Abstract
Clostridium difficile is a significant concern as a nosocomial pathogen, and genetic tools are important when analyzing the physiology of such organisms so that the underlying physiology/pathogenesis of the organisms can be studied. Here, we used TargeTron to investigate the role of selenoproteins in C. difficile Stickland metabolism and found that a TargeTron insertion into selD, encoding the selenophosphate synthetase that is essential for the specific incorporation of selenium into selenoproteins, results in a significant growth defect and a global loss of selenium incorporation. However, because of potential polar effects of the TargeTron insertion, we developed a CRISPR-Cas9 mutagenesis system for C. difficile. This system rapidly and efficiently introduces site-specific mutations into the C. difficile genome (20–50% mutation frequency). The selD CRISPR deletion mutant had a growth defect in protein-rich medium and mimicked the phenotype of a generated TargeTron selD mutation. Our findings suggest that Stickland metabolism could be a target for future antibiotic therapies and that the CRISPR-Cas9 system can introduce rapid and efficient modifications into the C. difficile genome.
Similar content being viewed by others
Introduction
Clostridioides difficile, more commonly known as Clostridium difficile 1,2, is a Gram-positive, anaerobic, spore-forming bacterium that is the major cause of antibiotic-associated diarrhea. Most-commonly, patients undergoing antibiotic treatment are at high risk for C. difficile infection (CDI) due to the disruption of the normal colonic microbiota by broad-spectrum antibiotics3,4,5. Spores, the metabolically-dormant form of C. difficile that can survive passage between hosts in the aerobic environment, are ingested by a host and germinate in response to small-molecule germinants (i.e., cholic acid, and its derivatives, and an amino acid, such as glycine) into a vegetative, toxin-producing cell6,7,8. These toxins, TcdA and TcdB, damage epithelial cells which results in the symptoms of CDI9,10,11.
Much of our understanding of C. difficile physiology has come in the last few years and coincided with the development of genetic tools for this organism12,13,14,15,16,17. The tools available to genetically manipulate C. difficile include: i) single-crossover integration of segregationally unstable plasmids15,16; ii) mobile, group II introns (TargeTron/ClosTron technology)14; iii) Allelic-Coupled Exchange using either the codA or pyrE systems12,13; and iv) Mariner transposition17,18. To date, the most widely used system is the TargeTron (or ClosTron) system which relies on the re-targeting of mobilizable group II introns and the use of retrotransposable activated markers (RAM)19. Though RAM markers allow for the easy identification of potential mutants, unfortunately, this system only creates insertion mutations resulting in potential polar effects on downstream genes. In addition, the inserted antibiotic resistance marker has led to perceived hypervirulence of mutant strains in the hamster model of CDI20,21.
Segregationally unstable plasmids can be used to create single insertions into the C. difficile genome15,16 or can be used as allelic exchange plasmids using codA counter selection or 5-fluoroorotic acid (FOA) in a pre-generated pyrE mutant12,13. Due to the segregationally unstable nature of the plasmids, daughter cells may not receive a copy of the plasmid and are killed by the antibiotic in the surrounding medium. Strains with single-integration events of the plasmid, due to homologous recombination, grow more rapidly. These ‘large’ colonies are then spread on nutrient-poor medium supplemented with 5-fluorocytosine (for codA-based plasmids) or FOA for the pyrE allelic exchange system12,13. Due to nutrient-poor media being a requirement for counter-selecting the integrated plasmids, mutations that generate slow-growth phenotypes or loss of metabolic pathways may result in difficulties in growth on such medium. Moreover, the pyrE system requires the correction of the pre-generated pyrE mutation before progressing experimentally – increasing the effort and time required to generate mutant strains13. On the other hand, advantages to this system are the ability to create single nucleotide mutations and clean deletions and the ability to integrate a single, chromosomal copy for complementation.
In a previous study, the TargeTron system was used to create insertions in genes (i.e., prdR, prdB and grdA) whose products are involved in Stickland metabolism22. Stickland reactions are a primary source of energy for a small group of anaerobic bacteria grown that typically use amino acids as their sole carbon and nitrogen sources23. During these reactions, one amino acid, such as alanine or leucine, is oxidatively decarboxylated or deaminated23. Subsequently, in the reductive branch, D-proline or glycine acts as electron acceptors. In the case of D-proline the amino acid ring is reduced and converted to δ-amino valeric acid24. In the case of glycine reductase, two selenoproteins are involved (GrdB and GrdA) and the glycine is deaminated in a process that results in acetyl-phosphate (and thus ATP) production. PrdB, GrdA and GrdB are the selenium-containing subunits of the respective reductases and their expression is regulated by PrdR22. These enzymes are important for growth in protein-rich medium supplemented with proline or glycine indicating that selenium-containing enzymes are important for C. difficile physiology22.
Selenophosphate synthetase (SelD, CDR20291_2388) has been shown to be necessary for the activation of selenium for specific incorporation into biological macromolecules in several bacterial model systems25. Selenium incorporation in C. difficile is likely to be dependent on a selenophosphate synthetase (SelD) which generates selenophosphate from selenide and inorganic phosphate that is incorporated into a serine-charged tRNA by selenocysteine synthase (SelA, CDR20291_2387)26. The selenocysteine is then incorporated into proteins such as PrdB and GrdA during translation with the aid of a selenocysteine-specific elongation factor, SelB (CDR20291_2386)26. SelD, SelA and SelB are encoded in a single genetic locus and likely are part of the same transcriptional unit. Selenium is used for other enzymatic processes as well and requires SelD to generate selenophosphate for these processes25. In order to test the importance of selenium-containing factors on C. difficile growth, we engineered a mutation in selD using the established TargeTron gene knock out system and found that selD is important for C. difficile growth and incorporation of selenium into proteins. To avoid any potential polar effects of the TargeTron system, we developed a CRISPR-Cas9 mutagenesis system and used this system to engineer a selD in-frame deletion mutation. Our results highlight the importance of selenoproteins in C. difficile physiology and suggest that these proteins could be used as targets for future antibiotic therapies. Moreover, this newly developed C. difficile genetic system can be used to rapidly and efficiently introduce mutations into the C. difficile genome.
Results
Generation of a TargeTron mutation in C. difficile JIR8094 selD
In order to investigate the role of selenoproteins on C. difficile physiology, we generated a TargeTron insertion into the selD gene of C. difficile strain JIR8094. C. difficile selD is the first gene in the operon and is upstream of the genes encoding a selenocysteine synthase (selA) and a selenocysteine-specific elongation factor (selB) (Fig. 1A). The parental strain, JIR8094, and the selD mutant, LB-CD7, grew to nearly equal levels in rich BHIS medium (Fig. 1B). We next tested the growth of the two strains in protein-rich conditions; tryptone is a rich source of amino nitrogen27 and we reasoned that this medium may favor Stickland metabolism. In either TY or TYG medium, growth of strain LB-CD7 was significantly decreased compared to that of its parent strain (Fig. 1C).
C. difficile selD::ermB has a defect in growth compared to the WT strain. (A) Genetic organization of the selD locus: selD (CDR20291_2388), selA (CDR20291_2387) and selB (CDR20291_2386). The location of the TargeTron insertion into selD is illustrated. (B) C. difficile JIR8094 (●) and C. difficile LB-CD7 (selD::ermB) (■) were grown in BHIS medium and growth was monitored over time. (C) C. difficile JIR8094 was grown in TYG (●) or TY (○) medium and C. difficile LB-CD7 was grown in TYG (■) or TY (□) medium and growth was monitored over time. In both graphs, data points represent the average from three independent experiments and error bars represent the standard deviation of the mean.
C. difficile selD::ermB does not incorporate selenium into proteins
The growth defect observed for strain LB-CD7 (selD::ermB) suggests that selenoproteins are important for growth. To confirm that the selD mutation led to a loss of selenium incorporation during protein synthesis, we measured the incorporation of radioactively labeled selenium in TY medium (Fig. 2). A 1:100 dilution of overnight cultures were added to 10 mL TY medium supplemented with approximately 10 µCi of 75Se in the form of selenite (50 nM cold). After 24 hours cultures were harvested by centrifugation, lysed by brief sonication and clarified cell extracts were analyzed by SDS-PAGE.
C. difficile selD::ermB does not incorporate selenium into selenoproteins C. difficile strains were grown in TY medium and 10 µCi of 75Se was added to the culture medium during growth. After 24 hours of growth, cultures were harvested, cells were lysed and samples from clarified lysates were separated by SDS-PAGE (15%). Radioactively-labeled protein was detected using phosphorimage analysis. GrdB, PrdB and GrdA are labeled based on previously published data24. The full, uncropped phosphorimage can be found in the supplemental information.
When separated by SDS-PAGE and analyzed using autoradiography, we detected three distinct bands in the parental strain JIR8094 (Fig. 2). These likely correspond to GrdB (largest), PrdB and GrdA (smallest band) based on a previous study24. In both prdB and prdR TargeTron mutants, the PrdB band was lost; PrdR is required for prdB expression28 (Fig. 2). Similarly, in the grdA TargeTron mutant, both GrdA and GrdB proteins are not present; due to the insertion of the group II intron into grdA there were polar effects on grdB. Significantly, in two separate selD::ermB isolates (LB-CD7), we observed a lack of radioactive signal, suggesting that this mutation prevents the incorporation of selenium into all three of these established proteins thereby limiting optimal growth (Fig. 2)24.
Generation of a CRISPR-Cas9 mutagenesis system for use in C. difficile
Because the generated TargeTron insertion into C. difficile selD is likely polar on the downstream genes (selA and selB), we sought to generate a non-polar deletion in selD to confirm its growth disadvantage in protein-rich medium. However, the current state of C. difficile genetics requires that strains be isolated under nutrient-poor conditions. Because the growth of the C. difficile selD deletion may behave similarly to the TargeTron insertion, we sought to develop a mutagenesis technique that permits the isolation of mutants under nutrient-rich conditions. The CRISPR-Cas9 system was an attractive target. To design a Cas9-producing plasmid (Fig. 3), we placed a wild-type cas9 gene, which was codon-optimized for expression in C. difficile, under the conditional expression of the tetracycline-inducible tetR promoter18. Also, based on the finding that non-homologous end-joining in C. cellulolyticum is inefficient29, we engineered cas9 D10A to determine if the native double-stranded break repair system in C. difficile is also inefficient; the Cas9D10A protein functions as a ‘nickase’ and does not introduce double-stranded breaks into the targeted DNA30.
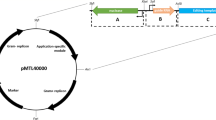
C. difficile CRISPR-Cas9 plasmid map. The pMTL84151 backbone, depicted in gray, consists of the pCD6 C. difficile (Gram-positive) replicon oriV, orfB and repA, the thiamphenicol resistance marker catP, the Gram-negative replicon colE1, and traJ for conjugal transfer from E. coli. The modifications to the backbone, depicted in various colors, consists of the insertions of the targeting region for homologous recombination, sgRNA under the expression of the gdh promoter, and the tetR promoter that regulates the expression of the S. pyogenes cas9 gene that was codon optimized for expression in C. difficile.
The expression of the single guide RNA (sgRNA) that directs Cas9 to the intended target site was placed under the control of the native glutamate dehydrogenase (gdh) promoter31,32. This promoter is constitutively expressed and allows for sufficient levels of the sgRNA to be present within the cell for Cas9 to act upon. We engineered the sgRNA to be produced as a single RNA molecule by fusing the crRNA and tracrRNA, as previously described33. Potential crRNA target sites were determined using an algorithm provided by the CRISPRscan.org website34 and sites were chosen within the first 200 bp of the gene.
The final portion of the C. difficile CRISPR-Cas9 plasmid is the donor region which is necessary to insert a desired mutation. This 2-kb region (one kb upstream of the targeted DNA and one kb downstream of the targeted DNA) surrounds the region to be deleted. The function of this donor region is to provide a template for the native DNA repair system to correct the Cas9-mediated double-stranded DNA (dsDNA) or single-stranded DNA (ssDNA) break.
Isolation of a CRISPR-Cas9 mediated pyrE mutation
To test the efficiency of the system in C. difficile, the first gene targeted for deletion was pyrE. The pyrE mutant strain is a uracil auxotroph and resistant to FOA-mediated toxicity. Therefore, mutations can easily be selected by incorporating FOA into growth medium. To engineer the deletion, we cloned 1-kb upstream of the pyrE start codon and 1-kb downstream of the stop codon in the mutagenesis plasmid (Fig. 4A). Next, a site near the 5′ end of the pyrE sequence was chosen for the crRNA. The resulting plasmid, pJK02, was introduced into C. difficile R20291 by conjugation from E. coli. The tetR promoter system was induced by aTet and mutants were isolated by selecting for those that were resistant to FOA. Colonies containing the mutation were confirmed by PCR amplification of the pyrE gene and surrounding DNA (Fig. 4B). A mutation in pyrE results in a 585-bp deletion within the 1.59 kb pyrE coding sequence (Fig. 4A and B). Subsequently, the phenotype of the pyrE mutant strain, C. difficile KNM5, was confirmed by plating on defined medium or medium supplemented with uracil (Fig. 4C and D). Only the wild-type strain was able to grow on medium without uracil supplementation. Surprisingly, we observed a pyrE deletion in the uninduced R20291 (pJK02) strain, which was to be used here as a control (Fig. 4B). This strain was also a uracil auxotroph (Fig. 4C and D). Based on results from a previous study35, we hypothesized that the tetR promoter has leaky expression and could lead to a small amount of transcription of cas9 in the absence of aTet. To confirm this, we extracted RNA from an uninduced culture and amplified a portion of cas9 (Fig. 4E). As expected, in the absence of induction, we observed cas9 transcript without amplification of contaminating DNA (Fig. S1). These results suggest that the tetR promoter is not tightly regulated and uncontrolled expression of cas9 led to a deletion of pyrE in the uninduced strain.
Isolating Cas9-mediated C. difficile pyrE mutants. (A) A deletion of the chromosomally-encoded pyrE gene was made by homologous recombination from a donor region located on the CRISPR-Cas9 plasmid during repair of a Cas9-mediated double-stranded DNA break. The location of the crRNA target region in pyrE is indicated by the cut DNA. Amplification of wildtype pyrE using primers 1 and 2 results in a 1.59 kb band on an agarose gel. (B) DNA was isolated from potential mutants. The region surrounding the pyrE gene was amplified from the chromosome, and the resulting DNA was separated on an agarose gel. The full, uncropped gel image can be found in the supplemental data. A clean deletion of pyrE is indicated by a faster-migrating DNA band. (C–D) C. difficile R20291, C. difficile R20291 pJK02, C. difficile KNM5 isolate 1 and C. difficile KNM5 isolate 2 were streaked onto either (C) CDMM supplemented with uracil or (D) CDMM alone, and incubated anaerobically for 4 days. (E) RT-PCR showing a comparison of C. difficile R20291 pMTL8151 and C. difficile R20291 pJK02 induced without aTet and those induced in the presence of aTet to turn on the expression of cas9. Also tested was the sgRNA and catP, as a positive control. The white dividing bar between R20291 pMTL84151 and R20291 pJK02 samples indicates an empty lane between samples. The white dividing bar between amplified genes indicates different gels. The full, uncropped gels can be found in the supplemental data.
Incorporation of FOA in the growth medium permits the selection against pyrE-containing strains, but not all mutations lend themselves to a selection process. Thus, we determined the efficiency of the CRISPR-Cas9-mediated pyrE mutations. C. difficile R20291 containing the CRISPR-Cas9 plasmid (pJK02) was induced and the resulting cells were plated on defined medium alone or medium supplemented with FOA. The number of colonies were enumerated and the efficiency of mutagenesis was calculated (Table 1). Surprisingly, wild-type Cas9 yielded a mutation frequency of approximately 50%. Moreover, this mutation frequency was dependent on the ability of Cas9 to introduce a dsDNA break at the target site. That is, the Cas9D10A protein yielded a much lower mutation frequency (~2 × 10−4, or 1 mutation in every 5,000 cells). This mutation frequency is above the empty vector control (~4 × 10−7), suggesting that C. difficile R20291 can use homology-directed repair to replace ssDNA breaks, but not efficiently. Importantly, the high mutation frequency of the wild-type Cas9 plasmid was not merely due to homologous recombination of the plasmid with the genome. If this were the case, the cas9 D10A plasmid should have yielded a similar frequency. These results suggest that the wild-type cas9-containing plasmid is best suited for introducing mutations into the C. difficile genome.
No off-target effects in C. difficile KNM5
To understand if the generated pyrE deletion mutants had other mutations in the genome, Illumina genome re-sequencing was performed for C. difficile KNM5 (ΔpyrE) and C. difficile R20291 (pMTL84151) (empty vector) after induction for the CRISPR-Cas9 system to account for any mutations that may have occurred due to the induction procedure (though C. difficile R20291 (pMTL84151) does not contain a region that could be used for homology directed repair, FOA-resistant colonies were observed on FOA-containing medium). As shown in Fig. 5, >300 reads were observed for every position across the genome (only the region surrounding pyrE is shown). However, at the pyrE gene, the reads drop drastically to ~20–30 reads, dropping further to undetectable levels until the end of the stop codon. The reads increase again after the stop codon; there were no other mutations in the KNM5 strains. Interestingly, the C. difficile R20291 (pMTL84151) FOA-resistant strain had a single-nucleotide polymorphism (SNP) in pyrE which resulted in a premature stop codon. These results suggest that there were no off-target effects generated by the CRISPR-Cas9 system and that it could be applied to other genes.
Deletion of selD using the CRISPR-Cas9 system
Because the TargeTron mutation results in the insertion of the group II intron into the selD gene, there are likely polar effects on the downstream selA and selB sequences. To generate a deletion in selD, we targeted the sgRNA to the selD gene and cloned the upstream and downstream regions for use in homology-directed repair. This plasmid was introduced into C. difficile R20291, the resulting strain was induced and cells were plated directly on BHIS medium (rich medium). Colonies that grew were tested directly for the desired mutation by PCR. Though the efficiency of mutation was lower compared to pyrE, we observed a frequency of ~1 selD deletion in every 5 colonies tested (~20% mutation frequency) (Table 2).
We then tested the growth phenotype of C. difficile KNM6 (ΔselD) and compared it to the growth observed for the wild-type R20291 strain. In the same experiment, we tested whether there were no polar effects on downstream genes in the operon, selA and selB, from this clean deletion of selD. To do this, we introduced selD with its native promoter on a multi-copy plasmid, C. difficile KNM6 pKM142. We included empty vector, pJS116, in wild-type R20291 as well as the mutant KNM6 strains. When grown in rich, BHIS medium (Fig. 6A), no difference is seen between the three strains, R20291 pJS116, KNM6 pJS116, and KNM6 pKM142. When grown in medium that may favor Stickland metabolism (TY or TYG medium), the wild-type strain with empty vector, R20291 pJS116, grew well while the mutant strain with empty vector, KNM6 pJS116, had a reduction in growth in comparison (Fig. 6B). When this strain was complemented by expressing selD, not the entire locus, KNM6 pKM142 grew to wild-type levels (Fig. 6B). This confirms there were no polar effects on the downstream genes, selA and selB, due to the deletion of selD.
C. difficile ΔselD has a moderate growth defect compared to the WT strain. (A) C. difficile R20291 pJS116 (empty vector) (●), C. difficile KNM6 (ΔselD) pJS116 (empty vector) (■) and C. difficile KNM6 pKM142 (selD complement) (▲) were grown in BHIS medium and growth was monitored over time. (B) C. difficile R20291 pJS116, KNM6 pJS116 and KNM6 pKM142 were grown in TYG (closed shapes, ●) or TY (open shapes, ○) medium and growth was monitored over time. Data points represent the average from two independent experiments and error bars represent the standard deviation of the mean.
Taken together, our results suggest that selenium incorporation into proteins is important for C. difficile growth and that CRISPR-Cas9 gene editing can be used to rapidly and efficiently introduce mutations with no polar effects into the C. difficile genome.
Discussion
CRISPRs were originally discovered in Escherichia coli and later in archaea and other bacteria, including C. difficile 36,37,38. The development of utilizing CRISPRs and Cas proteins has led to functional gene editing tools that are widely used. For gene editing in bacteria, the components necessary for this system include: a Cas protein, a guide RNA, and a region of donor DNA to make the desired mutation33,39. A short sequence proximal to the target sequence, which helps the CRISPR-cas9 system to distinguish between self and non-self-sequences, is called the protospacer adjacent motif (PAM) sequence40. S. pyogenes Cas9 recognizes the 5′–NGG–3′ PAM sequence40,41.
C. difficile encodes a native CRISPR-Cas system and belongs to the class I-B subtype38. The S. pyogenes Cas9, used in this study, belongs to the class II group. The classes are defined by their mechanisms and also the composition of Cas proteins33; thus, C. difficile has different Cas proteins than that of S. pyogenes. The CRISPR-Cas system of C. difficile is predicted to recognize a PAM sequence of 5′–CCW–3′, where “W” indicates either an adenine (A) or thymine (T)38. Due to the differences in PAM recognition sequences of these two CRISPR-Cas systems, we do not predict that the C. difficile CRISPR-Cas locus will interfere with this genetic tool.
We successfully developed the first application of a CRISPR-Cas9 system for genetic modification in C. difficile. Due to the problems inherent to each genetic system used in C. difficile, we wanted to create a plasmid containing the system which was simple and easy to modify for future use by others in the field. Towards this goal, the fragments to generate the homology region can be cloned in one step using Gibson Assembly; a gBlock containing the entirety of the sgRNA is also cloned into the plasmid using Gibson Assembly, each at unique restriction sites in the plasmid. Thus, a new mutagenesis plasmid can be generated in two consecutive cloning steps.
We used the pyrE gene as a starting point to optimize the system. By doing so, we were able to determine that C. difficile has a very poor ssDNA break repair system, as evident by the low efficiency of the Cas9D10A-mediated pyrE deletion. However, Cas9D10A had a greater frequency of FOA-resistant colonies than did C. difficile R20291 pMTL84151 (empty vector), suggesting that C. difficile can use homology-directed repair to correct ssDNA breaks, but not at an efficiency which could allow for the isolation of mutants without selective pressure. Thus, the cas9 D10A allele is not a viable option for this CRISPR-based genetic system.
Moving forward with the wild-type Cas9, a concern was whether the mutations were due simply to homologous recombination between the chromosome and the donor DNA on the plasmid or required the repair of the CRISPR-Cas9-mediated dsDNA break. The efficiency of FOA-resistant cells (pyrE mutants) was greater for strains containing wild-type Cas9 than for strains that contained the cas9 D10A allele. Thus, without the aid of the wild-type Cas9 nuclease, C. difficile cannot introduce the desired mutation by homologous recombination with a high enough efficiency to allow for isolation of a mutant without a selection.
Selenophosphate synthetase is an enzyme that uses ATP, water, inorganic phosphate and hydrogen selenide to generate selenophosphate26,42. Selenophosphate is used as a donor to generate selenocysteine-charged tRNAs by attaching selenide to serine-charged tRNAs, leading to incorporation of selenocysteine into selenoproteins (e.g., PrdB or GrdA)26,43. Previously, a prdB mutant was shown to have a decreased growth rate compared to the parent strain22. We had hypothesized that, because SelD is required for generating the precursor to selenocysteine-charged tRNAs, the CRISPR selD mutant would have a greater effect on growth rate than a prdB mutation due to the global reduction in selenoproteins. Indeed both the TargeTron mutant and the CRISPR-generated selD deletion had reduced growth in protein-rich medium (medium where Stickland metabolism is important for growth). We also show there were no polar effects on the downstream selA and selB genes from the clean deletion of selD. In the future, this mutant will help in studying the global effect of the disruption of selenium incorporation into proteins in C. difficile, not just growth and metabolism.
The mutation efficiencies for pyrE and selD were similar and both were well within testable limits. The crRNA chosen for pyrE began at the 36th nucleotide of the 585-bp pyrE gene and had a score in CRISPRscan.org of 30, which is low compared to the highest score of 62 for a crRNA for this gene. The crRNA chosen for selD had a score in CRISPRscan.org of 67, the highest listed, and started at the 183rd nucleotide of the 951-bp selD gene. From these values for the respective genes, there appears to be no pattern for how efficient the CRISPR-Cas9 system in C. difficile can make the mutation. The rules for choosing the optimal crRNA that will yield the highest efficiency for generating a mutation in C. difficile is still under investigation.
In summary, we have developed a functional CRISPR-Cas9 system for use in C. difficile. Because other systems rely on the integration of segregationally-unstable plasmids into the genome, an event that can take several passages and the eventual regeneration of a chromosomal deletion in pyrE, this CRISPR-based plasmid has the potential to rapidly generate mutations within the C. difficile genome. With future adjustments to this system, a larger range of mutations, insertions and even point mutations, could possibly be made in C. difficile which has been difficult or even impossible in the past.
Materials and Methods
Bacterial strains and growth conditions
C. difficile strains (Table S1) were routinely grown in an anaerobic atmosphere (10% H2, 5% CO2, 85% N2) at 37 °C in brain heart infusion medium supplemented with 5 g/L yeast extract and 0.1% L-cysteine (BHIS), as described previously7,44,45,46 or TY medium (3% tryptone, 2% yeast extract)47. For conjugation experiments, cells were plated on BHI agar medium, as described previously for E. coli-based conjugations48, or on TY medium for Bacillus subtilis-based conjugations. For selenium incorporation (see below), strains were grown in TY (1% tryptone, 0.5% yeast extract)24. Where indicated, growth was supplemented with taurocholate (TA; 0.1% w/v), thiamphenicol (10 µg/mL), kanamycin (50 µg/mL), D-cycloserine (250 µg/mL), erythromycin (5 µg/mL) and/or glucose (1% w/v) as needed. Induction of the CRISPR-Cas9 system was performed in TY medium47 supplemented with thiamphenicol (10 µg/mL) and anhydrous tetracycline (aTet; 100 ng/mL). A defined minimal medium for C. difficile growth (CDMM), described previously12,49, was used for selection of pyrE mutants by supplementation with 5-fluoroorotic acid (FOA; 2 mg/mL) and uracil (5 µg/mL). E. coli strains (Table S1) were routinely grown at 37 °C in LB medium. Strains were supplemented with chloramphenicol (20 µg/mL), kanamycin (50 µg/mL), and/or ampicillin (100 µg/mL) as needed. B. subtilis BS49 was routinely grown at 37 °C in BHIS broth or on LB agar plates. Strains were supplemented with chloramphenicol (2.5 µg/mL) and/or tetracycline (5 µg/mL).
Plasmid construction and molecular biology
The JIR8094 selD TargeTron insertion was created in several steps. First, plasmid pBL38 was constructed by retargeting of the group II intron from pCE24050, using the primers oLB70, oLB71, oLB72 and EBS-Universal, as outlined in the TargeTron user manual (Sigma-Aldrich), followed by initial cloning of the retargeted fragment in pCE240 digested with BsrGI and HindIII. The retargeted group II intron from pBL38 was then extracted by digestion with SfoI and SphI and cloned between the SphI and SnaBI sites of pMC12351, resulting in pBL54. Conjugation experiments between C. difficile and E. coli were carried out as described previously52. C. difficile transconjugants were selected on BHIS plates supplemented with D-cycloserine, kanamycin, and thiamphenicol and potential TargeTron mutants were identified by plating on erythromycin. Erythromycin-resistant colonies were screened for the insertion of the intron into C. difficile selD by PCR using primers specific for full-length C. difficile selD (oLB76 and oLB77). A positive clone, strain LB-CD7, was identified. To identify if the TargeTron system integrated at sites other than the selD gene, we sequenced the C. difficile LB-CD7 strain and the parent JIR8094 strain. No additional TargeTron insertions were found except the one in selD.
To construct the CRISPR-cas9 plasmid, the constitutively-expressed cwp2 promoter was chosen to drive expression of a sgRNA targeted to spoVAC. Oligonucleotides were designed and the fragments generated were stitched together by PCR SOEing with Phusion DNA polymerase (due to the nature of the A: T-rich sequence, the entire fragment could not be synthesized directly and thus had to be stitched together by PCR) (all oligonucleotide sequences can be found in Table S2). The first round of amplification was done with primer sets gRNA_1_for and gRNA_2_rev, gRNA_3_for and gRNA_4_rev, and gRNA_5_for and gRNA_6_rev. The resulting fragments, gRNA_1_2 and gRNA_3_4, were used along with primers gRNA_1_for and gRNA _4_rev in a PCR to yield fragment gRNA_1_4. Fragment gRNA_5_6 was expanded in a PCR using primers gRNA_5_for and gRNA_7_rev to yield fragment gRNA_5_7. The complete sgRNA was made by PCR sowing of fragments gRNA_1_4 and gRNA_5_7 using primers 5′gRNA and 3′gRNA. This DNA fragment was introduced into pJS116 (all plasmid descriptions can be found in Table S1) at the PmeI restriction site using Gibson Assembly53 and transformed into E. coli DH5α to generate pKM22. The tetR repressor gene along with the Ptet promoter for conditional expression of cas9 was amplified by PCR from pRPF21518 (Table S1) using primers 5′MTL_tetRprom and 3′tetR_Cas9. cas9 from Streptococcus pyogenes was codon optimized for expression in C. difficile by Thermo Fisher (Thermo Fisher Scientific, Waltham, MA). Codon-optimized cas9 was amplified using primers 5′tetR_CO_cas9 and 3′MTL_CO_cas9 from pMK-RQ-Bs-cas9. To introduce a D10A mutation into cas9, we used primer set 5′tetR_CO_cas9_D10A and 3′MTL_CO_cas9 for PCR amplification. The Ptet promoter and the wild-type cas9 and cas9 D10A alleles were introduced into pKM22 at the HindIII restriction site using Gibson Assembly to generate pKM46 and pKM48, respectively.
To facilitate future crRNA changes and increase the expression of the sgRNA, the stronger, constitutively-expressed gdh promoter was used to replace the cwp2 promoter. Also, KpnI and MluI restriction sites were added at the 5′ and 3′ ends of the sgRNA, respectively. 5′gdh and 3′gdh_gRNA were used to amplify the gdh promoter from C. difficile R20291. The sgRNA was amplified from pKM22 using primers 5′gRNA_gdh and 3′gRNA 2. The resulting two fragments were both inserted into the PmeI and BsrGI sites using Gibson Assembly and transformed into E. coli DH5α to generate pKM54 and pKM55.
In order to easily select for mutants, we designed a plasmid to target the pyrE gene. The donor region for homology directed repair was made such that 1-kb upstream and 1-kb downstream were stitched together to generate a clean deletion of pyrE. 1-kb upstream and 1-kb downstream of pyrE were separately amplified by PCR from C. difficile R20291 using primers 5′pyrE_UP and 3′pyrE_UP and 5′pyrE_DOWN and 3′pyrE_DOWN, respectively. The two resulting fragments were inserted into the NotI and XhoI restriction sites in pKM54 and pKM55 background by Gibson Assembly and transformed into E. coli DH5α to generate pKM64 and pKM65, respectively. A gBlock (Integrated DNA Technologies, Coralville, IA), pyrE_gRNA_gBlock, was designed which contained the sgRNA DNA sequence between, and including, the KpnI restriction site and the MluI site. This DNA fragment was introduced between the KpnI and MluI restriction sites of pKM64 and pKM65 to generate pKM71 and pKM72, respectively. To introduce the plasmids into C. difficile by conjugation with E. coli, the B. subtilis Tn916 oriT was replaced with the E. coli traJ gene. traJ was amplified from pMTL84151 by PCR using primers 5′traJ and 3′traJ. The resulting fragment was introduced into pKM71 and pKM72 using the ApaI restriction site by Gibson Assembly and transformed into E. coli DH5α to generate pJK02 (accession number MF782679) and pKM93 respectively.
The subsequent CRISPR-Cas9 plasmid targeting selD was made using pJK02. The homology regions for selD targeting were amplified by primer sets 5′MTL_selD_UP and 3′MTL_selD_UP and 5′MTL_selD_DN and 3′MTL_selD_DN. The resulting fragments were cloned by Gibson assembly into pJK02 at the NotI and XhoI restriction sites and transformed into E. coli DH5α resulting in pJS170. The gBlock for selD targeting sgRNA, CRISPR_selD_183, was introduced by ligation into the KpnI and MluI sites and transformed into E. coli DH5α resulting in pJS187. The selD targeting plasmid was modified by replacing traJ with oriT tn916 for B. subtilis conjugation by amplification from pJS116 using primers 5′Tn916ori and 3′Tn916ori. The resulting fragment was introduced into pJS187 by Gibson assembly at the ApaI site and transformed into E. coli DH5α resulting in pJS194.
To make a plasmid which would complement the selD mutation, selD along with 500 bp upstream to include the native promoter was amplified by primer sets 5′selD_comp and 3′selD_comp. The resulting fragment was cloned by Gibson assembly into pJS116 at the NotI and XhoI restriction sites and transformed into E. coli DH5α resulting in pKM142. The sequences of all plasmids were verified by DNA sequencing.
Conjugation for CRISPR-Cas9 and complementation plasmid insertion
All complete CRISPR-Cas9 and complement plasmids were transformed into either E. coli HB101 pRK24 or B. subtilis BS49 to be used as donor for conjugation with C. difficile. For E. coli conjugations, the strains were grown overnight at 37 °C in LB supplemented with ampicillin and chloramphenicol. C. difficile R20291 was grown anaerobically in TY medium overnight. Five hundred microliters of C. difficile overnight culture/mating was heated to either 52 °C for 5 minutes for an 8 hour conjugation or 50 °C for 15 minutes for a 24 hour conjugation, as described previously48. C. difficile cultures were removed from the heat block and let cool to 37 °C for 2 minutes. Meanwhile, 1 mL of E. coli HB101 pRK24 containing the CRISPR-Cas9 plasmid cultures were pelleted at 4,000 × g for 2 minutes and the supernatant was removed. The E. coli pellets were transferred to the anaerobic chamber and gently suspended in the heat shocked C. difficile sample. The resulting mix was plated onto pre-reduced BHI agar plates by spotting ten, 20 µL drops of culture. After either 8 or 24 hours, the growth was harvested by collecting in 1 mL pre-reduced TY broth. One hundred microliters of the resuspended growth was plated onto multiple BHIS agar plates supplemented with thiamphenicol, kanamycin, and D-cycloserine. Growth was monitored for 2 to 3 days. Individual colonies were restreaked for isolation and tested for insertion of plasmid by PCR amplification of the catP gene with primers 5′ catP 3 and 3′ catP 2.
For B. subtilis conjugation, C. difficile R20291 was grown anaerobically in BHIS broth overnight. The C. difficile overnight culture was diluted in fresh pre-reduced BHIS broth and grown anaerobically for 4 hours. Meanwhile, B. subtilis BS49 was grown aerobically at 37 °C in BHIS broth supplemented with tetracycline and chloramphenicol for 4 hours. One hundred microliters of each culture was plated on TY agar medium. After 24 hours, the growth was harvested by suspending in 2 mL pre-reduced BHIS broth. One hundred microliters of the resuspended growth was spread onto several BHIS agar plates supplemented with thiamphenicol, kanamycin, and D-cycloserine. C. difficile transconjugants were screened for the presence of Tn916 using tetracycline resistance, as described previously. Thiamphenicol-resistant, tetracycline-sensitive transconjugants were selected and used for further experiments.
Radiolabeling studies with 75Selenium
Selenium is taken up with high affinity and specifically incorporated into macromolecules through exposure of cells to 75Se in the form of selenite25. For these studies, a 1:100 dilution of overnight cultures were added to 10 mL TY medium supplemented with approximately 10 µCi 75Se in the form of selenite (corresponding to 50 nM cold). After 24 hours growth, nine milliliter cultures were grown overnight in 12 × 75 mm capped culture tubes in an atmosphere of 95% nitrogen and 5% hydrogen. Cells were harvested by centrifugation (5,000 × g for 5 minutes) and resuspended in a small amount (typically 0.2 mL) of lysis buffer (50 mM Tris, pH 8.0, 0.1 mM benzamidine, 0.5 mM EDTA). Cells were lysed by sonication (model 100 Fisher Scientific) for short 10 second bursts until lysis was seen. The crude cell lysates were further clarified by centrifugation (12,500 × g) for 10 minutes at 4 °C. Protein concentrations were determined by Bradford assay54 using albumin to generate a standard curve. Radiolabeled selenoproteins were separated by SDS-PAGE (15% resolving gel) and, after the gels were dried, selenoproteins were identified by phosphorimager analysis (Molecular Dynamics phosphorimager).
Induction of the CRISPR-Cas9 system and isolating mutants
C. difficile R20291 strains containing the pyrE-targeting plasmids were grown overnight in TY medium supplemented with thiamphenicol. In the morning, 250 µL of an overnight culture was diluted into 4.75 mL of fresh TY medium supplemented with thiamphenicol and aTet (100 ng/mL) and grown for 6 hours. Subsequently, cultures serially diluted and spread on CDMM medium supplemented with FOA and uracil. Colonies were isolated; DNA was extracted and tested for the desired mutation by PCR amplification of the target gene, 5′pyrE 2 and 3′pyrE 2 (Table S2). Once an isolate was confirmed, it was passaged ~3 times in BHIS liquid medium in order to lose the CRISPR-Cas9 plasmid. After pick-and-patch on BHIS agar with and without thiamphenicol, loss of plasmid was confirmed by PCR amplification of the catP gene using primer set 5′ catP 3 and 3′ catP 2. C. difficile R20291 strains containing the selD targeting plasmid was induced for 24 hours. Then ~10 µL of culture was spread onto BHIS medium. Colonies were tested as described above using primer sets 5′selD and 3′selD. Confirmed isolates were passaged on BHIS agar once in order to lose the CRISPR-Cas9 plasmid due to the slow growth of the mutant. Loss of plasmid was confirmed by PCR amplification of the catP gene using primer set 5′ catP 3 and 3′ catP 2.
RT-PCR
RNA was extracted from wild-type C. difficile R20291 pMTL84151 (empty vector) and C. difficile R20291 pJK02 induced for 30 minutes with or without aTet using a FastRNA Blue Kit (MP Biomedical). DNA contamination was eliminated by using a TURBO DNA-free Kit (Thermo Scientific) according to the standard protocol. cDNA was made using the SuperScript III First-Strand Synthesis System (Thermo Scientific) according to the protocol, including controls for each sample without the presence of reverse transcriptase. To determine if cas9 and the gRNA were being transcribed, the 5′ end of cas9, sgRNA, and catP were amplified from isolated cDNA in a PCR using primers sets 5′COcas9_RT and 3′COcas9_RT, 5′gRNA_RT and 3′gRNA_RT, and 5′catP_RT and 3′catP_RT and Taq DNA polymerase.
Illumina sequencing
High-quality, high-molecular weight genomic DNA from C. difficile R20291 (WT), aTet-induced C. difficile R20291 pMTL84151 (empty vector), two isolates of C. difficile KNM5, C. difficile LB-CD7, C. difficile KNM6 pJS116, and C. difficile KNM6 pKM142 was extracted as described previously52,55. The genomic DNA was submitted to Tufts University School of Medicine Genomics Core facility for Paired-End 50 Illumina re-sequencing as described previously7. Alignment and analysis of the sequences was performed using DNASTAR Lasergene program MegAlign Pro 14.
Determining mutation efficiencies
C. difficile R20291 strains containing the pyrE targeting CRISPR-Cas9 plasmids were induced as described above. Induced cultures were serially diluted and 100 µL was spread on CDMM supplemented with uracil and CDMM supplemented with FOA and uracil. After 4 days, colony forming units (CFUs) were counted for each dilution on each media and the total CFU/mL of the mutants (those on CDMM-FOA and uracil) and the total cell count (CDMM-uracil) were calculated.
C. difficile R20291 strains containing the selD targeting CRISPR-Cas9 plasmid was induced as described above. A loop containing ~10 µL of culture was spread onto rich BHIS agar medium. Individual colonies were isolated; DNA was extracted, and tested for the desired mutation by PCR amplification of the target genes using primers 5′selD and 3′selD (Table S2). These oligonucleotides only amplify DNA from the chromosome regardless of whether the CRISPR-Cas9 plasmid is present or not.
Statistical Analysis
Data points represent the mean from two or three independent experiments and, where indicated, error bars represent one standard deviation from the mean.
Availability of materials and data
The datasets generated during and/or analyzed during the current study are available in the NCBI SRA repository. The first accession number SRP115702 includes the following sequences and accession numbers: R20291 (SRX3104072), R20291 foaR (SRX3104073), and two KNM5 isolates (SRX3104071 and SRX3104074). The second accession number SRP119051 includes the following sequences and accession numbers: LB-CD7 (SRX3236353), KNM6 pJS116 (SRX3236354), and KNM6 pKM142 (SRX3236352). Finally, the pJK02 plasmid generated in this study is freely available to the scientific community.
References
Oren, A. & Garrity, G. M. Notification that new names of prokaryotes, new combinations, and new taxonomic opinions have appeared in volume 66, part 9, of the IJSEM. Int J Syst Evol Microbiol 66, 4921–4923, https://doi.org/10.1099/ijsem.0.001620 (2016).
Lawson, P. A., Citron, D. M., Tyrrell, K. L. & Finegold, S. M. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prevot 1938. Anaerobe 40, 95–99, https://doi.org/10.1016/j.anaerobe.2016.06.008 (2016).
Rupnik, M., Wilcox, M. H. & Gerding, D. N. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nature reviews. Microbiology 7, 526–536, https://doi.org/10.1038/nrmicro2164 (2009).
Smits, W. K., Lyras, D., Lacy, D. B., Wilcox, M. H. & Kuijper, E. J. Clostridium difficile infection. Nature Reviews Disease Primers 2, 16020, https://doi.org/10.1038/nrdp.2016.20 (2016).
Theriot, C. M. & Young, V. B. Interactions Between the Gastrointestinal Microbiome and Clostridium difficile. Annual review of microbiology 69, 445–461, https://doi.org/10.1146/annurev-micro-091014-104115 (2015).
Sorg, J. A. & Sonenshein, A. L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190, 2505–2512, https://doi.org/10.1128/jb.01765-07 (2008).
Francis, M. B., Allen, C. A., Shrestha, R. & Sorg, J. A. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS pathogens 9, e1003356, https://doi.org/10.1371/journal.ppat.1003356 (2013).
Paredes-Sabja, D., Shen, A. & Sorg, J. A. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends in microbiology 22, 406–416, https://doi.org/10.1016/j.tim.2014.04.003 (2014).
Martin-Verstraete, I., Peltier, J. & Dupuy, B. The Regulatory Networks That Control Clostridium difficile Toxin Synthesis. Toxins (Basel) 8, https://doi.org/10.3390/toxins8050153 (2016).
Poutanen, S. M. & Simor, A. E. Clostridium difficile-associated diarrhea in adults. CMAJ 171, 51–58, https://doi.org/10.1503/cmaj.1031189 (2004).
Aktories, K., Schwan, C. & Jank, T. Clostridium difficile Toxin Biology. Annual review of microbiology, https://doi.org/10.1146/annurev-micro-090816-093458 (2017).
Cartman, S. T., Kelly, M. L., Heeg, D., Heap, J. T. & Minton, N. P. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Applied and environmental microbiology 78, 4683–4690, https://doi.org/10.1128/AEM.00249-12 (2012).
Ng, Y. K. et al. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PloS one 8, e56051, https://doi.org/10.1371/journal.pone.0056051 (2013).
Heap, J. T., Pennington, O. J., Cartmant, S. T., Carter, G. P. & Minton, N. P. The ClosTron: A universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods. 79, 452–464 (2007).
Dineen, S. S., Villapakkam, A. C., Nordmant, J. T. & Sonenshein, A. L. Repression of Clostridium difficile toxin gene expression by CodY. Molecular Microbiology 61, 1335–1351 (2007).
O’Connor, J. R. et al. Construction and analysis of chromosomal Clostridium difficile mutants. Molecular Microbiology 61, 1335–1351, https://doi.org/10.1111/j.1365-2958.2006.05315.x (2006).
Cartman, S. T. & Minton, N. P. A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl. Environ. Microbiol., AEM. 02525-02509, https://doi.org/10.1128/aem.02525-09 (2009).
Dembek, M. et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio 6, e02383, https://doi.org/10.1128/mBio.02383-14 (2015).
Heap, J. T. et al. The ClosTron: Mutagenesis in Clostridium refined and streamlined. J Microbiol Methods 80, 49–55, https://doi.org/10.1016/j.mimet.2009.10.018 (2010).
Kelly, M. L. et al. Improving the reproducibility of the NAP1/B1/027 epidemic strain R20291 in the hamster model of infection. Anaerobe 39, 51–53, https://doi.org/10.1016/j.anaerobe.2016.02.011 (2016).
Bakker, D. et al. The HtrA-like protease CD3284 modulates virulence of Clostridium difficile. Infection and immunity 82, 4222–4232, https://doi.org/10.1128/IAI.02336-14 (2014).
Bouillaut, L., Self, W. T. & Sonenshein, A. L. Proline-dependent regulation of Clostridium difficile Stickland metabolism. J. Bacteriol. 195, 844–854, https://doi.org/10.1128/JB.01492-12 (2013).
Bouillaut, L., Dubois, T., Sonenshein, A. L. & Dupuy, B. Integration of metabolism and virulence in Clostridium difficile. Res Microbiol 166, 375–383, https://doi.org/10.1016/j.resmic.2014.10.002 (2015).
Jackson, S., Calos, M., Myers, A. & Self, W. T. Analysis of proline reduction in the nosocomial pathogen Clostridium difficile. Journal of Bacteriology 188, 8487–8495, https://doi.org/10.1128/JB.01370-06 (2006).
Self, W. T. Specific and nonspecific incorporation of selenium into macromolecules. Comprehensive Natural Products Ii: Chemistry and Biology, Vol 5: Amino Acids, Peptides andProteins, 121–148 (2010).
Srivastava, M., Mallard, C., Barke, T., Hancock, L. E. & Self, W. T. A selenium-dependent xanthine dehydrogenase triggers biofilm proliferation in Enterococcus faecalis through oxidant production. J Bacteriol 193, 1643–1652, https://doi.org/10.1128/JB.01063-10 (2011).
Power, D. A. & Zimbro, M. J. Difco & BBL manual: manual of microbiological culture media. 696 pages (Difco Laboratories, Division of Becton Dickinson and Co., 2003).
Bouillaut, L., Self, W. T. & Sonenshein, A. L. Proline-dependent regulation of Clostridium difficile Stickland metabolism. Journal of bacteriology 195, 844–854, https://doi.org/10.1128/JB.01492-12 (2013).
Xu, T. et al. Efficient Genome Editing in Clostridium cellulolyticum via CRISPR-Cas9 Nickase. Applied and environmental microbiology 81, 4423–4431, https://doi.org/10.1128/AEM.00873-15 (2015).
Shen, B. et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods 11, 399–402, https://doi.org/10.1038/nmeth.2857 (2014).
Mani, N. & Dupuy, B. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. PNAS 98, 5844–5849 (2001).
Mani, N., Dupuy, B. & Sonenshein, A. L. Isolation of RNA polymerase from Clostridium difficile and characterization of glutamate dehydrogenase and rRNA gene promoters in vitro and in vivo. Journal of bacteriology 188, 96–102, https://doi.org/10.1128/JB.188.1.96-102.2006 (2006).
Doudna, J. A. & Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096, https://doi.org/10.1126/science.1258096 (2014).
Moreno-Mateos, M. A. et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nature methods 12, 982–988, https://doi.org/10.1038/nmeth.3543 (2015).
Oliveira Paiva, A. M., Friggen, A. H., Hossein-Javaheri, S. & Smits, W. K. The Signal Sequence of the Abundant Extracellular Metalloprotease PPEP-1 Can Be Used to Secrete Synthetic Reporter Proteins in Clostridium difficile. ACS Synth Biol 5, 1376–1382, https://doi.org/10.1021/acssynbio.6b00104 (2016).
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M. & Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of bacteriology 169, 5429–5433 (1987).
Mojica, F. J., Diez-Villasenor, C., Soria, E. & Juez, G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Molecular microbiology 36, 244–246 (2000).
Boudry, P. et al. Function of the CRISPR-Cas System of the Human Pathogen Clostridium difficile. MBio 6, e01112–01115, https://doi.org/10.1128/mBio.01112-15 (2015).
Sternberg, S. H. & Doudna, J. A. Expanding the Biologist’s Toolkit with CRISPR-Cas9. Molecular cell 58, 568–574, https://doi.org/10.1016/j.molcel.2015.02.032 (2015).
Mojica, F. J., Diez-Villasenor, C., Garcia-Martinez, J. & Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740, https://doi.org/10.1099/mic.0.023960-0 (2009).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821, https://doi.org/10.1126/science.1225829 (2012).
Kim, I. Y., Veres, Z. & Stadtman, T. C. Escherichia coli mutant SELD enzymes. The cysteine 17 residue is essential for selenophosphate formation from ATP and selenide. J Biol Chem 267, 19650–19654 (1992).
Forchhammer, K. & Bock, A. Selenocysteine synthase from Escherichia coli. Analysis of the reaction sequence. J Biol Chem 266, 6324–6328 (1991).
Francis, M. B., Allen, C. A. & Sorg, J. A. Spore cortex hydrolysis precedes dipicolinic acid release during Clostridium difficile spore germination. J. Bacteriol. 197, 2276–2283, https://doi.org/10.1128/JB.02575-14 (2015).
Allen, C. A., Babakhani, F., Sears, P., Nguyen, L. & Sorg, J. A. Both fidaxomicin and vancomycin inhibit outgrowth of Clostridium difficile spores. Antimicrobial agents and chemotherapy 57, 664–667, https://doi.org/10.1128/AAC.01611-12 (2013).
Sorg, J. A. & Sonenshein, A. L. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J. Bacteriol. 191, 1115–1117, https://doi.org/10.1128/jb.01260-08 (2009).
Dupuy, B. & Sonenshein, A. L. Regulated transcription of Clostridium difficile toxin genes. Molecular Microbiology 27, 107–120 (1998).
Kirk, J. A. & Fagan, R. P. Heat shock increases conjugation efficiency in Clostridium difficile. Anaerobe 42, 1–5, https://doi.org/10.1016/j.anaerobe.2016.06.009 (2016).
Karlsson, S., Burman, L. G. & Akerlund, T. Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology 145, 1683–1693 (1999).
Ho, T. D. & Ellermeier, C. D. PrsW is required for colonization, resistance to antimicrobial peptides, and expression of extracytoplasmic function sigma factors in Clostridium difficile. Infect Immun 79, 3229–3238, https://doi.org/10.1128/IAI.00019-11 (2011).
McBride, S. M. & Sonenshein, A. L. Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect. Immun., IAI. 00731–00710, https://doi.org/10.1128/iai.00731-10 (2010).
Bouillaut, L., McBride, S. M. & Sorg, J. A. Genetic manipulation of Clostridium difficile. Curr Protoc Microbiol Chapter 9, Unit 9A 2, https://doi.org/10.1002/9780471729259.mc09a02s20 (2011).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods 6, 343–345, https://doi.org/10.1038/nmeth.1318 (2009).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry 72, 248–254 (1976).
Wren, B. W. & Tabaqchali, S. Restriction endonuclease DNA analysis of Clostridium difficile. Journal of clinical microbiology 25, 2402–2404 (1987).
Acknowledgements
We would like to thank Dr. Robert Fagan at The University of Sheffield for the generous gift of the plasmid, pRPF215, containing the tetracycline-inducible system and Dr. Linc Sonenshein for helpful advice. We would also like to thank members of the Sorg lab, Dr. Leif Smith, and members from Dr. Leif Smith′s lab at Texas A&M University for their helpful comments and suggestions during the preparation of this manuscript. This project was supported by awards 5R01AI116895 and 1U01AI124290 to J.A.S. from the National Institute of Allergy and Infectious Diseases and R01GM042219 from the National Institute of General Medical Sciences to Dr. Abraham L. Sonenshein. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID and NIGMS. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author information
Authors and Affiliations
Contributions
K.N.M., L.B., J.N.K., W.T.S. and J.A.S. performed the experiments. K.N.M., L.B., W.T.S. and J.A.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
McAllister, K.N., Bouillaut, L., Kahn, J.N. et al. Using CRISPR-Cas9-mediated genome editing to generate C. difficile mutants defective in selenoproteins synthesis. Sci Rep 7, 14672 (2017). https://doi.org/10.1038/s41598-017-15236-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-15236-5
This article is cited by
-
Clostridioides difficile ferrosome organelles combat nutritional immunity
Nature (2023)
-
Inhibition of selenoprotein synthesis is not the mechanism by which auranofin inhibits growth of Clostridioides difficile
Scientific Reports (2023)
-
Multiplex genome engineering in Clostridium beijerinckii NCIMB 8052 using CRISPR-Cas12a
Scientific Reports (2023)
-
Investigating plant–microbe interactions within the root
Archives of Microbiology (2022)
-
Application of different types of CRISPR/Cas-based systems in bacteria
Microbial Cell Factories (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.