–The Human Metabolome Database in 2013. Nucleic. Acids. Res. 41, D801–D807 (2013).
Dieterle, F. et al. NMR and MS methods for metabonomics. Methods. Mol. Biol. 691, 385–415 (2011).
Waters, N. J. The role of metabonomics at the interface between drug metabolism and safety assessment. Curr. Drug. Metab. 11, 686–692 (2010).
Nicholson, J. K., Everett, J. R. & Lindon, J. C. Longitudinal pharmacometabonomics for predicting patient responses to therapy: drug metabolism, toxicity and efficacy. Expert. Opin. Drug. Metab. Toxicol. 8, 135–139 (2012).
Sengupta, A., Ghosh, S., Sharma, S. & Sonawat, H. M. 1H NMR metabonomics indicates continued metabolic changes and sexual dimorphism post-parasite clearance in self-limiting murine malaria model. PLoS One. 8, e66954 (2013).
Zhao, L. et al. Identification of key metabolic changes in renal interstitial fibrosis rats using metabonomics and pharmacology. Sci. Rep. 6, 27194 (2016).
Li, Q. et al. Metabonomics study of intestinal transplantation using ultrahigh-performance liquid chromatography time-of-flight mass spectrometry. Digestion. 77, 122–130 (2008).
Chen, J. et al. Metabonomics study of the acute graft rejection in rat renal transplantation using reversed-phase liquid chromatography and hydrophilic interaction chromatography coupled with mass spectrometry. Mol. Biosyst. 8, 871–878 (2012).
Benahmed, M. A. et al. The assessment of the quality of the graft in an animal model for lung transplantation using the metabolomics 1H high-resolution magic angle spinning NMR spectroscopy. Magn. Reson. Med. 68, 1026–1038 (2012).
Serkova, N. J., Standiford, T. J. & Stringer, K. A. The emerging field of quantitative blood metabolomics for biomarker discovery in critical illnesses. Am. J. Respir. Crit. Care. Med. 184, 647–655 (2011).
Serkova, N. J. & Brown, M. S. Quantitative analysis in magnetic resonance spectroscopy: from metabolic profiling to in vivo biomarkers. Bioanalysis. 4, 321–341 (2012).
Serkova, N. J. et al. Early detection of graft failure using the blood metabolic profile of a liver recipient. Transplantation. 83, 517–521 (2007).
Calne, R. Challenges of organ transplantation. Transplant. Proc. 37, 1979–1983 (2005).
Perk, S. et al. A metabolic index of ischemic injury for perfusion-recovery of cadaveric rat livers. PLoS One. 6, e28518 (2011).
Nath, J. et al. Metabolic differences between cold stored and machine perfused porcine kidneys: A 1H NMR based study. Cryobiology. 74, 115–120 (2017).
Hyndman, M. E., Mullins, J. K. & Bivalacqua, T. J. Metabolomics and bladder cancer. Urol. Oncol. 29, 558–561 (2011).
Bodi, V. et al. Metabolomic profile of human myocardial ischemia by nuclear magnetic resonance spectroscopy of peripheral blood serum: a translational study based on transient coronary occlusion models. J. Am. Coll. Cardiol. 59, 1629–1641 (2012).
Abbott, P. P., Lindsey, E. S., Creech, O. Jr. & Dewitt, C. W. A technique for heart transplantation in the rat. Arch. Surg. 89, 645–652 (1964).
Ono, K. & Lindsey, E. S. Improved technique of heart transplantation in rats. J. Thorac. Cardiovasc. Surg. 57, 225–229 (1969).
Ponticelli, C. The mechanisms of acute transplant rejection revisited. J. Nephrol. 25, 150–158 (2012).
Wang, L. Y. et al. Protective effect of choline on ischemia-reperfusion injury in isolated rat heart. Chin. Pharmacol. Bull. 23, 600–603 (2007).
Bartke, N. & Hannun, Y. A. Bioactive sphingolipids: metabolism and function. J. Lipid. Res. 50, 91–96 (2009).
Livesey, G. & Brown, J. C. D-tagatose is a bulk sweetener with zero energy determined in rats. J. Nutr. 126, 1601–1609 (1996).
Zhang, L. & Liu, S.Y. Experimental study on F protein-induced immune tolerance in rats undergoing orthotopic liver transplantation. Tianjin: Tianjin Medical University, 2012.
Chen, H. et al. Studying the signaling role of 2-oxoglutaric acid using analogs that mimic the ketone and ketal forms of 2-oxoglutaric acid. Chem. Biol. 13, 849–56 (2006).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic. Acids. Res. 28, 27–30 (2000).
Hasan, R. I. & White, D. J. Modification of Heron’s technique for heterotopic cardiac transplantation in rats. J. Thorac. Cardiovasc. Surg. 107, 1168–1169 (1994).
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R. & Siuzdak, G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 (2006).
Wang, J. B. et al. Metabolomic profiling of autoimmune hepatitis: the diagnostic utility of nuclear magnetic resonance spectroscopy. J. Proteome. Res. 13, 3792–3801 (2014).
Eriksson, L. et al. Multivariate and Megavariate Data Analysis (eds Eriksson, L. et al.). 27–29 (Umetrics Academy, 2006).
Acknowledgements
This study was supported by the Natural Science Foundation of Fujian Province (grant no. 2014J01324).
Author information
Authors and Affiliations
Contributions
F.L. and Y.B.Y. conceived and designed the study. F.L., Y.O., C.Z.H., S.Z.L. and Y.B.Y. performed the experiments and the field survey. F.L. collected and analyzed the data. F.L. prepared the manuscript. Y.B.Y. provided valuable comments on the revision of the manuscript. F.L. revised and finalized the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lin, F., Ou, Y., Huang, CZ. et al. Metabolomics identifies metabolite biomarkers associated with acute rejection after heart transplantation in rats. Sci Rep 7, 15422 (2017). https://doi.org/10.1038/s41598-017-15761-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-15761-3
This article is cited by
-
Towards Allograft Longevity: Leveraging Omics Technologies to Improve Heart Transplant Outcomes
Current Heart Failure Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.